Cu Homeostasis in Bacteria: The Ins and Outs
Abstract
1. Introduction
2. Copper Import across the Outer and Inner Membranes in Bacteria
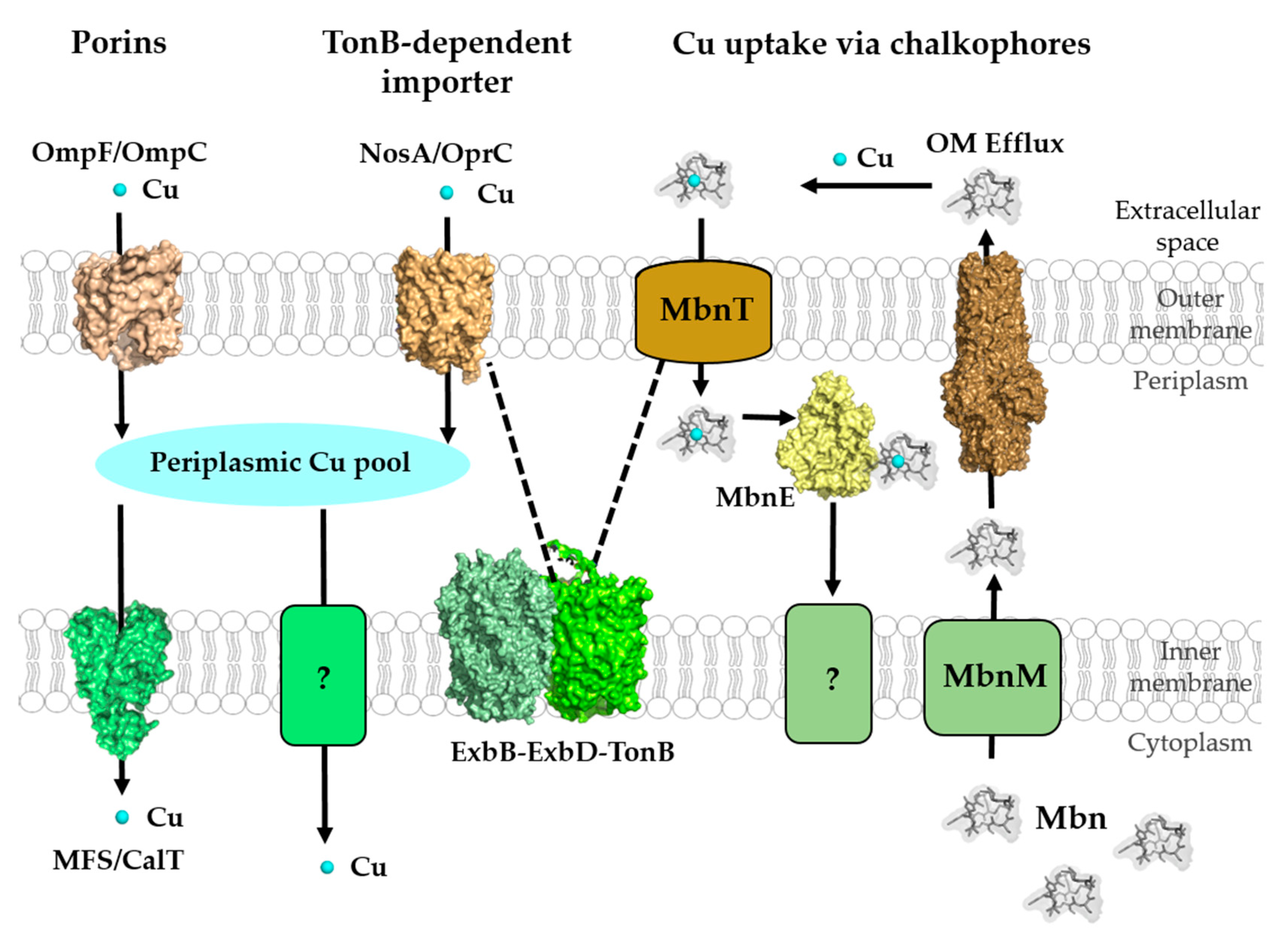
2.1. Cu-Uptake across the Outer Membrane of Gram-Negative Bacteria and Mycobacteria
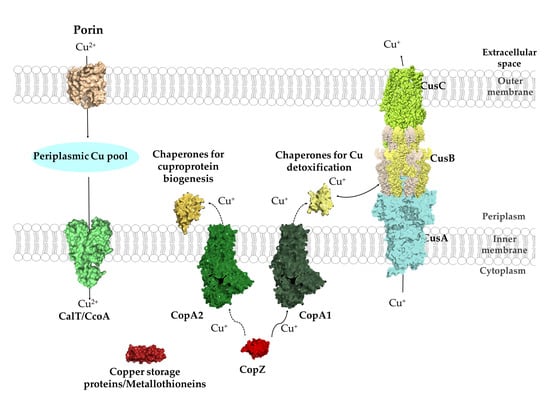
2.2. Cu Transit through the Periplasmic Space in Gram-Negative Bacteria
2.3. Cu Uptake across the Inner Membrane
The CcoA-Like Cu-Transporter (CalT) Family
2.4. Cu Uptake via Chalkophores: Methanobactins and Yersiniabactin
2.4.1. Methanobactin (Mbn)
2.4.2. Yersiniabactin (Ybt)
3. Reduction of Cu(II) to Cu(I) Is a Prerequisite for Cytoplasmic Copper Storage and for Re-Routing It to the Periplasm
4. The Cytosolic Cu Pool: Chaperones, Storage Proteins and Chemical Chelators
4.1. The CopZ-Like Chaperones
4.2. CupA
| Protein/Organism | Affinity for Cu(I) (Kd, M) | Reference |
|---|---|---|
| CopZ (B. subtilis) | 6 × 10−22 | [189] |
| N-MBS1 (E. coli) | 7 × 10−19 | [189] |
| N-MBS2 (E. coli) | 3 × 10−18 | [189] |
| TM-MBS1 (A. fulgidus) | 1 × 10−15 | [190] |
| TM-MBS2 (A. fulgidus) | 1 × 10−15 | [190] |
| CusF (E. coli) | 5 × 10−11 | [191] |
| PccA (R. capsulatus) | 8 × 10−16 | [47] |
| SenC (R. capsulatus) | 3 × 10−15 | [47] |
4.3. Metallothioneins
4.4. Copper Storage Proteins
4.5. Glutathione as Chemical Cu Chelator
5. Copper Export across the Cytoplasmic Membrane
5.1. P1B-Type ATPases
5.1.1. Structure of P1B-Type ATPases
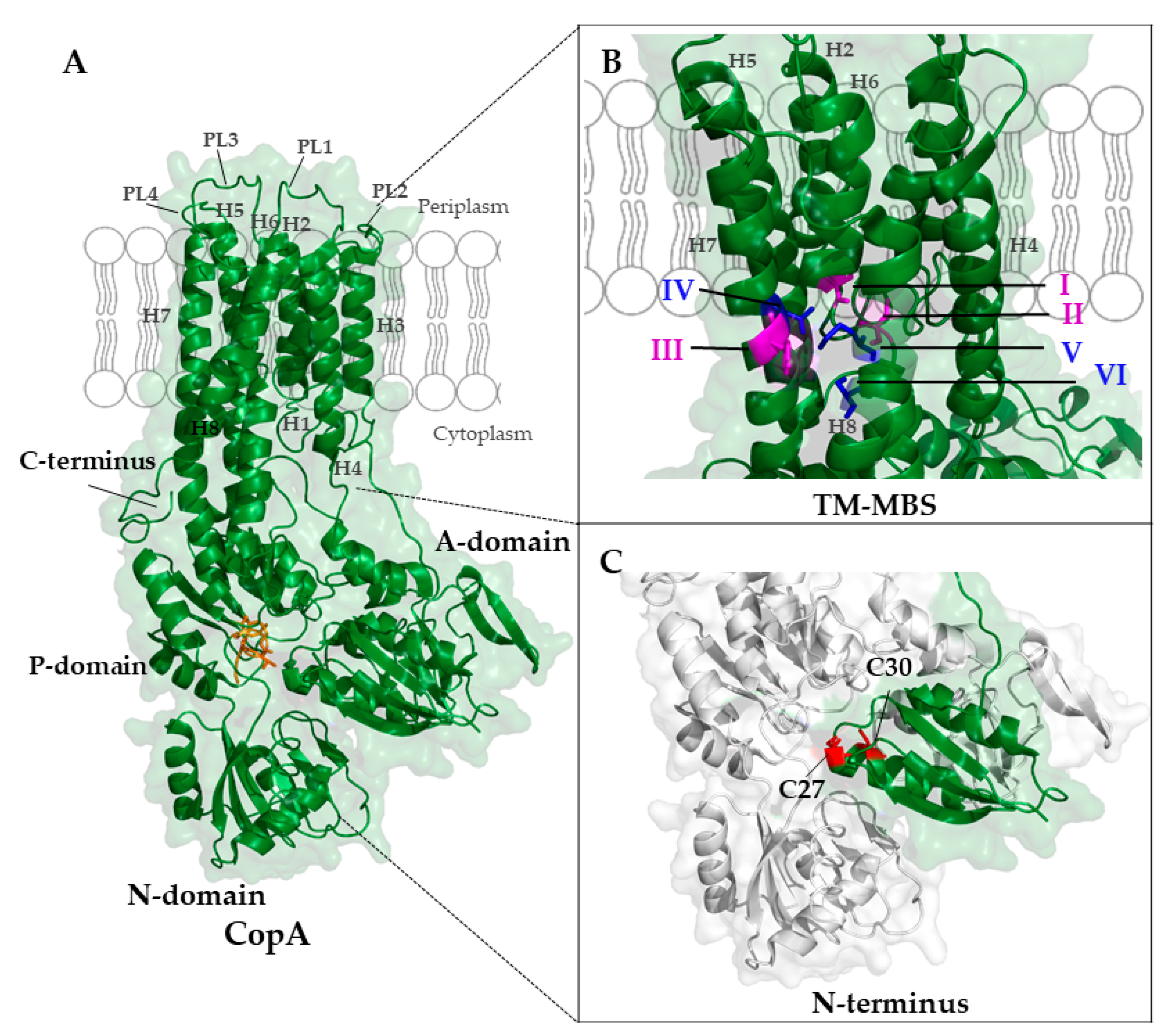
5.1.2. Mechanism of Cu Transfer by P1B-Type ATPases
5.2. Cu-Transporting RND Systems: The Cus-System
Structure and Mechanism of the CusCBA Complex
5.3. The Cop/Pco Systems
5.4. Regulation of Cu Export
6. Periplasmic Copper Chaperones and their Targets
6.1. CusF
6.2. CopI
6.3. Periplasmic Cu Chaperones for Cox Assembly: Sco1, PCuAC and Cox11
6.4. Copper Chaperones for Nitrous Oxide Reductase (NosZ): NosL, SenC2 and PCuAC
6.5. Copper Chaperones for Laccase-Like Multi-Copper Oxidases (MCO): CopG
6.6. CueP
7. Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Argüello, J.M.; Raimunda, D.; Padilla-Benavides, T. Mechanisms of copper homeostasis in bacteria. Front. Cell. Infect. Microbiol. 2013, 3, 73. [Google Scholar] [CrossRef]
- Giachino, A.; Waldron, K.J. Copper tolerance in bacteria requires the activation of multiple accessory pathways. Mol. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Z.; Petrusan, A.J.; Hooke, P.; Hinsa-Leasure, S.M. Reduction of bacterial burden by copper alloys on high-touch athletic center surfaces. Am. J. Infect. Control 2018, 46, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Besold, A.N.; Culbertson, E.M.; Culotta, V.C. The Yin and Yang of copper during infection. J. Biol. Inorg. Chem. 2016, 21, 137–144. [Google Scholar] [CrossRef]
- Ladomersky, E.; Khan, A.; Shanbhag, V.; Cavet, J.S.; Chan, J.; Weisman, G.A.; Petris, M.J. Host and Pathogen Copper-Transporting P-Type ATPases Function Antagonistically during Salmonella Infection. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [PubMed]
- Antoine, R.; Rivera-Millot, A.; Roy, G.; Jacob-Dubuisson, F. Relationships Between Copper-Related Proteomes and Lifestyles in β Proteobacteria. Front. Microbiol. 2019, 10, 2217. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.K.; Hoye, E.A.; Talaat, A.M. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 2008, 190, 2939–2946. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.K.; Abomoelak, B.; Hoye, E.A.; Steinberg, H.; Talaat, A.M. CtpV: A putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2010, 77, 1096–1110. [Google Scholar] [CrossRef] [PubMed]
- Wolschendorf, F.; Ackart, D.; Shrestha, T.B.; Hascall-Dove, L.; Nolan, S.; Lamichhane, G.; Wang, Y.; Bossmann, S.H.; Basaraba, R.J.; Niederweis, M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2011, 108, 1621–1626. [Google Scholar] [CrossRef]
- Price, E.E.; Boyd, J.M. Genetic Regulation of Metal Ion Homeostasis in Staphylococcus aureus. Trends Microbiol. 2020. [Google Scholar] [CrossRef]
- Begg, S.L. The role of metal ions in the virulence and viability of bacterial pathogens. Biochem. Soc. Trans. 2019, 47, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.; Ding, C. The Role of Copper Homeostasis at the Host-Pathogen Axis: From Bacteria to Fungi. Int. J. Mol. Sci. 2019, 20, 175. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.L.; Grass, G.; Rensing, C. Copper toxicity and the origin of bacterial resistance—New insights and applications. Met. Integr. Biometal Sci. 2011, 3, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Darwin, K.H. Copper homeostasis in Mycobacterium tuberculosis. Met. Integr. Biometal Sci. 2015, 7, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Braymer, J.J.; Giedroc, D.P. Recent developments in copper and zinc homeostasis in bacterial pathogens. Curr. Opin. Chem. Biol. 2014, 19, 59–66. [Google Scholar] [CrossRef]
- Foster, A.W.; Osman, D.; Robinson, N.J. Metal preferences and metallation. J. Biol. Chem. 2014, 289, 28095–28103. [Google Scholar] [CrossRef]
- Gaetke, L.M.; Chow, C.K. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 2003, 189, 147–163. [Google Scholar] [CrossRef]
- Stewart, L.J.; Thaqi, D.; Kobe, B.; McEwan, A.G.; Waldron, K.J.; Djoko, K.Y. Handling of nutrient copper in the bacterial envelope. Met. Integr. Biometal Sci. 2019, 11, 50–63. [Google Scholar] [CrossRef]
- Durand, A.; Azzouzi, A.; Bourbon, M.L.; Steunou, A.S.; Liotenberg, S.; Maeshima, A.; Astier, C.; Argentini, M.; Saito, S.; Ouchane, S. c-Type Cytochrome Assembly Is a Key Target of Copper Toxicity within the Bacterial Periplasm. mBio 2015, 6, e01007–e01015. [Google Scholar] [CrossRef]
- Steunou, A.S.; Durand, A.; Bourbon, M.L.; Babot, M.; Tambosi, R.; Liotenberg, S.; Ouchane, S. Cadmium and Copper Cross-Tolerance. Cu+ Alleviates Cd2+ Toxicity, and Both Cations Target Heme and Chlorophyll Biosynthesis Pathway in Rubrivivax gelatinosus. Front. Microbiol. 2020, 11, 893. [Google Scholar] [CrossRef]
- Hiniker, A.; Collet, J.F.; Bardwell, J.C. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J. Biol. Chem. 2005, 280, 33785–33791. [Google Scholar] [CrossRef] [PubMed]
- Decker, H.; Terwilliger, N. Cops and robbers: Putative evolution of copper oxygen-binding proteins. J. Exp. Biol. 2000, 203, 1777–1782. [Google Scholar] [PubMed]
- Ekici, S.; Yang, H.; Koch, H.-G.; Daldal, F. Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. mBio 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Cantini, F.; Ciofi-Baffoni, S. Cellular copper distribution: A mechanistic systems biology approach. Cell. Mol. Life Sci. CMLS 2010, 67, 2563–2589. [Google Scholar] [CrossRef] [PubMed]
- Rensing, C.; McDevitt, S.F. The copper metallome in prokaryotic cells. Met. Ions. Life Sci. 2013, 12, 417–450. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, G.; Pilon, M. Copper Delivery to Chloroplast Proteins and its Regulation. Front. Plant Sci. 2015, 6, 1250. [Google Scholar] [CrossRef]
- Canonica, F.; Klose, D.; Ledermann, R.; Sauer, M.M.; Abicht, H.K.; Quade, N.; Gossert, A.D.; Chesnov, S.; Fischer, H.M.; Jeschke, G.; et al. Structural basis and mechanism for metallochaperone-assisted assembly of the Cu(A) center in cytochrome oxidase. Sci. Adv. 2019, 5, eaaw8478. [Google Scholar] [CrossRef]
- Ekici, S.; Pawlik, G.; Lohmeyer, E.; Koch, H.-G.; Daldal, F. Biogenesis of cbb(3)-type cytochrome c oxidase in Rhodobacter capsulatus. Biochim. Biophys. Acta 2012, 1817, 898–910. [Google Scholar] [CrossRef]
- Kudva, R.; Denks, K.; Kuhn, P.; Vogt, A.; Muller, M.; Koch, H.G. Protein translocation across the inner membrane of Gram-negative bacteria: The Sec and Tat dependent protein transport pathways. Res. Microbiol. 2013, 164, 505–534. [Google Scholar] [CrossRef]
- Stolle, P.; Hou, B.; Brüser, T. The Tat Substrate CueO Is Transported in an Incomplete Folding State. J. Biol. Chem. 2016, 291, 13520–13528. [Google Scholar] [CrossRef]
- Koch, H.G.; Winterstein, C.; Saribas, A.S.; Alben, J.O.; Daldal, F. Roles of the ccoGHIS gene products in the biogenesis of the cbb(3)-type cytochrome c oxidase. J. Mol. Biol. 2000, 297, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Kulajta, C.; Thumfart, J.O.; Haid, S.; Daldal, F.; Koch, H.G. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. J. Mol. Biol. 2006, 355, 989–1004. [Google Scholar] [CrossRef] [PubMed]
- Khalfaoui-Hassani, B.; Verissimo, A.F.; Shroff, N.P.; Ekici, S.; Trasnea, P.-I.; Utz, M.; Koch, H.-G.; Daldal, F. Biogenesis of Cytochrome c Complexes: From Insertion of Redox Cofactors to Assembly of Different Subunits. In Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling; Cramer, W.A., Kallas, T., Eds.; Springer: Dordrecht, The Netherlands, 2016; pp. 527–554. [Google Scholar]
- Richter, O.M.; Ludwig, B. Cytochrome c oxidase—Structure, function, and physiology of a redox-driven molecular machine. Rev. Physiol. Biochem. Pharmacol. 2003, 147, 47–74. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.O.; Rosenzweig, A.C. A tale of two methane monooxygenases. J. Biol. Inorg. Chem. JBIC Publ. Soc. Biol. Inorg. Chem. 2017, 22, 307–319. [Google Scholar] [CrossRef]
- Roberts, S.A.; Weichsel, A.; Grass, G.; Thakali, K.; Hazzard, J.T.; Tollin, G.; Rensing, C.; Montfort, W.R. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 2766–2771. [Google Scholar] [CrossRef]
- Horrell, S.; Kekilli, D.; Strange, R.W.; Hough, M.A. Recent structural insights into the function of copper nitrite reductases. Met. Integr. Biometal Sci. 2017, 9, 1470–1482. [Google Scholar] [CrossRef]
