The Det.Belt Server: A Tool to Visualize and Estimate Amphipathic Solvent Belts around Membrane Proteins
Abstract
1. Introduction
2. Materials and Methods
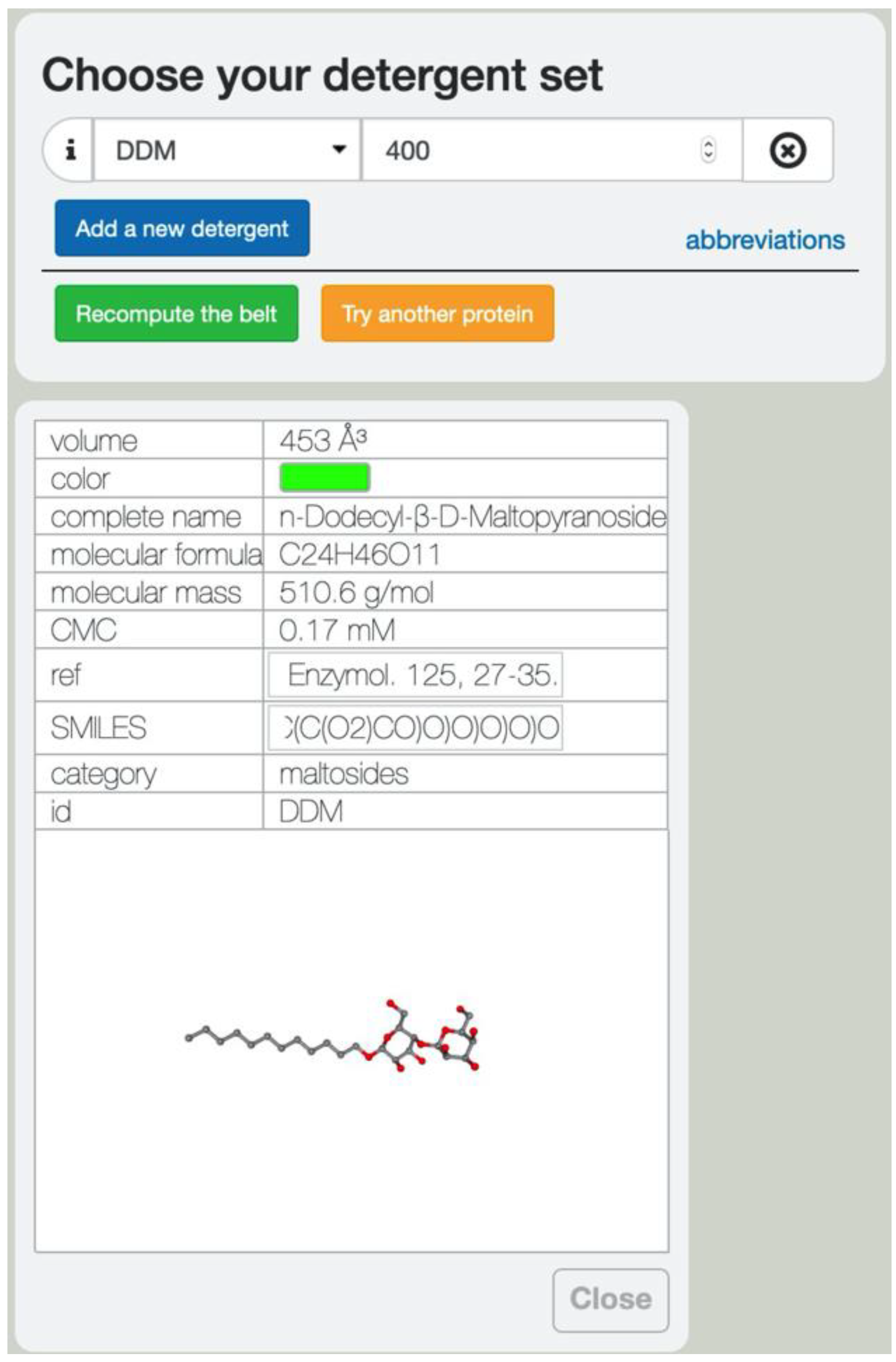
2.1. Server Input
2.2. Calculation of the Belt Dimensions
2.3. Server Implementation
3. Results
3.1. Det.Belt Web Interface
3.2. Detergent Belt Representation as a Hollow Cylinder
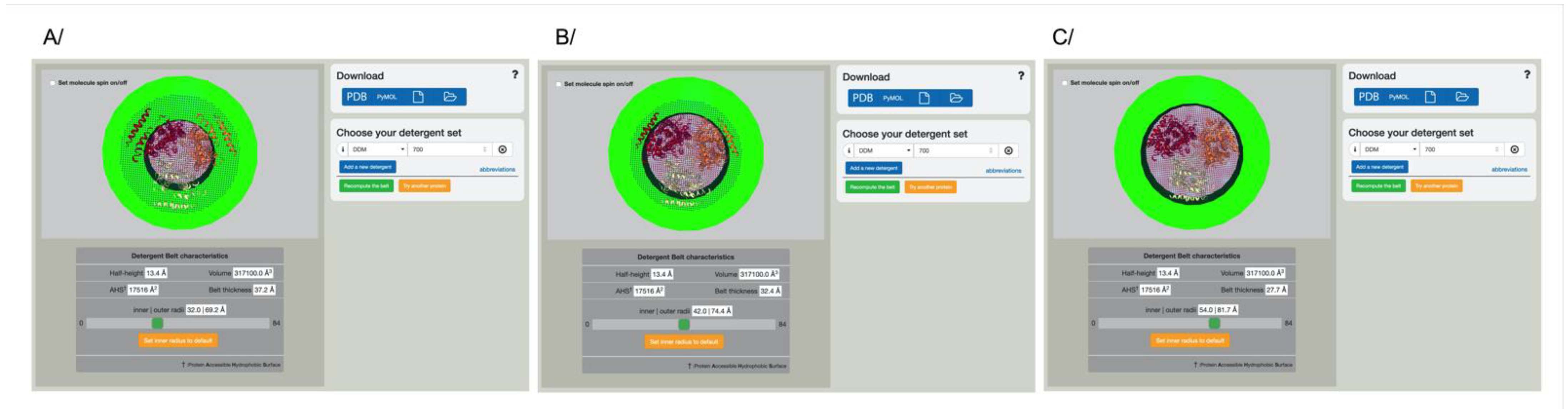
3.3. Grasping the Correct Amount of Detergent around Any Membrane Protein
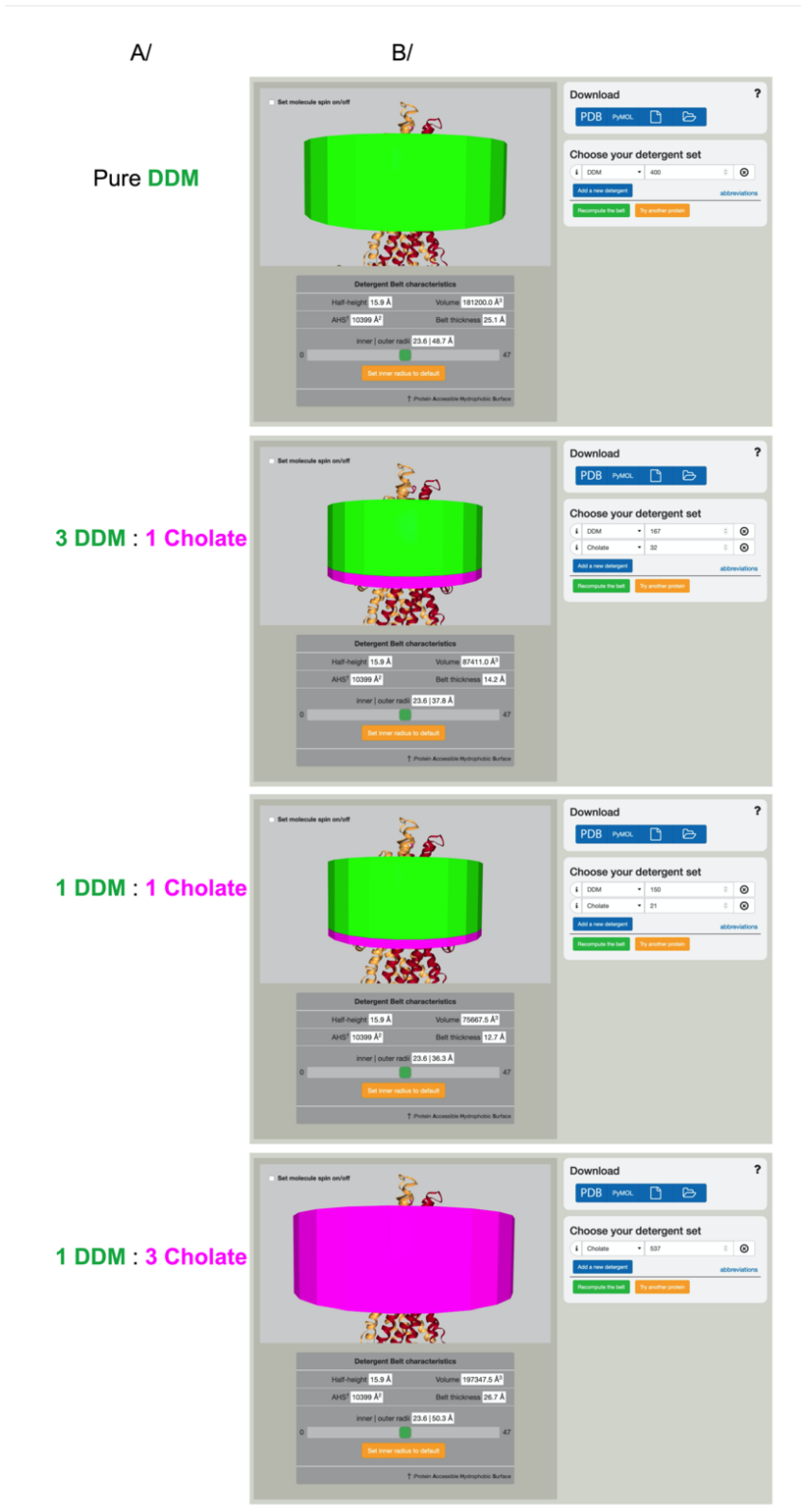
3.4. Detergent Mixtures
3.5. Application to Detergent Prediction
3.6. Detergent and Lipids Database
3.7. Special Cases
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Le Bon, C.; Michon, B.; Popot, J.-L.; Zoonens, M. Amphipathic environments for determining the structure of membrane proteins by single-particle electron cryo-microscopy. Q. Rev. Biophys. 2021, 54, 1–62. [Google Scholar] [CrossRef] [PubMed]
- Choy, B.C.; Cater, R.J.; Mancia, F.; Pryor, E.E. A 10-year meta-analysis of membrane protein structural biology: Detergents, membrane mimetics, and structure determination techniques. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cherezov, V. Chemical tools for membrane protein structural biology. Curr. Opin. Struct. Biol. 2019, 58, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.A.; Peuchmaur, M.; Magnard, S.; Haudecoeur, R.; Boyère, C.; Mounien, S.; Benammar, I.; Zampieri, V.; Igonet, S.; Chaptal, V.; et al. Glycosyl-Substituted Dicarboxylates as Detergents for the Extraction, Overstabilization, and Crystallization of Membrane Proteins. Angew. Chem. Int. Ed. 2018, 57, 2948–2952. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Bennett, B.C.; Hong, W.-X.; Fu, Y.; Baker, K.A.; Marcoux, J.; Robinson, C.; Ward, A.B.; Halpert, J.R.; Stevens, R.C.; et al. Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA 2013, 110, E1203–E1211. [Google Scholar] [CrossRef]
