Ultrafiltration Fractionation of Bovine Hemoglobin Hydrolysates: Prediction of Separation Performances for Optimal Enrichment in Antimicrobial Peptide
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
2.2. Hydrolysate Preparation
2.3. Yield and Enrichment Simulation Methodology
2.3.1. Membrane Calibration
2.3.2. Yield
2.3.3. Enrichment
2.4. Ultrafiltration Experiments
2.5. Hydrolysates and Fraction Reverse Phase HPLC Analyses
2.6. Hydrolysates and Fraction SE-HPLC Analysis
2.7. Neokyotorphin Relative Concentration Quantification by RP-HPLC/MS
2.8. Evaluation of Antimicrobial Activity
3. Results and Discussion
3.1. bHb Hydrolysates Characterization
3.2. NKT Enrichement by Ultrafiltration
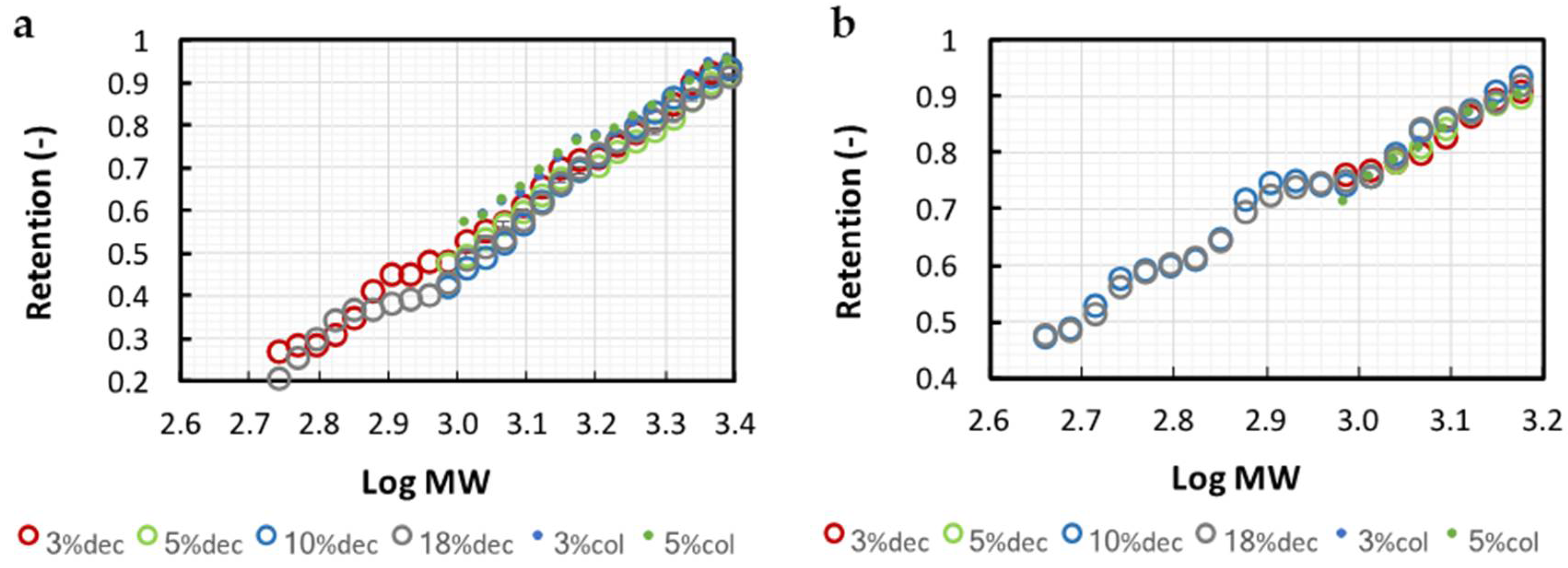
3.2.1. Membrane Calibrations
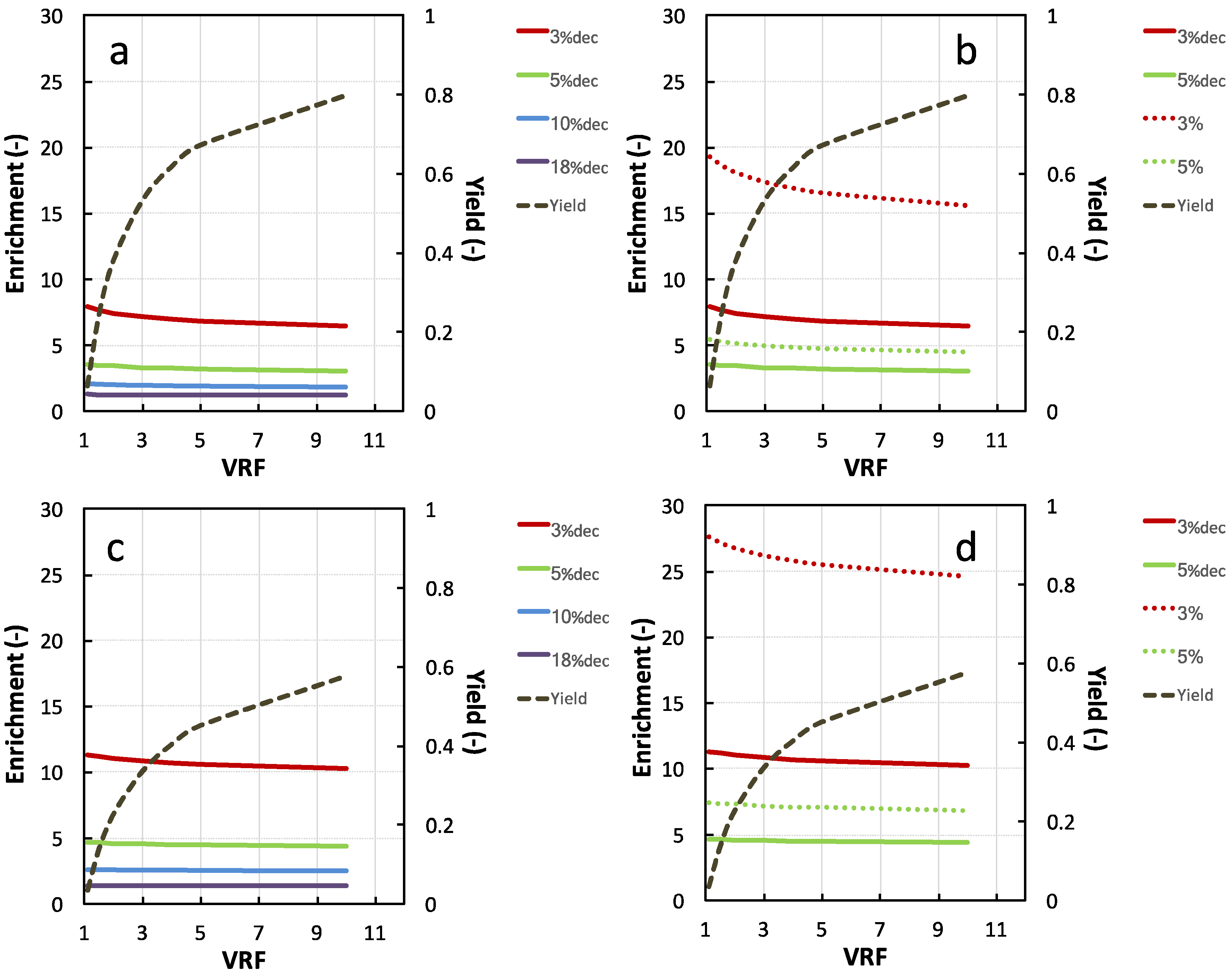
3.2.2. NKT Yield, Enrichment, and Purity Simulation
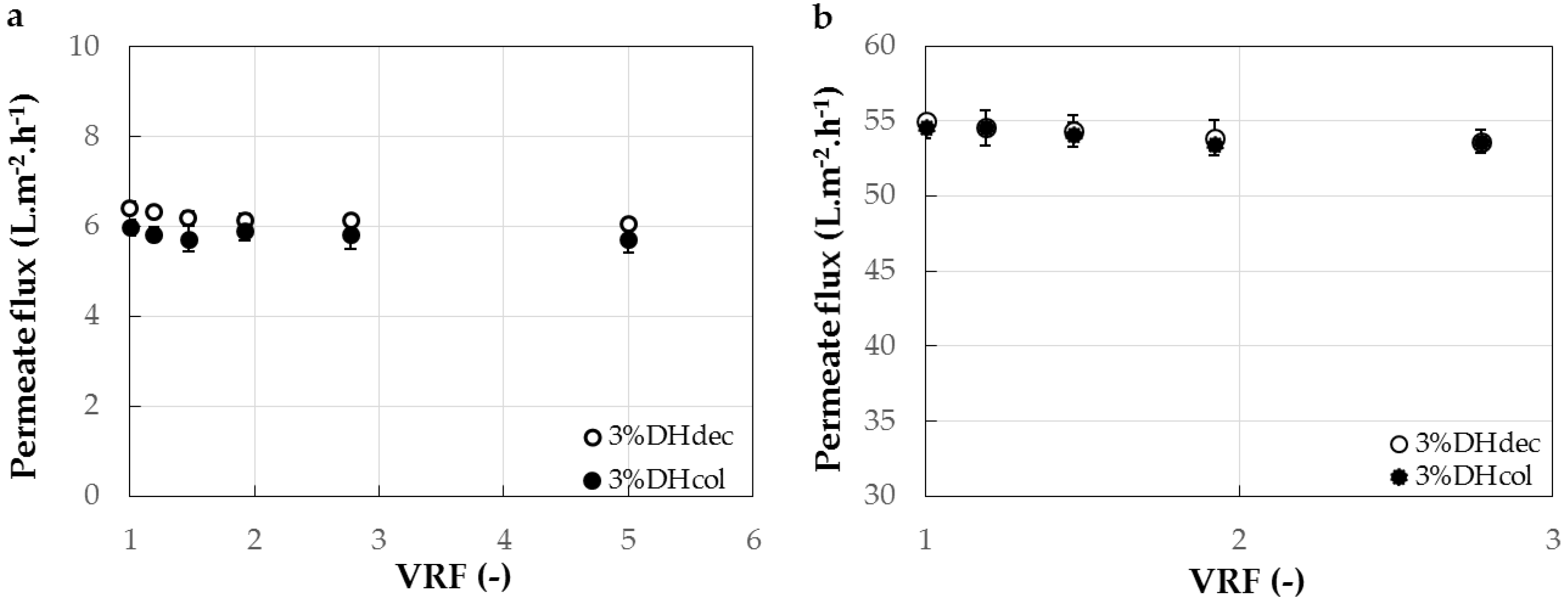
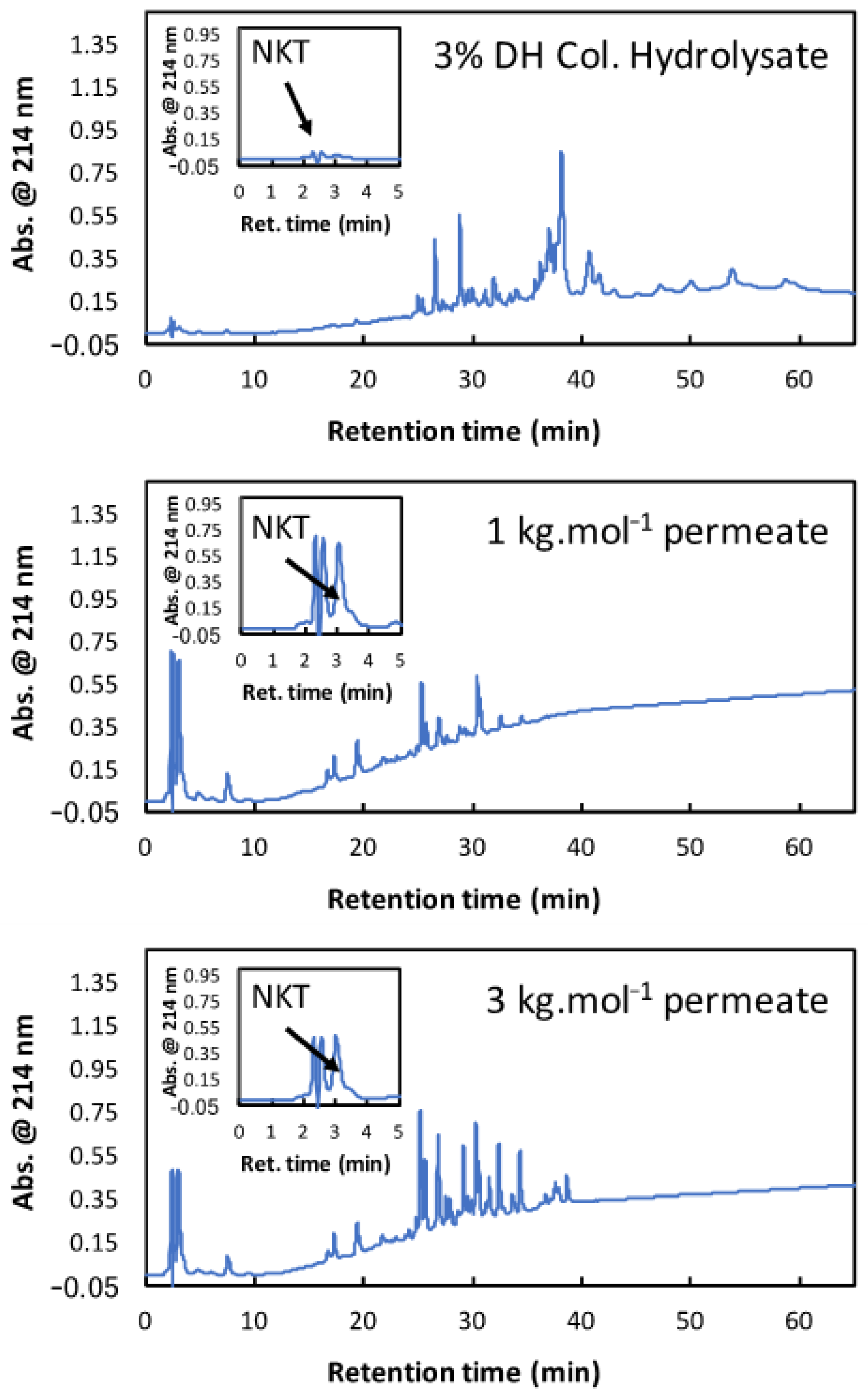
3.3. Experimental Fractionation with 1 kg·mol−1 and 3 kg·mol−1 Membranes
3.4. Evaluation of Peptide Fraction Antimicrobial Activity
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lynch, S.A.; Mullen, A.M.; O’Neill, E.E.; García, C.Á. Harnessing the potential of blood proteins as functional ingredients: A review of the state of the art in blood processing. Compr. Rev. Food Sci. Food Saf. 2017, 16, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Nedjar-Arroume, N.; Dubois-Delval, V.; Adje, E.Y.; Traisnel, J.; Krier, F.; Mary, P.; Guillochon, D. Bovine hemoglobin: An attractive source of antibacterial peptides. Peptides 2008, 29, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.P.S.; Hancock, R.E. The