- Palm-Espling, M.E.; Niemiec, M.S.; Wittung-Stafshede, P. Role of metal in folding and stability of copper proteins in vitro. Biochim. Biophys. Acta 2012, 1823, 1594–1603. [Google Scholar] [CrossRef]
- Fenlon, L.A.; Slauch, J.M. Cytoplasmic Copper Detoxification in Salmonella Can Contribute to SodC Metalation but Is Dispensable during Systemic Infection. J. Bacteriol. 2017, 199, e00437-17. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Wüst, A.; Prasser, B.; Müller, C.; Einsle, O. Functional assembly of nitrous oxide reductase provides insights into copper site maturation. Proc. Natl. Acad. Sci. USA 2019, 116, 12822–12827. [Google Scholar] [CrossRef]
- Buffoni, F.; Ignesti, G. The copper-containing amine oxidases: Biochemical aspects and functional role. Mol. Genet. Metab. 2000, 71, 559–564. [Google Scholar] [CrossRef]
- Hermann, B.; Kern, M.; La Pietra, L.; Simon, J.; Einsle, O. The octahaem MccA is a haem c-copper sulfite reductase. Nature 2015, 520, 706–709. [Google Scholar] [CrossRef] [PubMed]
- Kanteev, M.; Goldfeder, M.; Fishman, A. Structure-function correlations in tyrosinases. Protein Sci. Publ. Protein Soc. 2015, 24, 1360–1369. [Google Scholar] [CrossRef] [PubMed]
- McGuirl, M.A.; Bollinger, J.A.; Cosper, N.; Scott, R.A.; Dooley, D.M. Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. J. Biol. Inorg. Chem. JBIC Publ. Soc. Biol. Inorg. Chem. 2001, 6, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Bennett, S.P.; Soriano-Laguna, M.J.; Bradley, J.M.; Svistunenko, D.A.; Richardson, D.J.; Gates, A.J.; Le Brun, N.E. NosL is a dedicated copper chaperone for assembly of the Cu(Z) center of nitrous oxide reductase. Chem. Sci. 2019, 10, 4985–4993. [Google Scholar] [CrossRef] [PubMed]
- Lohmeyer, E.; Schröder, S.; Pawlik, G.; Trasnea, P.-I.; Peters, A.; Daldal, F.; Koch, H.-G. The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb3-type cytochrome oxidase in Rhodobacter capsulatus. Biochim. Biophys. Acta 2012, 1817, 2005–2015. [Google Scholar] [CrossRef] [PubMed]
- Trasnea, P.I.; Andrei, A.; Marckmann, D.; Utz, M.; Khalfaoui-Hassani, B.; Selamoglu, N.; Daldal, F.; Koch, H.G. A Copper Relay System Involving Two Periplasmic Chaperones Drives cbb3-Type Cytochrome c Oxidase Biogenesis in Rhodobacter capsulatus. ACS Chem. Biol. 2018, 13, 1388–1396. [Google Scholar] [CrossRef]
- Serventi, F.; Youard, Z.A.; Murset, V.; Huwiler, S.; Buhler, D.; Richter, M.; Luchsinger, R.; Fischer, H.M.; Brogioli, R.; Niederer, M.; et al. Copper starvation-inducible protein for cytochrome oxidase biogenesis in Bradyrhizobium japonicum. J. Biol. Chem. 2012, 287, 38812–38823. [Google Scholar] [CrossRef]
- Canonica, F.; Hennecke, H.; Glockshuber, R. Biochemical pathway for the biosynthesis of the Cu(A) center in bacterial cytochrome c oxidase. FEBS Lett. 2019, 593, 2977–2989. [Google Scholar] [CrossRef]
- Abicht, H.K.; Scharer, M.A.; Quade, N.; Ledermann, R.; Mohorko, E.; Capitani, G.; Hennecke, H.; Glockshuber, R. How periplasmic thioredoxin TlpA reduces bacterial copper chaperone ScoI and cytochrome oxidase subunit II (CoxB) prior to metallation. J. Biol. Chem. 2014, 289, 32431–32444. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Katsari, E.; Katsaros, N.; Kubicek, K.; Mangani, S. A copper(I) protein possibly involved in the assembly of CuA center of bacterial cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2005, 102, 3994–3999. [Google Scholar] [CrossRef]
- Trasnea, P.-I.; Utz, M.; Khalfaoui-Hassani, B.; Lagies, S.; Daldal, F.; Koch, H.-G. Cooperation between two periplasmic copper chaperones is required for full activity of the cbb3-type cytochrome c oxidase and copper homeostasis in Rhodobacter capsulatus. Mol. Microbiol. 2016, 100, 345–361. [Google Scholar] [CrossRef]
- Thompson, A.K.; Gray, J.; Liu, A.; Hosler, J.P. The roles of Rhodobacter sphaeroides copper chaperones PCu(A)C and Sco (PrrC) in the assembly of the copper centers of the aa(3)-type and the cbb(3)-type cytochrome c oxidases. Biochim. Biophys. Acta 2012, 1817, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Rubino, J.T.; Franz, K.J. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. J. Inorg. Biochem. 2012, 107, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Arguello, J.M.; Gonzalez-Guerrero, M.; Raimunda, D. Bacterial transition metal P(1B)-ATPases: Transport mechanism and roles in virulence. Biochemistry 2011, 50, 9940–9949. [Google Scholar] [CrossRef] [PubMed]
- Mattle, D.; Zhang, L.; Sitsel, O.; Pedersen, L.T.; Moncelli, M.R.; Tadini-Buoninsegni, F.; Gourdon, P.; Rees, D.C.; Nissen, P.; Meloni, G. A sulfur-based transport pathway in Cu+-ATPases. EMBO Rep. 2015, 16, 728–740. [Google Scholar] [CrossRef]
- Gourdon, P.; Liu, X.Y.; Skjorringe, T.; Morth, J.P.; Moller, L.B.; Pedersen, B.P.; Nissen, P. Crystal structure of a copper-transporting PIB-type ATPase. Nature 2011, 475, 59–64. [Google Scholar] [CrossRef]
- Osman, D.; Cavet, J.S. Copper homeostasis in bacteria. Adv. Appl. Microbiol. 2008, 65, 217–247. [Google Scholar] [CrossRef]
- Schweigel-Röntgen, M. The families of zinc (SLC30 and SLC39) and copper (SLC31) transporters. Curr. Top. Membr. 2014, 73, 321–355. [Google Scholar] [CrossRef]
- Dancis, A.; Yuan, D.S.; Haile, D.; Askwith, C.; Eide, D.; Moehle, C.; Kaplan, J.; Klausner, R.D. Molecular characterization of a copper transport protein in S. cerevisiae: An unexpected role for copper in iron transport. Cell 1994, 76, 393–402. [Google Scholar] [CrossRef]
- Öhrvik, H.; Thiele, D.J. The role of Ctr1 and Ctr2 in mammalian copper homeostasis and platinum-based chemotherapy. J. Trace Elem. Med. Biol. 2015, 31, 178–182. [Google Scholar] [CrossRef]
- Petris, M.J. The SLC31 (Ctr) copper transporter family. Pflügers Arch. 2004, 447, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Logeman, B.L.; Zhang, X.; Liu, Y.; Thiele, D.J.; Yuan, P. X-ray structures of the high-affinity copper transporter Ctr1. Nat. Commun. 2019, 10, 1386. [Google Scholar] [CrossRef] [PubMed]
- Dyla, M.; Kjærgaard, M.; Poulsen, H.; Nissen, P. Structure and Mechanism of P-Type ATPase Ion Pumps. Annu. Rev. Biochem. 2020, 89, 583–603. [Google Scholar] [CrossRef] [PubMed]
- Kanamaru, K.; Kashiwagi, S.; Mizuno, T. A copper-transporting P-type ATPase found in the thylakoid membrane of the cyanobacterium Synechococcus species PCC7942. Mol. Microbiol. 1994, 13, 369–377. [Google Scholar] [CrossRef]
- Tottey, S.; Rich, P.R.; Rondet, S.A.M.; Robinson, N.J. Two Menkes-type ATPases Supply Copper for Photosynthesis inSynechocystis PCC 6803. J. Biol. Chem. 2001, 276, 19999–20004. [Google Scholar] [CrossRef]
- Solioz, M. Chapter 11—Copper Disposition in Bacteria. In Clinical and Translational Perspectives on WILSON DISEASE; Kerkar, N., Roberts, E.A., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 101–113. [Google Scholar]
- Solioz, M.; Stoyanov, J.V. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 2003, 27, 183–195. [Google Scholar] [CrossRef]
- Badarau, A.; Dennison, C. Thermodynamics of copper and zinc distribution in the cyanobacterium Synechocystis PCC 6803. Proc. Natl. Acad. Sci. USA 2011, 108, 13007–13012. [Google Scholar] [CrossRef]
- Raimunda, D.; Gonzalez-Guerrero, M.; Leeber, B.W., 3rd; Arguello, J.M. The transport mechanism of bacterial Cu+-ATPases: Distinct efflux rates adapted to different function. Biometals 2011, 24, 467–475. [Google Scholar] [CrossRef]
- Robinson, N.J.; Winge, D.R. Copper metallochaperones. Annu. Rev. Biochem. 2010, 79, 537–562. [Google Scholar] [CrossRef]
- Teitzel, G.M.; Geddie, A.; De Long, S.K.; Kirisits, M.J.; Whiteley, M.; Parsek, M.R. Survival and Growth in the Presence of Elevated Copper: Transcriptional Profiling of Copper-Stressed Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7242. [Google Scholar] [CrossRef]
- Almárcegui, R.J.; Navarro, C.A.; Paradela, A.; Albar, J.P.; von Bernath, D.; Jerez, C.A. New Copper Resistance Determinants in the Extremophile Acidithiobacillus ferrooxidans: A Quantitative Proteomic Analysis. J. Proteome Res. 2014, 13, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Selamoglu, N.; Önder, Ö.; Öztürk, Y.; Khalfaoui-Hassani, B.; Blaby-Haas, C.E.; Garcia, B.A.; Koch, H.-G.; Daldal, F. Comparative differential cuproproteomes of Rhodobacter capsulatus reveal novel copper homeostasis related proteins. Metallomics 2020, 12, 572–591. [Google Scholar] [CrossRef] [PubMed]
- Lutkenhaus, J.F. Role of a major outer membrane protein in Escherichia coli. J. Bacteriol. 1977, 131, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Nikaido, H.; Williams, K.E. Silver-resistant mutants of Escherichia coli display active efflux of Ag+ and are deficient in porins. J. Bacteriol. 1997, 179, 6127–6132. [Google Scholar] [CrossRef] [PubMed]
- Niederweis, M.; Danilchanka, O.; Huff, J.; Hoffmann, C.; Engelhardt, H. Mycobacterial outer membranes: In search of proteins. Trends Microbiol. 2010, 18, 109–116. [Google Scholar] [CrossRef]
- Alderwick, L.J.; Harrison, J.; Lloyd, G.S.; Birch, H.L. The Mycobacterial Cell Wall—Peptidoglycan and Arabinogalactan. Cold Spring Harbor Perspect. Med. 2015, 5, a021113. [Google Scholar] [CrossRef]
- Speer, A.; Rowland, J.L.; Haeili, M.; Niederweis, M.; Wolschendorf, F. Porins Increase Copper Susceptibility of Mycobacterium tuberculosis. J. Bacteriol. 2013, 195, 5133. [Google Scholar] [CrossRef]
- Faller, M.; Niederweis, M.; Schulz, G.E. The Structure of a Mycobacterial Outer-Membrane Channel. Science 2004, 303, 1189. [Google Scholar] [CrossRef]
- Niederweis, M.; Ehrt, S.; Heinz, C.; Klöcker, U.; Karosi, S.; Swiderek, K.M.; Riley, L.W.; Benz, R. Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol. Microbiol. 1999, 33, 933–945. [Google Scholar] [CrossRef]