- Matar-Merheb, R.; Rhimi, M.; Leydier, A.; Huché, F.; Galián, C.; Desuzinges-Mandon, E.; Ficheux, D.; Flot, D.; Aghajari, N.; Kahn, R.; et al. Structuring Detergents for Extracting and Stabilizing Functional Membrane Proteins. PLoS ONE 2011, 6, e18036. [Google Scholar] [CrossRef]
- Chae, P.S.; Rasmussen, S.G.F.; Rana, R.R.; Gotfryd, K.; Chandra, R.; Goren, M.A.; Kruse, A.C.; Nurva, S.; Loland, C.; Pierre, Y.; et al. Maltose–neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat. Methods 2010, 7, 1003–1008. [Google Scholar] [CrossRef]
- Marconnet, A.; Michon, B.; Le Bon, C.; Giusti, F.; Tribet, C.; Zoonens, M. Solubilization and Stabilization of Membrane Proteins by Cycloalkane-Modified Amphiphilic Polymers. Biomacromolecules 2020, 21, 3459–3467. [Google Scholar] [CrossRef] [PubMed]
- Horsey, A.J.; Briggs, D.A.; Holliday, N.; Briddon, S.J.; Kerr, I.D. Application of fluorescence correlation spectroscopy to study substrate binding in styrene maleic acid lipid copolymer encapsulated ABCG2. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183218. [Google Scholar] [CrossRef] [PubMed]
- Teo, A.C.K.; Lee, S.C.; Pollock, N.L.; Stroud, Z.; Hall, S.; Thakker, A.; Pitt, A.R.; Dafforn, T.R.; Spickett, C.M.; Roper, D.I. Analysis of SMALP co-extracted phospholipids shows distinct membrane environments for three classes of bacterial membrane protein. Sci. Rep. 2019, 9, 1813. [Google Scholar] [CrossRef]
- Le Bon, C.; Marconnet, A.; Masscheleyn, S.; Popot, J.-L.; Zoonens, M. Folding and stabilizing membrane proteins in amphipol A8-35. Methods 2018, 147, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Dörr, J.M.; Scheidelaar, S.; Koorengevel, M.C.; Dominguez, J.J.; Schäfer, M.; van Walree, C.A.; Killian, J.A. The styrene–maleic acid copolymer: A versatile tool in membrane research. Eur. Biophys. J. 2016, 45, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Prince, C.C.; Jia, Z. Detergent quantification in membrane protein samples and its application to crystallization experiments. Amino Acids 2013, 45, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Le Maire, M.; Arnou, B.; Olesen, C.; Georgin, D.; Ebel, C.; Møller, J.V. Gel chromatography and analytical ultracentrifugation to determine the extent of detergent binding and ag-gregation, and Stokes radius of membrane proteins using sarcoplasmic reticulum Ca2+-ATPase as an example. Nat. Protoc. 2008, 3, 1782–1795. [Google Scholar] [CrossRef] [PubMed]