relationship between peptide structure and antibacterial activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Lopes, S.C.; Fedorov, A.; Castanho, M.A. Lipidic membranes are potential “catalysts” in the ligand activity of the multifunctional pentapeptide neokyotorphin. Chembiochem 2005, 6, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Catiau, L.; Traisnel, J.; Delval-Dubois, V.; Chihib, N.E.; Guillochon, D.; Nedjar-Arroume, N. Minimal antimicrobial peptidic sequence from hemoglobin alpha-chain: KYR. Peptides 2011, 32, 633–638. [Google Scholar] [CrossRef]
- Przybylski, R.; Firdaous, L.; Châtaigné, G.; Dhulster, P.; Nedjar, N. Production of an antimicrobial peptide derived from slaughterhouse by-product and its potential application on meat as preservative. Food Chem. 2016, 211, 306–313. [Google Scholar] [CrossRef]
- Vanhoute, M.; Firdaous, L.; Bazinet, L.; Froidevaux, R.; Lecouturier, D.; Guillochon, D.; Dhulster, P. Effect of haem on the fractionation of bovine haemoglobin peptic hydrolysate by electrodialysis with ultrafiltration membranes. J. Membr. Sci. 2010, 365, 16–24. [Google Scholar] [CrossRef]
- Aubes-Dafau, I.; Capdevielle, J.; Seris, J.L.; Combes, D. Bitter peptide from hemoglobin hydrolysate: Isolation and characterization. FEBS Lett. 1995, 364, 115–119. [Google Scholar]
- Dhulster, P.; Kapel, R.; Froidevaux, R.; Nedjar-Arroume, N.; Fertin-Bazus, A.; Choisnard, L.; Guillochon, D. Advancement in intermediate opioid peptide production in an enzymatic membrane reactor assisted by solvent extraction. Desalination 2002, 148, 221–226. [Google Scholar] [CrossRef]