- Shah, S.; Dalecki, A.G.; Malalasekera, A.P.; Crawford, C.L.; Michalek, S.M.; Kutsch, O.; Sun, J.; Bossmann, S.H.; Wolschendorf, F. 8-Hydroxyquinolines Are Boosting Agents of Copper-Related Toxicity in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2016, 60, 5765. [Google Scholar] [CrossRef]
- Carreira, C.; Pauleta, S.R.; Moura, I. The catalytic cycle of nitrous oxide reductase—The enzyme that catalyzes the last step of denitrification. J. Inorg. Biochem. 2017, 177, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Pomowski, A.; Zumft, W.G.; Kroneck, P.M.H.; Einsle, O. N2O binding at a [4Cu:2S] copper–sulphur cluster in nitrous oxide reductase. Nature 2011, 477, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Mokhele, K.; Tang, Y.J.; Clark, M.A.; Ingraham, J.L. A Pseudomonas stutzeri outer membrane protein inserts copper into N2O reductase. J. Bacteriol. 1987, 169, 5721. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Abdelal, A.H.; Clark, M.A.; Ingraham, J.L. Molecular characterization of nosA, a Pseudomonas stutzeri gene encoding an outer membrane protein required to make copper-containing N2O reductase. J. Bacteriol. 1991, 173, 5406. [Google Scholar] [CrossRef] [PubMed]
- Schalk, I.J.; Mislin, G.L.; Brillet, K. Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Curr. Top. Membr. 2012, 69, 37–66. [Google Scholar] [CrossRef]
- Schalk, I.J.; Hannauer, M.; Braud, A. New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 2011, 13, 2844–2854. [Google Scholar] [CrossRef]
- Schalk, I.J.; Cunrath, O. An overview of the biological metal uptake pathways in Pseudomonas aeruginosa. Environ. Microbiol. 2016, 18, 3227–3246. [Google Scholar] [CrossRef]
- Rechnitzer, H.; Brzuszkiewicz, E.; Strittmatter, A.; Liesegang, H.; Lysnyansky, I.; Daniel, R.; Gottschalk, G.; Rottem, S. Genomic features and insights into the biology of Mycoplasma fermentans. Microbiology 2011, 157, 760–773. [Google Scholar] [CrossRef]
- Lee, H.S.; Hancock, R.E.; Ingraham, J.L. Properties of a Pseudomonas stutzeri outer membrane channel-forming protein (NosA) required for production of copper-containing N2O reductase. J. Bacteriol. 1989, 171, 2096. [Google Scholar] [CrossRef]
- Wunsch, P.; Herb, M.; Wieland, H.; Schiek, U.M.; Zumft, W.G. Requirements for CuA and Cu-S Center Assembly of Nitrous Oxide Reductase Deduced from Complete Periplasmic Enzyme Maturation in the Nondenitrifier Pseudomonas putida. J. Bacteriol. 2003, 185, 887. [Google Scholar] [CrossRef]
- Chillappagari, S.; Miethke, M.; Trip, H.; Kuipers, O.P.; Marahiel, M.A. Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J. Bacteriol. 2009, 191, 2362–2370. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, H.; Nakae, T. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 1996, 142, 2137–2144. [Google Scholar] [CrossRef] [PubMed]
- Quintana, J.; Novoa-Aponte, L.; Argüello, J.M. Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. J. Biol. Chem. 2017, 292, 15691–15704. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, T.; Chen, G.; Pu, Q.; Liu, Q.; Zhang, Y.; Xu, L.; Wu, M.; Liang, H. A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog. 2019, 15, e1008198. [Google Scholar] [CrossRef]
- Bhamidimarri, S.P.; Young, T.R.; Shanmugam, M.; Soderholm, S.; Belzunces, B.; Baslé, A.; Skylaris, C.; Bumann, D.; Khalid, S.; van den Berg, B. Acquisition of ionic copper by a bacterial outer membrane protein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mermod, M.; Magnani, D.; Solioz, M.; Stoyanov, J.V. The copper-inducible ComR (YcfQ) repressor regulates expression of ComC (YcfR), which affects copper permeability of the outer membrane of Escherichia coli. Biometals 2012, 25, 33–43. [Google Scholar] [CrossRef]
- Egler, M.; Grosse, C.; Grass, G.; Nies, D.H. Role of the Extracytoplasmic Function Protein Family Sigma Factor RpoE in Metal Resistance of Escherichia coli. J. Bacteriol. 2005, 187, 2297. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Y.; Jing, C. Transcriptome analysis of silver, palladium, and selenium stresses in Pantoea sp. IMH. Chemosphere 2018, 208, 50–58. [Google Scholar] [CrossRef]
- Zheng, M.; Wang, X.; Templeton, L.J.; Smulski, D.R.; LaRossa, R.A.; Storz, G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 2001, 183, 4562–4570. [Google Scholar] [CrossRef]
- Richmond, C.S.; Glasner, J.D.; Mau, R.; Jin, H.; Blattner, F.R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999, 27, 3821–3835. [Google Scholar] [CrossRef]
- Dennison, C.; David, S.; Lee, J. Bacterial copper storage proteins. J. Biol. Chem. 2018, 293, 4616–4627. [Google Scholar] [CrossRef] [PubMed]
- Pittman, M.S.; Robinson, H.C.; Poole, R.K. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 2005, 280, 32254–32261. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Benavides, T.; George Thompson, A.M.; McEvoy, M.M.; Arguello, J.M. Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: The interaction of Escherichia coli CopA and CusF. J. Biol. Chem. 2014, 289, 20492–20501. [Google Scholar] [CrossRef] [PubMed]
- Dennison, C. The Coordination Chemistry of Copper Uptake and Storage for Methane Oxidation. Chemistry 2019, 25, 74–86. [Google Scholar] [CrossRef]
- Lee, J.; Dennison, C. Cytosolic Copper Binding by a Bacterial Storage Protein and Interplay with Copper Efflux. Int. J. Mol. Sci. 2019, 20, 4144. [Google Scholar] [CrossRef]
- Quistgaard, E.M.; Löw, C.; Guettou, F.; Nordlund, P. Understanding transport by the major facilitator superfamily (MFS): Structures pave the way. Nat. Rev. Mol. Cell Biol. 2016, 17, 123–132. [Google Scholar] [CrossRef]
- Ekici, S.; Turkarslan, S.; Pawlik, G.; Dancis, A.; Baliga, N.S.; Koch, H.-G.; Daldal, F. Intracytoplasmic copper homeostasis controls cytochrome c oxidase production. mBio 2014, 5, e01055-13. [Google Scholar] [CrossRef]
- Khalfaoui-Hassani, B.; Wu, H.; Blaby-Haas, C.E.; Zhang, Y.; Sandri, F.; Verissimo, A.F.; Koch, H.G.; Daldal, F. Widespread Distribution and Functional Specificity of the Copper Importer CcoA: Distinct Cu Uptake Routes for Bacterial Cytochrome c Oxidases. mBio 2018, 9. [Google Scholar] [CrossRef]
- Beaudoin, J.; Ekici, S.; Daldal, F.; Ait-Mohand, S.; Guérin, B.; Labbé, S. Copper transport and regulation in Schizosaccharomyces pombe. Biochem. Soc. Trans. 2013, 41, 1679–1686. [Google Scholar] [CrossRef]
- Arguello, J.M.; Eren, E.; Gonzalez-Guerrero, M. The structure and function of heavy metal transport P1B-ATPases. Biometals 2007, 20, 233–248. [Google Scholar] [CrossRef]
- Khalfaoui-Hassani, B.; Verissimo, A.F.; Koch, H.-G.; Daldal, F. Uncovering the Transmembrane Metal Binding Site of the Novel Bacterial Major Facilitator Superfamily-Type Copper Importer CcoA. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Blaby-Haas, C.E.; Steimle, S.; Verissimo, A.F.; Garcia-Angulo, V.A.; Koch, H.-G.; Daldal, F.; Khalfaoui-Hassani, B. Cu Transport by the Extended Family of CcoA-like Transporters (CalT) in Proteobacteria. Sci. Rep. 2019, 9, 1208. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Preciado, A.; Torres, A.G.; Merino, E.; Bonomi, H.R.; Goldbaum, F.A.; García-Angulo, V.A. Extensive Identification of Bacterial Riboflavin Transporters and Their Distribution across Bacterial Species. PLoS ONE 2015, 10, e0126124. [Google Scholar] [CrossRef]
- Vitreschak, A.G.; Rodionov, D.A.; Mironov, A.A.; Gelfand, M.S. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002, 30, 3141–3151. [Google Scholar] [CrossRef] [PubMed]
- Kenney, G.E.; Rosenzweig, A.C. Chalkophores. Annu. Rev. Biochem. 2018, 87, 645–676. [Google Scholar] [CrossRef]
- Anttila, J.; Heinonen, P.; Nenonen, T.; Pino, A.; Iwaï, H.; Kauppi, E.; Soliymani, R.; Baumann, M.; Saksi, J.; Suni, N.; et al. Is coproporphyrin III a copper-acquisition compound in Paracoccus denitrificans? Biochim. Biophys. Acta 2011, 1807, 311–318. [Google Scholar] [CrossRef]
- Byers, B.R.; Powell, M.V.; Lankford, C.E. Iron-chelating hydroxamic acid (schizokinen) active in initiation of cell division in Bacillus megaterium. J. Bacteriol. 1967, 93, 286–294. [Google Scholar] [CrossRef]
- Song, L.; Zhang, Y.; Chen, W.; Gu, T.; Zhang, S.Y.; Ji, Q. Mechanistic insights into staphylopine-mediated metal acquisition. Proc. Natl. Acad. Sci. USA 2018, 115, 3942–3947. [Google Scholar] [CrossRef]
- Fitch, M.W.; Graham, D.W.; Arnold, R.G.; Agarwal, S.K.; Phelps, P.; Speitel, G.E.; Georgiou, G. Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 1993, 59, 2771. [Google Scholar] [CrossRef]
- Kenney, G.E.; Rosenzweig, A.C. Methanobactins: Maintaining copper homeostasis in methanotrophs and beyond. J. Biol. Chem. 2018, 293, 4606–4615. [Google Scholar] [CrossRef]
- Kim, H.J.; Graham, D.W.; DiSpirito, A.A.; Alterman, M.A.; Galeva, N.; Larive, C.K.; Asunskis, D.; Sherwood, P.M.A. Methanobactin, a Copper-Acquisition Compound from Methane-Oxidizing Bacteria. Science 2004, 305, 1612. [Google Scholar] [CrossRef] [PubMed]
- Behling, L.A.; Hartsel, S.C.; Lewis, D.E.; DiSpirito, A.A.; Choi, D.W.; Masterson, L.R.; Veglia, G.; Gallagher, W.H. NMR, mass spectrometry and chemical evidence reveal a different chemical structure for methanobactin that contains oxazolone rings. J. Am. Chem. Soc. 2008, 130, 12604–12605. [Google Scholar] [CrossRef] [PubMed]
- Krentz, B.D.; Mulheron, H.J.; Semrau, J.D.; DiSpirito, A.A.; Bandow, N.L.; Haft, D.H.; Vuilleumier, S.; Murrell, J.C.; McEllistrem, M.T.; Hartsel, S.C.; et al. A Comparison of Methanobactins from Methylosinus trichosporium OB3b and Methylocystis Strain SB2 Predicts Methanobactins Are Synthesized from Diverse Peptide Precursors Modified To Create a Common Core for Binding and Reducing Copper Ions. Biochemistry 2010, 49, 10117–10130. [Google Scholar] [CrossRef] [PubMed]