- Møller, J.V.; Le Maire, M. Detergent binding as a measure of hydrophobic surface area of integral membrane proteins. J. Biol. Chem. 1993, 268, 18659–18672. [Google Scholar] [CrossRef]
- Chaptal, V.; Delolme, F.; Kilburg, A.; Magnard, S.; Montigny, C.; Picard, M.; Prier, C.; Monticelli, L.; Bornert, O.; Agez, M.; et al. Quantification of Detergents Complexed with Membrane Proteins. Sci. Rep. 2017, 7, 41751. [Google Scholar] [CrossRef]
- Pebay-Peyroula, E.; Garavito, R.; Rosenbusch, J.; Zulauf, M.; Timmins, P. Detergent structure in tetragonal crystals of OmpF porin. Structure 1995, 3, 1051–1059. [Google Scholar] [CrossRef][Green Version]
- Mio, K.; Sato, C. Lipid environment of membrane proteins in cryo-EM based structural analysis. Biophys. Rev. 2018, 10, 307–316. [Google Scholar] [CrossRef]
- Schulz, S.; Wilkes, M.; Mills, D.J.; Meier, T. Molecular architecture of the N-type ATPase rotor ring from Burkholderia pseudomallei. EMBO Rep. 2017, 18, 526–535. [Google Scholar] [CrossRef]
- Zampieri, V.; Gobet, A.; Robert, X.; Falson, P.; Chaptal, V. Nanodisc, amphipol or detergent belts in cryoEM reconstructions of membrane proteins are similar and cor-respond to a common ordered solvent layer. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ravaud, S.; Cao, M.-A.D.; Jidenko, M.; Ebel, C.; Le Maire, M.; Jault, J.-M.; Di Pietro, A.; Haser, R.; Aghajari, N. The ABC transporter BmrA from Bacillus subtilis is a functional dimer when in a detergent-solubilized state. Biochem. J. 2006, 395, 345–353. [Google Scholar] [CrossRef]
- Pogozheva, I.D.; Tristram-Nagle, S.; Mosberg, H.I.; Lomize, A.L. Structural adaptations of proteins to different biological membranes. Biochim. Biophys. Acta Biomembr. 2013, 1828, 2592–2608. [Google Scholar] [CrossRef] [PubMed]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef] [PubMed]