- Lebrun, F.; Bazus, A.; Dhulster, P.; Guillochon, D. Influence of molecular interactions on ultrafiltration of a bovine hemoglobin hydrolysate with an organic membrane. J. Membr. Sci. 1998, 146, 113–124. [Google Scholar] [CrossRef]
- Agyei, D.; Ongkudon, C.M.; Wei, C.Y.; Chan, A.S.; Danquah, M.K. Bioprocess challenges to the isolation and purification of bioactive peptides. Food Bioprod. Process. 2016, 98, 244–256. [Google Scholar] [CrossRef]
- He, R.; Girgih, A.T.; Malomo, S.A.; Ju, X.; Aluko, R.E. Antioxidant activities of enzymatic rapeseed protein hydrolysates and the membrane ultrafiltration fractions. J. Funct. Foods 2013, 5, 219–227. [Google Scholar] [CrossRef]
- Onuh, J.O.; Girgih, A.T.; Aluko, R.E.; Aliani, M. In vitro antioxidant properties of chicken skin enzymatic protein hydrolysates and membrane fractions. Food Chem. 2014, 150, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Picot, L.; Ravallec, R.; Fouchereau-Péron, M.; Vandanjon, L.; Jaouen, P.; Chaplain-Derouiniot, M.; Bergé, J.P. Impact of ultrafiltration and nanofiltration of an industrial fish protein hydrolysate on its bioactive properties. J. Sci. Food Agric. 2010, 90, 1819–1826. [Google Scholar] [CrossRef]
- Ranamukhaarachchi, S.; Meissner, L.; Moresoli, C. Production of antioxidant soy protein hydrolysates by sequential ultrafiltration and nanofiltration. J. Membr. Sci. 2013, 429, 81–87. [Google Scholar] [CrossRef]