- El Ghazouani, A.; Baslé, A.; Gray, J.; Graham, D.W.; Firbank, S.J.; Dennison, C. Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 8400–8404. [Google Scholar] [CrossRef] [PubMed]
- El Ghazouani, A.; Baslé, A.; Firbank, S.J.; Knapp, C.W.; Gray, J.; Graham, D.W.; Dennison, C. Copper-Binding Properties and Structures of Methanobactins from Methylosinus trichosporium OB3b. Inorg. Chem. 2011, 50, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Hakemian, A.S.; Tinberg, C.E.; Kondapalli, K.C.; Telser, J.; Hoffman, B.M.; Stemmler, T.L.; Rosenzweig, A.C. The Copper Chelator Methanobactin from Methylosinus trichosporium OB3b Binds Copper(I). J. Am. Chem. Soc. 2005, 127, 17142–17143. [Google Scholar] [CrossRef] [PubMed]
- Semrau, J.D.; Jagadevan, S.; DiSpirito, A.A.; Khalifa, A.; Scanlan, J.; Bergman, B.H.; Freemeier, B.C.; Baral, B.S.; Bandow, N.L.; Vorobev, A.; et al. Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs. Environ. Microbiol. 2013, 15, 3077–3086. [Google Scholar] [CrossRef]
- Kenney, G.E.; Rosenzweig, A.C. Genome mining for methanobactins. BMC Biol. 2013, 11, 17. [Google Scholar] [CrossRef]
- Dassama, L.M.K.; Kenney, G.E.; Rosenzweig, A.C. Methanobactins: From genome to function. Metallomics 2017, 9, 7–20. [Google Scholar] [CrossRef]
- Dassama, L.M.K.; Kenney, G.E.; Ro, S.Y.; Zielazinski, E.L.; Rosenzweig, A.C. Methanobactin transport machinery. Proc. Natl. Acad. Sci. USA 2016, 113, 13027–13032. [Google Scholar] [CrossRef]
- Kuroda, T.; Tsuchiya, T. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 763–768. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Szewczyk, P.; Karyakin, A.; Evin, M.; Hong, W.-X.; Zhang, Q.; Chang, G. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 2010, 467, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, R.; Kenney, G.E.; Rosenzweig, A.C. Dual pathways for copper uptake by methanotrophic bacteria. J. Biol. Chem. 2011, 286, 37313–37319. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Farhan Ul Haque, M.; Baral, B.S.; Turpin, E.A.; Bandow, N.L.; Kremmer, E.; Flatley, A.; Zischka, H.; DiSpirito, A.A.; Semrau, J.D. A TonB-Dependent Transporter Is Responsible for Methanobactin Uptake by Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2016, 82, 1917. [Google Scholar] [CrossRef]
- Carniel, E. The Yersinia high-pathogenicity island: An iron-uptake island. Microbes Infect. 2001, 3, 561–569. [Google Scholar] [CrossRef]
- Perry, R.D.; Fetherston, J.D. Yersiniabactin iron uptake: Mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011, 13, 808–817. [Google Scholar] [CrossRef]
- Chaturvedi, K.S.; Hung, C.S.; Giblin, D.E.; Urushidani, S.; Austin, A.M.; Dinauer, M.C.; Henderson, J.P. Cupric Yersiniabactin Is a Virulence-Associated Superoxide Dismutase Mimic. ACS Chem. Biol. 2014, 9, 551–561. [Google Scholar] [CrossRef]
- Chaturvedi, K.S.; Hung, C.S.; Crowley, J.R.; Stapleton, A.E.; Henderson, J.P. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 2012, 8, 731–736. [Google Scholar] [CrossRef]
- Chaturvedi, K.S.; Henderson, J.P. Pathogenic adaptations to host-derived antibacterial copper. Front. Cell. Infect. Microbiol. 2014, 4, 3. [Google Scholar] [CrossRef]
- Koh, E.-I.; Robinson, A.E.; Bandara, N.; Rogers, B.E.; Henderson, J.P. Copper import in Escherichia coli by the yersiniabactin metallophore system. Nat. Chem. Biol. 2017, 13, 1016–1021. [Google Scholar] [CrossRef]
- Rakin, A.; Saken, E.; Harmsen, D.; Heesemann, J. The pesticin receptor of Yersinia enterocolitica: A novel virulence factor with dual function. Mol. Microbiol. 1994, 13, 253–263. [Google Scholar] [CrossRef]
- Schubert, S.; Rakin, A.; Heesemann, J. The Yersinia high-pathogenicity island (HPI): Evolutionary and functional aspects. Int. J. Med. Microbiol. 2004, 294, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Boal, A.K.; Rosenzweig, A.C. Structural biology of copper trafficking. Chem. Rev. 2009, 109, 4760–4779. [Google Scholar] [CrossRef] [PubMed]
- Aliaga, M.E.; Lopez-Alarcon, C.; Bridi, R.; Speisky, H. Redox-implications associated with the formation of complexes between copper ions and reduced or oxidized glutathione. J. Inorg. Biochem. 2016, 154, 78–88. [Google Scholar] [CrossRef]
- Hatori, Y.; Inouye, S.; Akagi, R. Thiol-based copper handling by the copper chaperone Atox1. IUBMB Life 2017, 69, 246–254. [Google Scholar] [CrossRef]
- Kosman, D.J. The teleos of metallo-reduction and metallo-oxidation in eukaryotic iron and copper trafficking. Metallomics 2018, 10, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Matson, D.M.; Ariöz, C.; Wittung-Stafshede, P. Extended functional repertoire for human copper chaperones. Biomol. Concepts 2016, 7, 29–39. [Google Scholar] [CrossRef]
- Finegold, A.A.; Shatwell, K.P.; Segal, A.W.; Klausner, R.D.; Dancis, A. Intramembrane Bis-Heme Motif for Transmembrane Electron Transport Conserved in a Yeast Iron Reductase and the Human NADPH Oxidase. J. Biol. Chem. 1996, 271, 31021–31024. [Google Scholar] [CrossRef]
- Knutson, M.D. Steap proteins: Implications for iron and copper metabolism. Nutr. Rev. 2007, 65, 335–340. [Google Scholar]
- Ohgami, R.S.; Campagna, D.R.; McDonald, A.; Fleming, M.D. The Steap proteins are metalloreductases. Blood 2006, 108, 1388–1394. [Google Scholar] [CrossRef]
- Wyman, S.; Simpson, R.J.; McKie, A.T.; Sharp, P.A. Dcytb (Cybrd1) functions as both a ferric and a cupric reductase in vitro. FEBS Lett. 2008, 582, 1901–1906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Krause, K.H.; Xenarios, I.; Soldati, T.; Boeckmann, B. Evolution of the ferric reductase domain (FRD) superfamily: Modularity, functional diversification, and signature motifs. PLoS ONE 2013, 8, e58126. [Google Scholar] [CrossRef] [PubMed]
- Attar, N.; Campos, O.A.; Vogelauer, M.; Cheng, C.; Xue, Y.; Schmollinger, S.; Salwinski, L.; Mallipeddi, N.V.; Boone, B.A.; Yen, L.; et al. The histone H3-H4 tetramer is a copper reductase enzyme. Science 2020, 369, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, J.; Luger, K. The secret life of histones. Science 2020, 369, 33. [Google Scholar] [CrossRef]
- Marckmann, D.; Trasnea, P.I.; Schimpf, J.; Winterstein, C.; Andrei, A.; Schmollinger, S.; Blaby-Haas, C.E.; Friedrich, T.; Daldal, F.; Koch, H.G. The cbb 3-type cytochrome oxidase assembly factor CcoG is a widely distributed cupric reductase. Proc. Natl. Acad. Sci. USA 2019, 116, 21166–21175. [Google Scholar] [CrossRef]
- Helbig, K.; Bleuel, C.; Krauss, G.J.; Nies, D.H. Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 2008, 190, 5431–5438. [Google Scholar] [CrossRef]
- Pawlik, G.; Kulajta, C.; Sachelaru, I.; Schroder, S.; Waidner, B.; Hellwig, P.; Daldal, F.; Koch, H.G. The putative assembly factor CcoH is stably associated with the cbb3-type cytochrome oxidase. J. Bacteriol. 2010, 192, 6378–6389. [Google Scholar] [CrossRef]
- Preisig, O.; Zufferey, R.; Hennecke, H. The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Arch. Microbiol. 1996, 165, 297–305. [Google Scholar] [CrossRef]
- Koch, H.G.; Hwang, O.; Daldal, F. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J. Bacteriol. 1998, 180, 969–978. [Google Scholar] [CrossRef]
- Volentini, S.I.; Farias, R.N.; Rodriguez-Montelongo, L.; Rapisarda, V.A. Cu(II)-reduction by Escherichia coli cells is dependent on respiratory chain components. Biometals 2011, 24, 827–835. [Google Scholar] [CrossRef]
- Abicht, H.K.; Gonskikh, Y.; Gerber, S.D.; Solioz, M. Non-enzymic copper reduction by menaquinone enhances copper toxicity in Lactococcus lactis IL1403. Microbiology 2013, 159, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Andreazza, R.; Okeke, B.C.; Pieniz, S.; Brandelli, A.; Lambais, M.R.; Camargo, F.A.O. Bioreduction of Cu(II) by Cell-Free Copper Reductase from a Copper Resistant Pseudomonas sp. NA. Biol. Trace Elem. Res. 2011, 143, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Neubert, M.J.; Dahlmann, E.A.; Ambrose, A.; Johnson, M.D.L. Copper Chaperone CupA and Zinc Control CopY Regulation of the Pneumococcal cop Operon. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Bruce, K.E.; Wu, H.; Giedroc, D.P. The S2 Cu(i) site in CupA from Streptococcus pneumoniae is required for cellular copper resistance. Metallomics 2016, 8, 61–70. [Google Scholar] [CrossRef]
- Sazinsky, M.H.; LeMoine, B.; Orofino, M.; Davydov, R.; Bencze, K.Z.; Stemmler, T.L.; Hoffman, B.M.; Arguello, J.M.; Rosenzweig, A.C. Characterization and structure of a Zn2+ and [2Fe-2S]-containing copper chaperone from Archaeoglobus fulgidus. J. Biol. Chem. 2007, 282, 25950–25959. [Google Scholar] [CrossRef]
- Palumaa, P. Copper chaperones. The concept of conformational control in the metabolism of copper. FEBS Lett. 2013, 587, 1902–1910. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Kozyreva, T.; Zovo, K.; Palumaa, P. Affinity gradients drive copper to cellular destinations. Nature 2010, 465, 645–648. [Google Scholar] [CrossRef]
- Odermatt, A.; Solioz, M. Two trans-acting metalloregulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. J. Biol. Chem. 1995, 270, 4349–4354. [Google Scholar] [CrossRef]
- Wimmer, R.; Herrmann, T.; Solioz, M.; Wuthrich, K. NMR structure and metal interactions of the CopZ copper chaperone. J. Biol. Chem. 1999, 274, 22597–22603. [Google Scholar] [CrossRef]
- Utz, M.; Andrei, A.; Milanov, M.; Trasnea, P.I.; Marckmann, D.; Daldal, F.; Koch, H.G. The Cu chaperone CopZ is required for Cu homeostasis in Rhodobacter capsulatus and influences cytochrome cbb3 oxidase assembly. Mol. Microbiol. 2019, 111, 764–783. [Google Scholar] [CrossRef]