- Kleywegt, G.J.; Jones, T.A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Echols, N.; Headd, J.J.; Hung, L.-W.; Jain, S.; Kapral, G.J.; Kunstleve, R.W.G.; et al. The Phenix software for automated determination of macromolecular structures. Methods 2011, 55, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.; Thornton, J. Nacess V2.1.1—Solvent Acessible Area Calculations. Available online: http://www.bioinf.manchester.ac.uk/naccess/ (accessed on 15 December 2020).
- PyMOL. The PyMOL Molecular Graphics System; Version 1.5.0.4; Schrödinger, LLC: New York, NY, USA, 2010. [Google Scholar]
- WwPDB Consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef]
- Penel, S.; Pebay-Peyroula, E.; Rosenbusch, J.; Rummel, G.; Schirmer, T.; Timmins, P. Detergent binding in trigonal crystals of OmpF porin from Escherichia coli. Biochimie 1998, 80, 543–551. [Google Scholar] [CrossRef]
- Roth, M.; Arnoux, B.; Ducruix, A.; Reiss-Husson, F. Structure of the detergent phase and protein-detergent interactions in crystals of the wild-type (strain Y) Rhodobacter sphaeroides photochemical reaction center. Biochemistry 1991, 30, 9403–9413. [Google Scholar] [CrossRef]
- Wolfe, A.J.; Si, W.; Zhang, Z.; Blanden, A.R.; Hsueh, Y.-C.; Gugel, J.F.; Pham, B.; Chen, M.; Loh, S.N.; Rozovsky, S.; et al. Quantification of Membrane Protein-Detergent Complex Interactions. J. Phys. Chem. B 2017, 121, 10228–10241. [Google Scholar] [CrossRef]
- Perlmutter, J.D.; Popot, J.-L.; Sachs, J.N. Molecular Dynamics Simulations of a Membrane Protein/Amphipol Complex. J. Membr. Biol. 2014, 247, 883–895. [Google Scholar] [CrossRef]
- Etzkorn, M.; Zoonens, M.; Catoire, L.J.; Popot, J.-L.; Hiller, S. How Amphipols Embed Membrane Proteins: Global Solvent Accessibility and Interaction with a Flexible Protein Terminus. J. Membr. Biol. 2014, 247, 965–970. [Google Scholar] [CrossRef]