- Saidi, S.; Deratani, A.; Belleville, M.P.; Amar, R.B. Production and fractionation of tuna by-product protein hydrolysate by ultrafiltration and nanofiltration: Impact on interesting peptides fractions and nutritional properties. Food Res. Int. 2014, 65, 453–461. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, M.; Yang, X.; Jiang, Y. Improvement on functional properties of wheat gluten by enzymatic hydrolysis and ultrafiltration. J. Cereal Sci. 2006, 44, 93–100. [Google Scholar] [CrossRef]
- Yu, G.C.; Li, J.T.; He, H.U.I.; Huang, W.h.; Zhang, W.J. Ultrafiltration preparation of potent bioactive corn peptide as alcohol metabolism stimulator in vivo and study on its mechanism of action. J. Food Biochem. 2013, 37, 161–167. [Google Scholar] [CrossRef]
- Bourseau, P.; Vandanjon, L.; Jaouen, P.; Chaplain-Derouiniot, M.; Masse, A.; Guérard, F.; Berge, J.P. Fractionation of fish protein hydrolysates by ultrafiltration and nanofiltration: Impact on peptidic populations. Desalination 2009, 244, 303–320. [Google Scholar] [CrossRef]
- Chabeaud, A.; Vandanjon, L.; Bourseau, P.; Jaouen, P.; Chaplain-Derouiniot, M.; Guérard, F. Performances of ultrafiltration membranes for fractionating a fish protein hydrolysate: Application to the refining of bioactive peptidic fractions. Sep. Purif. Technol. 2009, 66, 463–471. [Google Scholar] [CrossRef]
- Kapel, R.; Klingenberg, F.; Framboisier, X.; Dhulster, P.; Marc, I. An original use of size exclusion-HPLC for predicting the performances of batch ultrafiltration implemented to enrich a complex protein hydrolysate in a targeted bioactive peptide. J. Membr. Sci. 2011, 383, 26–34. [Google Scholar] [CrossRef]
- Wh, C.; Ji, M.; Fw, F. Standardizing a method for clinical hemoglobinometry. US Armed Forces Med. J. 1954, 5, 693–703. [Google Scholar]