- Lu, Z.H.; Dameron, C.T.; Solioz, M. The Enterococcus hirae paradigm of copper homeostasis: Copper chaperone turnover, interactions, and transactions. Biometals 2003, 16, 137–143. [Google Scholar] [PubMed]
- Peuser, V.; Glaeser, J.; Klug, G. The RSP_2889 gene product of Rhodobacter sphaeroides is a CueR homologue controlling copper-responsive genes. Microbiology 2011, 157, 3306–3313. [Google Scholar] [CrossRef] [PubMed]
- Drees, S.L.; Klinkert, B.; Helling, S.; Beyer, D.F.; Marcus, K.; Narberhaus, F.; Lubben, M. One gene, two proteins: Coordinated production of a copper chaperone by differential transcript formation and translational frameshifting in Escherichia coli. Mol. Microbiol. 2017, 106, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Meydan, S.; Klepacki, D.; Karthikeyan, S.; Margus, T.; Thomas, P.; Jones, J.E.; Khan, Y.; Briggs, J.; Dinman, J.D.; Vázquez-Laslop, N.; et al. Programmed Ribosomal Frameshifting Generates a Copper Transporter and a Copper Chaperone from the Same Gene. Mol. Cell 2017, 65, 207–219. [Google Scholar] [CrossRef]
- Kay, K.L.; Zhou, L.; Tenori, L.; Bradley, J.M.; Singleton, C.; Kihlken, M.A.; Ciofi-Baffoni, S.; Le Brun, N.E. Kinetic analysis of copper transfer from a chaperone to its target protein mediated by complex formation. Chem. Commun. 2017, 53, 1397–1400. [Google Scholar] [CrossRef]
- Singleton, C.; Hearnshaw, S.; Zhou, L.; Le Brun, N.E.; Hemmings, A.M. Mechanistic insights into Cu(I) cluster transfer between the chaperone CopZ and its cognate Cu(I)-transporting P-type ATPase, CopA. Biochem. J. 2009, 424, 347–356. [Google Scholar] [CrossRef]
- Singleton, C.; Le Brun, N.E. Atx1-like chaperones and their cognate P-type ATPases: Copper-binding and transfer. Biometals 2007, 20, 275–289. [Google Scholar] [CrossRef]
- Gonzalez-Guerrero, M.; Arguello, J.M. Mechanism of Cu+-transporting ATPases: Soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc. Natl. Acad. Sci. USA 2008, 105, 5992–5997. [Google Scholar] [CrossRef]
- Kihlken, M.A.; Leech, A.P.; Le Brun, N.E. Copper-mediated dimerization of CopZ, a predicted copper chaperone from Bacillus subtilis. Biochem. J. 2002, 368, 729–739. [Google Scholar] [CrossRef]
- Hearnshaw, S.; West, C.; Singleton, C.; Zhou, L.; Kihlken, M.A.; Strange, R.W.; Le Brun, N.E.; Hemmings, A.M. A tetranuclear Cu(I) cluster in the metallochaperone protein CopZ. Biochemistry 2009, 48, 9324–9326. [Google Scholar] [CrossRef]
- Kay, K.L.; Hamilton, C.J.; Le Brun, N.E. Mass spectrometry of B. subtilis CopZ: Cu(i)-binding and interactions with bacillithiol. Metallomics 2016, 8, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Carmel-Harel, O.; Storz, G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 2000, 54, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Novoa-Aponte, L.; Ramírez, D.; Argüello, J.M. The interplay of the metallosensor CueR with two distinct CopZ chaperones defines copper homeostasis in Pseudomonas aeruginosa. J. Biol. Chem. 2019, 294, 4934–4945. [Google Scholar] [CrossRef]
- Fu, Y.; Tsui, H.C.; Bruce, K.E.; Sham, L.T.; Higgins, K.A.; Lisher, J.P.; Kazmierczak, K.M.; Maroney, M.J.; Dann, C.E., 3rd; Winkler, M.E.; et al. A new structural paradigm in copper resistance in Streptococcus pneumoniae. Nat. Chem. Biol. 2013, 9, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.D.; Kehl-Fie, T.E.; Klein, R.; Kelly, J.; Burnham, C.; Mann, B.; Rosch, J.W. Role of copper efflux in pneumococcal pathogenesis and resistance to macrophage-mediated immune clearance. Infect. Immun. 2015, 83, 1684–1694. [Google Scholar] [CrossRef]
- Davidi, D.; Longo, L.M.; Jabłońska, J.; Milo, R.; Tawfik, D.S. A Bird’s-Eye View of Enzyme Evolution: Chemical, Physicochemical, and Physiological Considerations. Chem. Rev. 2018, 118, 8786–8797. [Google Scholar] [CrossRef]
- Drees, S.L.; Beyer, D.F.; Lenders-Lomscher, C.; Lubben, M. Distinct functions of serial metal-binding domains in the Escherichia coli P1B-ATPase CopA. Mol. Microbiol. 2015, 97, 423–438. [Google Scholar] [CrossRef]
- Gonzalez-Guerrero, M.; Hong, D.; Arguello, J.M. Chaperone-mediated Cu+ delivery to Cu+ transport ATPases: Requirement of nucleotide binding. J. Biol. Chem. 2009, 284, 20804–20811. [Google Scholar] [CrossRef]
- Kittleson, J.T.; Loftin, I.R.; Hausrath, A.C.; Engelhardt, K.P.; Rensing, C.; McEvoy, M.M. Periplasmic metal-resistance protein CusF exhibits high affinity and specificity for both CuI and AgI. Biochemistry 2006, 45, 11096–11102. [Google Scholar] [CrossRef]
- Vasák, M. Advances in metallothionein structure and functions. J. Trace Elem. Med. Biol. 2005, 19, 13–17. [Google Scholar] [CrossRef]
- Calvo, J.; Jung, H.; Meloni, G. Copper metallothioneins. IUBMB Life 2017, 69, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Bremner, I. Oxygen free radicals and metallothionein. Free Radic. Biol. Med. 1993, 14, 325–337. [Google Scholar] [CrossRef]
- Turner, J.S.; Robinson, N.J. Cyanobacterial metallothioneins: Biochemistry and molecular genetics. J. Ind. Microbiol. 1995, 14, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Blindauer, C.A. Bacterial metallothioneins: Past, present, and questions for the future. J. Biol. Inorg. Chem. 2011, 16, 1011–1024. [Google Scholar] [CrossRef] [PubMed]
- Ziller, A.; Fraissinet-Tachet, L. Metallothionein diversity and distribution in the tree of life: A multifunctional protein. Metallomics 2018, 10, 1549–1559. [Google Scholar] [CrossRef]
- Olafson, R.W.; Abel, K.; Sim, R.G. Prokaryotic metallothionein: Preliminary characterization of a blue-green alga heavy metal-binding protein. Biochem. Biophys. Res. Commun. 1979, 89, 36–43. [Google Scholar] [CrossRef]
- Blindauer, C.A.; Harrison, M.D.; Parkinson, J.A.; Robinson, A.K.; Cavet, J.S.; Robinson, N.J.; Sadler, P.J. A metallothionein containing a zinc finger within a four-metal cluster protects a bacterium from zinc toxicity. Proc. Natl. Acad. Sci. USA 2001, 98, 9593–9598. [Google Scholar] [CrossRef]
- Gold, B.; Deng, H.; Bryk, R.; Vargas, D.; Eliezer, D.; Roberts, J.; Jiang, X.; Nathan, C. Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat. Chem. Biol. 2008, 4, 609–616. [Google Scholar] [CrossRef]
- Vita, N.; Platsaki, S.; Baslé, A.; Allen, S.J.; Paterson, N.G.; Crombie, A.T.; Murrell, J.C.; Waldron, K.J.; Dennison, C. A four-helix bundle stores copper for methane oxidation. Nature 2015, 525, 140–143. [Google Scholar] [CrossRef]
- DiSpirito, A.A.; Semrau, J.D.; Murrell, J.C.; Gallagher, W.H.; Dennison, C.; Vuilleumier, S. Methanobactin and the Link between Copper and Bacterial Methane Oxidation. Microbiol. Mol. Biol. Rev. 2016, 80, 387–409. [Google Scholar] [CrossRef]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 2008, 63, 183–229. [Google Scholar] [CrossRef] [PubMed]
- Vita, N.; Landolfi, G.; Basle, A.; Platsaki, S.; Lee, J.; Waldron, K.J.; Dennison, C. Bacterial cytosolic proteins with a high capacity for Cu(I) that protect against copper toxicity. Sci. Rep. 2016, 6, 39065. [Google Scholar] [CrossRef] [PubMed]
- Straw, M.L.; Chaplin, A.K.; Hough, M.A.; Paps, J.; Bavro, V.N.; Wilson, M.T.; Vijgenboom, E.; Worrall, J.A.R. A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans. Metallomics 2018, 10, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.; Chakraborty, K.; Shukla, A. Cellular copper homeostasis: Current concepts on its interplay with glutathione homeostasis and its implication in physiology and human diseases. Metallomics 2017, 9, 1376–1388. [Google Scholar] [CrossRef] [PubMed]
- Kachur, A.V.; Koch, C.J.; Biaglow, J.E. Mechanism of copper-catalyzed oxidation of glutathione. Free Radic. Res. 1998, 28, 259–269. [Google Scholar] [CrossRef]
- Corazza, A.; Harvey, I.; Sadler, P.J. 1H,13C-NMR and X-ray absorption studies of copper(I) glutathione complexes. Eur. J. Biochem. 1996, 236, 697–705. [Google Scholar] [CrossRef]
- Morgan, M.T.; Nguyen, L.A.H.; Hancock, H.L.; Fahrni, C.J. Glutathione limits aquacopper(I) to sub-femtomolar concentrations through cooperative assembly of a tetranuclear cluster. J. Biol. Chem. 2017, 292, 21558–21567. [Google Scholar] [CrossRef]
- Saporito-Magriñá, C.M.; Musacco-Sebio, R.N.; Andrieux, G.; Kook, L.; Orrego, M.T.; Tuttolomondo, M.V.; Desimone, M.F.; Boerries, M.; Borner, C.; Repetto, M.G. Copper-induced cell death and the protective role of glutathione: The implication of impaired protein folding rather than oxidative stress. Metallomics 2018, 10, 1743–1754. [Google Scholar] [CrossRef]
- Freedman, J.H.; Ciriolo, M.R.; Peisach, J. The role of glutathione in copper metabolism and toxicity. J. Biol. Chem. 1989, 264, 5598–5605. [Google Scholar]
- Potter, A.J.; Trappetti, C.; Paton, J.C. Streptococcus pneumoniae uses glutathione to defend against oxidative stress and metal ion toxicity. J. Bacteriol. 2012, 194, 6248–6254. [Google Scholar] [CrossRef]
- Stewart, L.J.; Ong, C.-l.Y.; Zhang, M.M.; Brouwer, S.; McIntyre, L.; Davies, M.R.; Walker, M.J.; McEwan, A.G.; Waldron, K.J.; Djoko, K.Y. A role for glutathione in buffering excess intracellular copper in Streptococcus pyogenes. bioRxiv 2020. [Google Scholar] [CrossRef]
- Poole, R.K.; Cozens, A.G.; Shepherd, M. The CydDC family of transporters. Res. Microbiol. 2019, 170, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Lutsenko, S. Copper trafficking to the secretory pathway. Metallomics 2016, 8, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Arguello, J.M.; Raimunda, D.; Gonzalez-Guerrero, M. Metal transport across biomembranes: Emerging models for a distinct chemistry. J. Biol. Chem. 2012, 287, 13510–13517. [Google Scholar] [CrossRef]