- Breyton, C.; Javed, W.; Vermot, A.; Arnaud, C.-A.; Hajjar, C.; Dupuy, J.; Petit-Hartlein, I.; Le Roy, A.; Martel, A.; Thépaut, M.; et al. Assemblies of lauryl maltose neopentyl glycol (LMNG) and LMNG-solubilized membrane proteins. Biochim. Biophys. Acta Biomembr. 2019, 1861, 939–957. [Google Scholar] [CrossRef]
- Le Maire, M.; Champeil, P.; Møller, J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta Biomembr. 2000, 1508, 86–111. [Google Scholar] [CrossRef]
- Chaptal, V.; Zampieri, V.; Wiseman, B.; Orelle, C.; Martin, J.; Nguyen, K.-A.; Magnard, S.; Gobet, A.; Di Cesare, M.; Javed, W.; et al. Drug-bound and -free outward-facing structures of a multidrug ABC exporter point to a swing mechanism. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ostuni, M.A.; Iatmanen, S.; Teboul, D.; Robert, J.-C.; Lacapere, J.-J. Characterization of Membrane Protein Preparations: Measurement of Detergent Content and Ligand Binding After Proteoliposomes Reconstitution. Methods Mol. Biol. 2010, 654, 3–18. [Google Scholar]
- Geertsma, E.R.; Mahmood, N.A.B.N.; Schuurman-Wolters, G.K.; Poolman, B. Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 2008, 3, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Ollivon, M.; Lesieur, S.; Grabielle-Madelmont, C.; Paternostre, M. Vesicle reconstitution from lipid–detergent mixed micelles. Biochim. Biophys. Acta Biomembr. 2000, 1508, 34–50. [Google Scholar] [CrossRef]
- Kragh-Hansen, U.; le Maire, M.; Møller, J.V. The Mechanism of Detergent Solubilization of Liposomes and Protein-Containing Membranes. Biophys. J. 1998, 75, 2932–2946. [Google Scholar] [CrossRef]
- Rigaud, J.L.; Levy, D.; Mosser, G.; Lambert, O. Detergent removal by non-polar polystyrene beads. Applications to membrane protein reconstitution and two-dimensional crystallization. Eur. Biophys. J. 1998, 27, 305–319. [Google Scholar] [CrossRef]
- Zoonens, M.; Popot, J.L. Amphipols for each season. J. Membr. Biol. 2014, 247, 759–796. [Google Scholar] [CrossRef]
- Stieger, B.; Steiger, J.; Locher, K.P. Membrane lipids and transporter function. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166079. [Google Scholar] [CrossRef] [PubMed]