- Church, F.C.; Porter, D.h.; Catignani, G.L.; Swaisgood, H.E. An o-phthalaldehyde spectrophotometric assay for proteinases. Anal. Biochem. 1985, 146, 343–348. [Google Scholar] [CrossRef]
- Bodin, A.; Framboisier, X.; Alonso, D.; Marc, I.; Kapel, R. Size-exclusion HPLC as a sensitive and calibrationless method for complex peptide mixtures quantification. J. Chromatogr. B 2015, 1006, 71–79. [Google Scholar] [CrossRef]
- Sila, A.; Hedhili, K.; Przybylski, R.; Ellouz-Chaabouni, S.; Dhulster, P.; Bougatef, A.; Nedjar-Arroume, N. Antibacterial activity of new peptides from barbel protein hydrolysates and mode of action via a membrane damage mechanism against Listeria monocytogenes. J. Funct. Foods 2014, 11, 322–329. [Google Scholar] [CrossRef]
- Dubois, V.; Nedjar-Arroume, N.; Guillochon, D. Influence of pH on the appearance of active peptides in the course of peptic hydrolysis of bovine haemoglobin. Prep. Biochem. Biotechnol. 2005, 35, 85–102. [Google Scholar] [CrossRef]
- Linderstrom-Lang, K. The initial phases of the enzymatic degradation of proteins. Bull. Société Chim. Biol. 1953, 35, 100–116. [Google Scholar]
- Przybylski, R.; Bazinet, L.; Kouach, M.; Goossens, J.F.; Dhulster, P.; Firdaous, L.; Nedjar-Arroume, N. Slaughterhouse By-Product Valorization: Hydrolysis Degree Modification for Higher Antimicrobial Recovery by Electroseparation. Waste Biomass Valorization 2020, 1–13. [Google Scholar] [CrossRef]
- Gourley, L.; Gauthie, S.F.; Pouliot, Y.; Mollé, D.; Léonil, J.; Maubois, J.L. Identification of casein peptides interacting with polysulfone ultrafiltration membranes. Le Lait 1998, 78, 633–646. [Google Scholar] [CrossRef]