- Rosenzweig, A.C.; Arguello, J.M. Toward a molecular understanding of metal transport by P(1B)-type ATPases. Curr. Top. Membr. 2012, 69, 113–136. [Google Scholar] [CrossRef]
- Delmar, J.A.; Su, C.C.; Yu, E.W. Bacterial multidrug efflux transporters. Annu. Rev. Biophys. 2014, 43, 93–117. [Google Scholar] [CrossRef]
- Hernández-Montes, G.; Argüello, J.M.; Valderrama, B. Evolution and diversity of periplasmic proteins involved in copper homeostasis in gamma proteobacteria. BMC Microbiol. 2012, 12, 249. [Google Scholar] [CrossRef]
- Purohit, R.; Ross, M.O.; Batelu, S.; Kusowski, A.; Stemmler, T.L.; Hoffman, B.M.; Rosenzweig, A.C. Cu(+)-specific CopB transporter: Revising P(1B)-type ATPase classification. Proc. Natl. Acad. Sci. USA 2018, 115, 2108–2113. [Google Scholar] [CrossRef]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef]
- Smith, A.T.; Smith, K.P.; Rosenzweig, A.C. Diversity of the metal-transporting P1B-type ATPases. J. Biol. Inorg. Chem. 2014, 19, 947–960. [Google Scholar] [CrossRef]
- Smith, A.T.; Barupala, D.; Stemmler, T.L.; Rosenzweig, A.C. A new metal binding domain involved in cadmium, cobalt and zinc transport. Nat. Chem. Biol. 2015, 11, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Zielazinski, E.L.; Gonzalez-Guerrero, M.; Subramanian, P.; Stemmler, T.L.; Arguello, J.M.; Rosenzweig, A.C. Sinorhizobium meliloti Nia is a P(1B-5)-ATPase expressed in the nodule during plant symbiosis and is involved in Ni and Fe transport. Metallomics 2013, 5, 1614–1623. [Google Scholar] [CrossRef] [PubMed]
- Rangarh, P.; Kohli, N. Neuroimaging findings in Menkes disease: A rare neurodegenerative disorder. BMJ Case Rep. 2018, 2018, bcr2017223858. [Google Scholar] [CrossRef] [PubMed]
- Tümer, Z. An Overview and Update of ATP7A Mutations Leading to Menkes Disease and Occipital Horn Syndrome. Hum. Mutat. 2013, 34, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Bandmann, O.; Weiss, K.H.; Kaler, S.G. Wilson’s disease and other neurological copper disorders. Lancet Neurol. 2015, 14, 103–113. [Google Scholar] [CrossRef]
- Czonkowska, A.; Litwin, T.; Dusek, P.; Ferenci, P.; Lutsenko, S.; Medici, V.; Rybakowski, J.K.; Weiss, K.H.; Schilsky, M.L. Wilson disease. Nat. Rev. Dis. Primers 2018, 4, 21. [Google Scholar] [CrossRef]
- González-Guerrero, M.; Raimunda, D.; Cheng, X.; Argüello, J.M. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 2010, 78, 1246–1258. [Google Scholar] [CrossRef]
- Peters, A.; Kulajta, C.; Pawlik, G.; Daldal, F.; Koch, H.G. Stability of the cbb3-type cytochrome oxidase requires specific CcoQ-CcoP interactions. J. Bacteriol. 2008, 190, 5576–5586. [Google Scholar] [CrossRef]
- Schurig-Briccio, L.A.; Gennis, R.B. Characterization of the PIB-Type ATPases present in Thermus thermophilus. J. Bacteriol. 2012, 194, 4107–4113. [Google Scholar] [CrossRef]
- Wu, C.C.; Rice, W.J.; Stokes, D.L. Structure of a copper pump suggests a regulatory role for its metal-binding domain. Structure 2008, 16, 976–985. [Google Scholar] [CrossRef]
- Padilla-Benavides, T.; McCann, C.J.; Arguello, J.M. The mechanism of Cu+ transport ATPases: Interaction with CU+ chaperones and the role of transient metal-binding sites. J. Biol. Chem. 2013, 288, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Calderone, V.; Della-Malva, N.; Felli, I.C.; Neri, S.; Pavelkova, A.; Rosato, A. Copper(I)-mediated protein-protein interactions result from suboptimal interaction surfaces. Biochem. J. 2009, 422, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Sazinsky, M.H.; Agarwal, S.; Arguello, J.M.; Rosenzweig, A.C. Structure of the actuator domain from the Archaeoglobus fulgidus Cu+-ATPase. Biochemistry 2006, 45, 9949–9955. [Google Scholar] [CrossRef] [PubMed]
- Sazinsky, M.H.; Mandal, A.K.; Arguello, J.M.; Rosenzweig, A.C. Structure of the ATP binding domain from the Archaeoglobus fulgidus Cu+-ATPase. J. Biol. Chem. 2006, 281, 11161–11166. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Hong, D.; Desai, N.K.; Sazinsky, M.H.; Arguello, J.M.; Rosenzweig, A.C. Structure and interactions of the C-terminal metal binding domain of Archaeoglobus fulgidus CopA. Proteins 2010, 78, 2450–2458. [Google Scholar] [CrossRef]
- Singleton, C.; Banci, L.; Ciofi-Baffoni, S.; Tenori, L.; Kihlken, M.A.; Boetzel, R.; Le Brun, N.E. Structure and Cu(I)-binding properties of the N-terminal soluble domains of Bacillus subtilis CopA. Biochem. J. 2008, 411, 571–579. [Google Scholar] [CrossRef]
- Allen, G.S.; Wu, C.C.; Cardozo, T.; Stokes, D.L. The architecture of CopA from Archeaoglobus fulgidus studied by cryo-electron microscopy and computational docking. Structure 2011, 19, 1219–1232. [Google Scholar] [CrossRef]
- Apell, H.J. How do P-type ATPases transport ions? Bioelectrochemistry 2004, 63, 149–156. [Google Scholar] [CrossRef]
- Mandal, A.K.; Arguello, J.M. Functional roles of metal binding domains of the Archaeoglobus fulgidus Cu+-ATPase CopA. Biochemistry 2003, 42, 11040–11047. [Google Scholar] [CrossRef]
- Arguello, J.M.; Patel, S.J.; Quintana, J. Bacterial Cu+-ATPases: Models for molecular structure-function studies. Metallomics 2016, 8, 906–914. [Google Scholar] [CrossRef]
- Ralle, M.; Lutsenko, S.; Blackburn, N.J. Copper transfer to the N-terminal domain of the Wilson disease protein (ATP7B): X-ray absorption spectroscopy of reconstituted and chaperone-loaded metal binding domains and their interaction with exogenous ligands. J. Inorg. Biochem. 2004, 98, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Huster, D.; Lutsenko, S. The distinct roles of the N-terminal copper-binding sites in regulation of catalytic activity of the Wilson’s disease protein. J. Biol. Chem. 2003, 278, 32212–32218. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.H.; Lee, W.; Nokhrin, S.; Dmitriev, O.Y. The Structure of Metal Binding Domain 1 of the Copper Transporter ATP7B Reveals Mechanism of a Singular Wilson Disease Mutation. Sci. Rep. 2018, 8, 581. [Google Scholar] [CrossRef] [PubMed]
- Bartee, M.Y.; Ralle, M.; Lutsenko, S. The loop connecting metal-binding domains 3 and 4 of ATP7B is a target of a kinase-mediated phosphorylation. Biochemistry 2009, 48, 5573–5581. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Lutsenko, S. Human copper transporters: Mechanism, role in human diseases and therapeutic potential. Future Med. Chem. 2009, 1, 1125–1142. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Cantini, F.; Massagni, C.; Migliardi, M.; Rosato, A. An NMR study of the interaction of the N-terminal cytoplasmic tail of the Wilson disease protein with copper(I)-HAH1. J. Biol. Chem. 2009, 284, 9354–9360. [Google Scholar] [CrossRef] [PubMed]
- Singleton, W.C.; McInnes, K.T.; Cater, M.A.; Winnall, W.R.; McKirdy, R.; Yu, Y.; Taylor, P.E.; Ke, B.X.; Richardson, D.R.; Mercer, J.F.; et al. Role of glutaredoxin1 and glutathione in regulating the activity of the copper-transporting P-type ATPases, ATP7A and ATP7B. J. Biol. Chem. 2010, 285, 27111–27121. [Google Scholar] [CrossRef]
- Gonzalez-Guerrero, M.; Eren, E.; Rawat, S.; Stemmler, T.L.; Arguello, J.M. Structure of the two transmembrane Cu+ transport sites of the Cu+ -ATPases. J. Biol. Chem. 2008, 283, 29753–29759. [Google Scholar] [CrossRef]
- Andersson, M.; Mattle, D.; Sitsel, O.; Klymchuk, T.; Nielsen, A.M.; Møller, L.B.; White, S.H.; Nissen, P.; Gourdon, P. Copper-transporting P-type ATPases use a unique ion-release pathway. Nat. Struct. Mol. Biol. 2014, 21, 43–48. [Google Scholar] [CrossRef]
- Chandrangsu, P.; Rensing, C.; Helmann, J.D. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017, 15, 338–350. [Google Scholar] [CrossRef]
- Singh, S.K.; Grass, G.; Rensing, C.; Montfort, W.R. Cuprous oxidase activity of CueO from Escherichia coli. J. Bacteriol. 2004, 186, 7815–7817. [Google Scholar] [CrossRef] [PubMed]
- Wiethaus, J.; Wildner, G.F.; Masepohl, B. The multicopper oxidase CutO confers copper tolerance to Rhodobacter capsulatus. FEMS Microbiol. Lett. 2006, 256, 67–74. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buhler, D.; Rossmann, R.; Landolt, S.; Balsiger, S.; Fischer, H.M.; Hennecke, H. Disparate pathways for the biogenesis of cytochrome oxidases in Bradyrhizobium japonicum. J. Biol. Chem. 2010, 285, 15704–15713. [Google Scholar] [CrossRef] [PubMed]
- Mealman, T.D.; Blackburn, N.J.; McEvoy, M.M. Metal export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of Escherichia coli. Curr. Top. Membr. 2012, 69, 163–196. [Google Scholar] [CrossRef] [PubMed]
- Outten, F.W.; Huffman, D.L.; Hale, J.A.; O’Halloran, T.V. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 2001, 276, 30670–30677. [Google Scholar] [CrossRef]
- Long, F.; Su, C.C.; Lei, H.T.; Bolla, J.R.; Do, S.V.; Yu, E.W. Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2012, 367, 1047–1058. [Google Scholar] [CrossRef]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 2002, 419, 587–593. [Google Scholar] [CrossRef]
- Sennhauser, G.; Bukowska, M.A.; Briand, C.; Grütter, M.G. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 2009, 389, 134–145. [Google Scholar] [CrossRef]
- Long, F.; Su, C.C.; Zimmermann, M.T.; Boyken, S.E.; Rajashankar, K.R.; Jernigan, R.L.; Yu, E.W. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 2010, 467, 484–488. [Google Scholar] [CrossRef]
- Lei, H.T.; Bolla, J.R.; Bishop, N.R.; Su, C.C.; Yu, E.W. Crystal structures of CusC review conformational changes accompanying folding and transmembrane channel formation. J. Mol. Biol. 2014, 426, 403–411. [Google Scholar] [CrossRef]
- Atilgan, A.R.; Durell, S.R.; Jernigan, R.L.; Demirel, M.C.; Keskin, O.; Bahar, I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001, 80, 505–515. [Google Scholar] [CrossRef]