- Popot, J.L. Membrane Proteins in Aqueous Solutions: From Detergents to Amphipols; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Stetsenko, A.; Guskov, A. An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization. Crystals 2017, 7, 197. [Google Scholar] [CrossRef]
- Abel, S.; Marchi, M.; Solier, J.; Finet, S.; Brillet, K.; Bonneté, F. Structural insights into the membrane receptor ShuA in DDM micelles and in a model of gram-negative bacteria outer membrane as seen by SAXS and MD simulations. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183504. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ghosh, S.; Jana, S.; Robertson, N.; Tate, C.G.; Vaidehi, N. How Do Branched Detergents Stabilize GPCRs in Micelles? Biochemistry 2020, 59, 2125–2134. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zampieri, V.; Hilpert, C.; Garnier, M.; Gestin, Y.; Delolme, S.; Martin, J.; Falson, P.; Launay, G.; Chaptal, V. The Det.Belt Server: A Tool to Visualize and Estimate Amphipathic Solvent Belts around Membrane Proteins. Membranes 2021, 11, 459. https://doi.org/10.3390/membranes11070459
Zampieri V, Hilpert C, Garnier M, Gestin Y, Delolme S, Martin J, Falson P, Launay G, Chaptal V. The Det.Belt Server: A Tool to Visualize and Estimate Amphipathic Solvent Belts around Membrane Proteins. Membranes. 2021; 11(7):459. https://doi.org/10.3390/membranes11070459
Chicago/Turabian StyleZampieri, Veronica, Cécile Hilpert, Mélanie Garnier, Yannick Gestin, Sébastien Delolme, Juliette Martin, Pierre Falson, Guillaume Launay, and Vincent Chaptal. 2021. "The Det.Belt Server: A Tool to Visualize and Estimate Amphipathic Solvent Belts around Membrane Proteins" Membranes 11, no. 7: 459. https://doi.org/10.3390/membranes11070459
APA StyleZampieri, V., Hilpert, C., Garnier, M., Gestin, Y., Delolme, S., Martin, J., Falson, P., Launay, G., & Chaptal, V. (2021). The Det.Belt Server: A Tool to Visualize and Estimate Amphipathic Solvent Belts around Membrane Proteins. Membranes, 11(7), 459. https://doi.org/10.3390/membranes11070459