- Bouhallab, S.; Henry, G. Transmission of a hydrophobic peptide through an inorganic ultrafiltration membrane: Effect of membrane support. J. Membr. Sci. 1995, 104, 73–79. [Google Scholar] [CrossRef]
- Saidi, S.; Deratani, A.; Amar, R.B.; Belleville, M.P. Fractionation of a tuna dark muscle hydrolysate by a two-step membrane process. Sep. Purif. Technol. 2013, 108, 28–36. [Google Scholar] [CrossRef]
- Kapel, R.; Froidevaux, R.; Nedjar-Arroume, N.; Fertin-Bazus, A.; Dhulster, P.; Guillochon, D. Continuous production of a peptidic fraction containing the intermediate opioid peptide LVV-haemorphin-7 (LVVh-7) by peptic hydrolysis of bovine haemoglobin in a continuous membrane reactor. Biotechnol. Appl. Biochem. 2003, 37, 317–324. [Google Scholar] [CrossRef] [PubMed]

| Molecular Weight Cut-Off Membrane | 1 kg·mol−1 Permeate | 3 kg·mol−1 Permeate | ||
|---|---|---|---|---|
| Colored or decolored hydrolysate (3%DH) | Decolored | Colored | Decolored | Colored |
| Initial NKT purity (%) | 2.6 | 2.5 | 2.6 | 2.5 |
| Calc. NKT Enrich. (-) | 10.7 | 25.5 | 7.2 | 17.4 |
| Exp. NKT Enrich. (-) | 9.7 | 28.7 | 6.8 | 15.7 |
| Calc. NKT purity (%) | 27.9 | 62.5 | 18.8 | 42.6 |
| Exp. NKT purity (%) | 25.3 | 70.3 | 17.7 | 38.5 |
| Calc. NKT Yield. (%) | 46 | 46 | 54 | 54 |
| Exp. NKT Yield. (%) | 52 | 44 | 55 | 55 |
| Exp. productivity | 1.2 | 0.5 | 21.2 | 9.8 |
| (g.m−2·h−1) | ||||
| Sample | Colored or Decolored Hydrolysate (3%DH) | Molecular Weight Cut-off | Gram-Positive Bacteria | Gram-Negative Bacteria | ||
|---|---|---|---|---|---|---|
| Microccocus luteus ATCC 4698 | Listeria innocua ATCC 33090 | Escherichia coli ATCC 25922 | Salmonella enteretidis ATCC 13076 | |||