- Murakami, S. Multidrug efflux transporter, AcrB--the pumping mechanism. Curr. Opin. Struct. Biol. 2008, 18, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Su, C.C.; Yang, F.; Long, F.; Reyon, D.; Routh, M.D.; Kuo, D.W.; Mokhtari, A.K.; Van Ornam, J.D.; Rabe, K.L.; Hoy, J.A.; et al. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J. Mol. Biol. 2009, 393, 342–355. [Google Scholar] [CrossRef] [PubMed]
- Bagai, I.; Liu, W.; Rensing, C.; Blackburn, N.J.; McEvoy, M.M. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J. Biol. Chem. 2007, 282, 35695–35702. [Google Scholar] [CrossRef] [PubMed]
- Su, C.C.; Long, F.; Zimmermann, M.T.; Rajashankar, K.R.; Jernigan, R.L.; Yu, E.W. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 2011, 470, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Ucisik, M.N.; Chakravorty, D.K.; Merz, K.M., Jr. Structure and dynamics of the N-terminal domain of the Cu(I) binding protein CusB. Biochemistry 2013, 52, 6911–6923. [Google Scholar] [CrossRef]
- Meir, A.; Abdelhai, A.; Moskovitz, Y.; Ruthstein, S. EPR Spectroscopy Targets Structural Changes in the E. coli Membrane Fusion CusB upon Cu(I) Binding. Biophys. J. 2017, 112, 2494–2502. [Google Scholar] [CrossRef][Green Version]
- Chacón, K.N.; Perkins, J.; Mathe, Z.; Alwan, K.; Ho, E.N.; Ucisik, M.N.; Merz, K.M.; Blackburn, N.J. Trapping intermediates in metal transfer reactions of the CusCBAF export pump of Escherichia coli. Commun. Biol. 2018, 1, 192. [Google Scholar] [CrossRef]
- Kulathila, R.; Kulathila, R.; Indic, M.; van den Berg, B. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy metal efflux pump. PLoS ONE 2011, 6, e15610. [Google Scholar] [CrossRef]
- Brown, N.L.; Barrett, S.R.; Camakaris, J.; Lee, B.T.O.; Rouch, D.A. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 1995, 17, 1153–1166. [Google Scholar] [CrossRef]
- Cha, J.S.; Cooksey, D.A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. USA 1991, 88, 8915–8919. [Google Scholar] [CrossRef] [PubMed]
- Rensing, C.; Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003, 27, 197–213. [Google Scholar] [CrossRef]
- Lee, S.M.; Grass, G.; Rensing, C.; Barrett, S.R.; Yates, C.J.; Stoyanov, J.V.; Brown, N.L. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 2002, 295, 616–620. [Google Scholar] [CrossRef]
- Tetaz, T.J.; Luke, R.K. Plasmid-controlled resistance to copper in Escherichia coli. J. Bacteriol. 1983, 154, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.S.; Cooksey, D.A. Copper Hypersensitivity and Uptake in Pseudomonas syringae Containing Cloned Components of the Copper Resistance Operon. Appl. Environ. Microbiol. 1993, 59, 1671–1674. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.L.; Neu, H.M.; Alamneh, Y.A.; Reddinger, R.M.; Jacobs, A.C.; Singh, S.; Abu-Taleb, R.; Michel, S.L.J.; Zurawski, D.V.; Merrell, D.S. Characterization of Acinetobacter baumannii Copper Resistance Reveals a Role in Virulence. Front. Microbiol. 2020, 11, 16. [Google Scholar] [CrossRef]
- Mellano, M.A.; Cooksey, D.A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J. Bacteriol. 1988, 170, 2879–2883. [Google Scholar] [CrossRef]
- Kenney, G.E.; Sadek, M.; Rosenzweig, A.C. Copper-responsive gene expression in the methanotroph Methylosinus trichosporium OB3b. Metallomics 2016, 8, 931–940. [Google Scholar] [CrossRef]
- Lawton, T.J.; Kenney, G.E.; Hurley, J.D.; Rosenzweig, A.C. The CopC Family: Structural and Bioinformatic Insights into a Diverse Group of Periplasmic Copper Binding Proteins. Biochemistry 2016, 55, 2278–2290. [Google Scholar] [CrossRef]
- Gu, W.; Farhan Ul Haque, M.; Semrau, J.D. Characterization of the role of copCD in copper uptake and the ‘copper-switch’ in Methylosinus trichosporium OB3b. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Arnesano, F.; Banci, L.; Bertini, I.; Mangani, S.; Thompsett, A.R. A redox switch in CopC: An intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc. Natl. Acad. Sci. USA 2003, 100, 3814–3819. [Google Scholar] [CrossRef] [PubMed]
- Djoko, K.Y.; Xiao, Z.; Wedd, A.G. Copper Resistance in E. coli: The Multicopper Oxidase PcoA Catalyzes Oxidation of Copper(I) in CuICuII-PcoC. Chembiochem 2008, 9, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Koay, M.; Maher, M.J.; Xiao, Z.; Wedd, A.G. Intermolecular Transfer of Copper Ions from the CopC Protein of Pseudomonas syringae. Crystal Structures of Fully Loaded CuICuII Forms. J. Am. Chem. Soc. 2006, 128, 5834–5850. [Google Scholar] [CrossRef] [PubMed]
- Munson, G.P.; Lam, D.L.; Outten, F.W.; O’Halloran, T.V. Identification of a Copper-Responsive Two-Component System on the Chromosome of Escherichia coli K-12. J. Bacteriol. 2000, 182, 5864. [Google Scholar] [CrossRef]
- Rouch, D.A.; Brown, N.L. Copper-inducible transcriptional regulation at two promoters in the Escherichia coli copper resistance determinant pco. Microbiology 1997, 143, 1191–1202. [Google Scholar] [CrossRef]
- Zimmermann, M.; Udagedara, S.R.; Sze, C.M.; Ryan, T.M.; Howlett, G.J.; Xiao, Z.; Wedd, A.G. PcoE—A metal sponge expressed to the periplasm of copper resistance Escherichia coli. Implication of its function role in copper resistance. J. Inorg. Biochem. 2012, 115, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Matsui, K.; Lo, J.-F.; Silver, S. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 1999, 5, 183–188. [Google Scholar] [CrossRef]
- Capdevila, D.A.; Edmonds, K.A.; Giedroc, D.P. Metallochaperones and metalloregulation in bacteria. Essays Biochem. 2017, 61, 177–200. [Google Scholar] [CrossRef]
- Jordan, M.R.; Wang, J.; Capdevila, D.A.; Giedroc, D.P. Multi-metal nutrient restriction and crosstalk in metallostasis systems in microbial pathogens. Curr. Opin. Microbiol. 2020, 55, 17–25. [Google Scholar] [CrossRef]
- Baksh, K.A.; Zamble, D.B. Allosteric control of metal-responsive transcriptional regulators in bacteria. J. Biol. Chem. 2020, 295, 1673–1684. [Google Scholar] [CrossRef]
- Rademacher, C.; Masepohl, B. Copper-responsive gene regulation in bacteria. Microbiology 2012, 158, 2451–2464. [Google Scholar] [CrossRef]
- Rademacher, C.; Moser, R.; Lackmann, J.W.; Klinkert, B.; Narberhaus, F.; Masepohl, B. Transcriptional and posttranscriptional events control copper-responsive expression of a Rhodobacter capsulatus multicopper oxidase. J. Bacteriol. 2012, 194, 1849–1859. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Outten, C.E.; Outten, F.W.; O’Halloran, T.V. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J. Biol. Chem. 1999, 274, 37517–37524. [Google Scholar] [CrossRef] [PubMed]
- Bittner, L.M.; Kraus, A.; Schäkermann, S.; Narberhaus, F. The Copper Efflux Regulator CueR Is Subject to ATP-Dependent Proteolysis in Escherichia coli. Front. Mol. Biosci. 2017, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Marcus, S.A.; Sidiropoulos, S.W.; Steinberg, H.; Talaat, A.M. CsoR Is Essential for Maintaining Copper Homeostasis in Mycobacterium tuberculosis. PLoS ONE 2016, 11, e0151816. [Google Scholar] [CrossRef] [PubMed]
- Higgins, K.A.; Giedroc, D. Insights into Protein Allostery in the CsoR/RcnR Family of Transcriptional Repressors. Chem. Lett. 2014, 43, 20–25. [Google Scholar] [CrossRef]
- Liu, T.; Ramesh, A.; Ma, Z.; Ward, S.K.; Zhang, L.; George, G.N.; Talaat, A.M.; Sacchettini, J.C.; Giedroc, D.P. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 2007, 3, 60–68. [Google Scholar] [CrossRef]
- Smaldone, G.T.; Helmann, J.D. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 2007, 153, 4123–4128. [Google Scholar] [CrossRef]
- Cantini, F.; Banci, L.; Solioz, M. The copper-responsive repressor CopR of Lactococcus lactis is a ‘winged helix’ protein. Biochem. J. 2009, 417, 493–499. [Google Scholar] [CrossRef]
- Portmann, R.; Poulsen, K.R.; Wimmer, R.; Solioz, M. CopY-like copper inducible repressors are putative ‘winged helix’ proteins. Biometals 2006, 19, 61–70. [Google Scholar] [CrossRef][Green Version]
- Portmann, R.; Magnani, D.; Stoyanov, J.V.; Schmechel, A.; Multhaup, G.; Solioz, M. Interaction kinetics of the copper-responsive CopY repressor with the cop promoter of Enterococcus hirae. J. Biol. Inorg. Chem. 2004, 9, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Cobine, P.A.; George, G.N.; Jones, C.E.; Wickramasinghe, W.A.; Solioz, M.; Dameron, C.T. Copper transfer from the Cu(I) chaperone, CopZ, to the repressor, Zn(II)CopY: Metal coordination environments and protein interactions. Biochemistry 2002, 41, 5822–5829. [Google Scholar] [CrossRef] [PubMed]
- Cobine, P.; Wickramasinghe, W.A.; Harrison, M.D.; Weber, T.; Solioz, M.; Dameron, C.T. The Enterococcus hirae copper chaperone CopZ delivers copper(I) to the CopY repressor. FEBS Lett. 1999, 445, 27–30. [Google Scholar] [CrossRef]
- Saha, R.P.; Samanta, S.; Patra, S.; Sarkar, D.; Saha, A.; Singh, M.K. Metal homeostasis in bacteria: The role of ArsR-SmtB family of transcriptional repressors in combating varying metal concentrations in the environment. Biometals 2017, 30, 459–503. [Google Scholar] [CrossRef] [PubMed]
- Osman, D.; Cavet, J.S. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat. Prod. Rep. 2010, 27, 668–680. [Google Scholar] [CrossRef]
- Jung, J.; Lee, S.J. Biochemical and Biodiversity Insights into Heavy Metal Ion-Responsive Transcription Regulators for Synthetic Biological Heavy Metal Sensors. J. Microbiol. Biotechnol. 2019, 29, 1522–1542. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Chen, X.; Ma, Z.; Shokes, J.; Hemmingsen, L.; Scott, R.A.; Giedroc, D.P. A Cu(I)-sensing ArsR family metal sensor protein with a relaxed metal selectivity profile. Biochemistry 2008, 47, 10564–10575. [Google Scholar] [CrossRef]