| NKT | Standard | / | 5.85 * | 0.65 * | 5.85 * | 3.25 * |
| Hydrolysate | Colored | / | 256.2 ± 0.00 | 128.1 ± 0.00 | 53.38 ± 15.1 | 128.1 ± 0.00 |
| Permeate | Colored | 1 kg·mol−1 | 2.06 ± 0.00 | 16.43 ± 0.00 | 8.21 ± 0.00 | 8.21 ± 0.00 |
| Permeate | Colored | 3 kg·mol−1 | 47.93 ± 0.00 | 39.94 ± 11.3 | 5.99 ± 0.00 | 9.98 ± 1.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beaubier, S.; Przybylski, R.; Bodin, A.; Nedjar, N.; Dhulster, P.; Kapel, R. Ultrafiltration Fractionation of Bovine Hemoglobin Hydrolysates: Prediction of Separation Performances for Optimal Enrichment in Antimicrobial Peptide. Membranes 2021, 11, 73. https://doi.org/10.3390/membranes11020073
Beaubier S, Przybylski R, Bodin A, Nedjar N, Dhulster P, Kapel R. Ultrafiltration Fractionation of Bovine Hemoglobin Hydrolysates: Prediction of Separation Performances for Optimal Enrichment in Antimicrobial Peptide. Membranes. 2021; 11(2):73. https://doi.org/10.3390/membranes11020073
Chicago/Turabian StyleBeaubier, Sophie, Rémi Przybylski, Alice Bodin, Naïma Nedjar, Pascal Dhulster, and Romain Kapel. 2021. "Ultrafiltration Fractionation of Bovine Hemoglobin Hydrolysates: Prediction of Separation Performances for Optimal Enrichment in Antimicrobial Peptide" Membranes 11, no. 2: 73. https://doi.org/10.3390/membranes11020073
APA StyleBeaubier, S., Przybylski, R., Bodin, A., Nedjar, N., Dhulster, P., & Kapel, R. (2021). Ultrafiltration Fractionation of Bovine Hemoglobin Hydrolysates: Prediction of Separation Performances for Optimal Enrichment in Antimicrobial Peptide. Membranes, 11(2), 73. https://doi.org/10.3390/membranes11020073