- Affandi, T.; McEvoy, M.M. Mechanism of metal ion-induced activation of a two-component sensor kinase. Biochem. J. 2019, 476, 115–135. [Google Scholar] [CrossRef] [PubMed]
- Djoko, K.Y.; Franiek, J.A.; Edwards, J.L.; Falsetta, M.L.; Kidd, S.P.; Potter, A.J.; Chen, N.H.; Apicella, M.A.; Jennings, M.P.; McEwan, A.G. Phenotypic characterization of a copA mutant of Neisseria gonorrhoeae identifies a link between copper and nitrosative stress. Infect. Immun. 2012, 80, 1065–1071. [Google Scholar] [CrossRef]
- McEwan, A.G.; Djoko, K.Y.; Chen, N.H.; Couñago, R.L.; Kidd, S.P.; Potter, A.J.; Jennings, M.P. Novel bacterial MerR-like regulators their role in the response to carbonyl and nitrosative stress. Adv. Microb. Physiol. 2011, 58, 1–22. [Google Scholar] [CrossRef]
- Bondarczuk, K.; Piotrowska-Seget, Z. Molecular basis of active copper resistance mechanisms in Gram-negative bacteria. Cell Biol. Toxicol. 2013, 29, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Rensing, C.; McEvoy, M.M. Chaperone-mediated copper handling in the periplasm. Nat. Prod. Rep. 2010, 27, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Loftin, I.R.; Franke, S.; Roberts, S.A.; Weichsel, A.; Héroux, A.; Montfort, W.R.; Rensing, C.; McEvoy, M.M. A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry 2005, 44, 10533–10540. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Davis, A.V.; Balakrishnan, G.; Stasser, J.P.; Staehlin, B.M.; Focia, P.; Spiro, T.G.; Penner-Hahn, J.E.; O’Halloran, T.V. Cu(I) recognition via cation-pi and methionine interactions in CusF. Nat. Chem. Biol. 2008, 4, 107–109. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Miura, T. Copper(I) stabilization by cysteine/tryptophan motif in the extracellular domain of Ctr4. J. Inorg. Biochem. 2016, 159, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Azzouzi, A.; Steunou, A.-S.; Durand, A.; Khalfaoui-Hassani, B.; Bourbon, M.-I.; Astier, C.; Bollivar, D.W.; Ouchane, S. Coproporphyrin III excretion identifies the anaerobic coproporphyrinogen III oxidase HemN as a copper target in the Cu+-ATPase mutant copA? of Rubrivivax gelatinosus. Mol. Microbiol. 2013, 88, 339–351. [Google Scholar] [CrossRef]
- Glerum, D.M.; Shtanko, A.; Tzagoloff, A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 20531–20535. [Google Scholar] [CrossRef]
- Cobine, P.A.; Pierrel, F.; Winge, D.R. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta 2006, 1763, 759–772. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Cavallaro, G.; Ciofi-Baffoni, S. Seeking the determinants of the elusive functions of Sco proteins. FEBS J. 2011, 278, 2244–2262. [Google Scholar] [CrossRef]
- Buggy, J.; Bauer, C.E. Cloning and characterization of senC, a gene involved in both aerobic respiration and photosynthesis gene expression in Rhodobacter capsulatus. J. Bacteriol. 1995, 177, 6958–6965. [Google Scholar] [CrossRef]
- Ekim Kocabey, A.; Kost, L.; Gehlhar, M.; Rödel, G.; Gey, U. Mitochondrial Sco proteins are involved in oxidative stress defense. Redox Biol. 2019, 21, 101079. [Google Scholar] [CrossRef] [PubMed]
- Borsetti, F.; Tremaroli, V.; Michelacci, F.; Borghese, R.; Winterstein, C.; Daldal, F.; Zannoni, D. Tellurite effects on Rhodobacter capsulatus cell viability and superoxide dismutase activity under oxidative stress conditions. Res. Microbiol. 2005, 156, 807–813. [Google Scholar] [CrossRef] [PubMed]
- McEwan, A.G.; Lewin, A.; Davy, S.L.; Boetzel, R.; Leech, A.; Walker, D.; Wood, T.; Moore, G.R. PrrC from Rhodobacter sphaeroides, a homologue of eukaryotic Sco proteins, is a copper-binding protein and may have a thiol-disulfide oxidoreductase activity. FEBS Lett. 2002, 518, 10–16. [Google Scholar] [CrossRef]
- Balatri, E.; Banci, L.; Bertini, I.; Cantini, F.; Ciofi-Baffoni, S. Solution structure of Sco1: A thioredoxin-like protein Involved in cytochrome c oxidase assembly. Structure 2003, 11, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Calderone, V.; Ciofi-Baffoni, S.; Mangani, S.; Martinelli, M.; Palumaa, P.; Wang, S. A hint for the function of human Sco1 from different structures. Proc. Natl. Acad. Sci. USA 2006, 103, 8595–8600. [Google Scholar] [CrossRef] [PubMed]
- Abriata, L.A.; Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Gkazonis, P.; Spyroulias, G.A.; Vila, A.J.; Wang, S. Mechanism of Cu(A) assembly. Nat. Chem. Biol. 2008, 4, 599–601. [Google Scholar] [CrossRef]
- Horng, Y.C.; Cobine, P.A.; Maxfield, A.B.; Carr, H.S.; Winge, D.R. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J. Biol. Chem. 2004, 279, 35334–35340. [Google Scholar] [CrossRef]
- Hiser, L.; Di Valentin, M.; Hamer, A.G.; Hosler, J.P. Cox11p is required for stable formation of the Cu(B) and magnesium centers of cytochrome c oxidase. J. Biol. Chem. 2000, 275, 619–623. [Google Scholar] [CrossRef]
- Tzagoloff, A.; Nobrega, M.; Gorman, N.; Sinclair, P. On the functions of the yeast COX10 and COX11 gene products. Biochem. Mol. Biol. Int. 1993, 31, 593–598. [Google Scholar]
- Carr, H.S.; Maxfield, A.B.; Horng, Y.C.; Winge, D.R. Functional analysis of the domains in Cox11. J. Biol. Chem. 2005, 280, 22664–22669. [Google Scholar] [CrossRef]
- Carr, H.S.; George, G.N.; Winge, D.R. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 2002, 277, 31237–31242. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Cantini, F.; Ciofi-Baffoni, S.; Gonnelli, L.; Mangani, S. Solution structure of Cox11, a novel type of beta-immunoglobulin-like fold involved in CuB site formation of cytochrome c oxidase. J. Biol. Chem. 2004, 279, 34833–34839. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.; Simon, J. Bacterial nitrous oxide respiration: Electron transport chains and copper transfer reactions. Adv. Microb. Physiol. 2019, 75, 137–175. [Google Scholar] [CrossRef] [PubMed]
- Kroneck, P.M.H. Walking the seven lines: Binuclear copper A in cytochrome c oxidase and nitrous oxide reductase. J. Biol. Inorg. Chem. 2018, 23, 27–39. [Google Scholar] [CrossRef]
- Schneider, L.K.; Wüst, A.; Pomowski, A.; Zhang, L.; Einsle, O. No laughing matter: The unmaking of the greenhouse gas dinitrogen monoxide by nitrous oxide reductase. Met. Ions. Life Sci. 2014, 14, 177–210. [Google Scholar] [CrossRef]
- Heikkilä, M.P.; Honisch, U.; Wunsch, P.; Zumft, W.G. Role of the Tat ransport system in nitrous oxide reductase translocation and cytochrome cd1 biosynthesis in Pseudomonas stutzeri. J. Bacteriol. 2001, 183, 1663–1671. [Google Scholar] [CrossRef]
- Dreusch, A.; Riester, J.; Kroneck, P.M.; Zumft, W.G. Mutation of the conserved Cys165 outside of the CuA domain destabilizes nitrous oxide reductase but maintains its catalytic activity. Evidence for disulfide bridges and a putative protein disulfide isomerase gene. Eur. J. Biochem. 1996, 237, 447–453. [Google Scholar] [CrossRef]
- Dreusch, A.; Bürgisser, D.M.; Heizmann, C.W.; Zumft, W.G. Lack of copper insertion into unprocessed cytoplasmic nitrous oxide reductase generated by an R20D substitution in the arginine consensus motif of the signal peptide. Biochim. Biophys. Acta 1997, 1319, 311–318. [Google Scholar] [CrossRef][Green Version]
- Taubner, L.M.; McGuirl, M.A.; Dooley, D.M.; Copié, V. Structural studies of Apo NosL, an accessory protein of the nitrous oxide reductase system: Insights from structural homology with MerB, a mercury resistance protein. Biochemistry 2006, 45, 12240–12252. [Google Scholar] [CrossRef]
- Zückert, W.R. Secretion of bacterial lipoproteins: Through the cytoplasmic membrane, the periplasm and beyond. Biochim. Biophys. Acta 2014, 1843, 1509–1516. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Gates, A.J.; Appia-Ayme, C.; Rowley, G.; Richardson, D.J. Copper control of bacterial nitrous oxide emission and its impact on vitamin B12-dependent metabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 19926–19931. [Google Scholar] [CrossRef] [PubMed]
- Hoegger, P.J.; Kilaru, S.; James, T.Y.; Thacker, J.R.; Kües, U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006, 273, 2308–2326. [Google Scholar] [CrossRef] [PubMed]
- Bello, M.; Valderrama, B.; Serrano-Posada, H.; Rudiño-Piñera, E. Molecular dynamics of a thermostable multicopper oxidase from Thermus thermophilus HB27: Structural differences between the apo and holo forms. PLoS ONE 2012, 7, e40700. [Google Scholar] [CrossRef] [PubMed]
- Hausrath, A.C.; Ramirez, N.A.; Ly, A.T.; McEvoy, M.M. The bacterial copper-resistance protein CopG contains a cysteine-bridged tetranuclear copper cluster. J. Biol. Chem. 2020. [Google Scholar] [CrossRef]
- Pontel, L.B.; Soncini, F.C. Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol. Microbiol. 2009, 73, 212–225. [Google Scholar] [CrossRef]
- Osman, D.; Patterson, C.J.; Bailey, K.; Fisher, K.; Robinson, N.J.; Rigby, S.E.; Cavet, J.S. The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 2013, 87, 466–477. [Google Scholar] [CrossRef]
- Yoon, B.Y.; Kim, Y.H.; Kim, N.; Yun, B.Y.; Kim, J.S.; Lee, J.H.; Cho, H.S.; Lee, K.; Ha, N.C. Structure of the periplasmic copper-binding protein CueP from Salmonella enterica serovar Typhimurium. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1867–1875. [Google Scholar] [CrossRef]
- Osman, D.; Waldron, K.J.; Denton, H.; Taylor, C.M.; Grant, A.J.; Mastroeni, P.; Robinson, N.J.; Cavet, J.S. Copper Homeostasis in Salmonella Is Atypical and Copper-CueP Is a Major Periplasmic Metal Complex. J. Biol. Chem. 2010, 285, 25259–25268. [Google Scholar] [CrossRef]
- Xiao, Z.; Wedd, A.G. The challenges of determining metal-protein affinities. Nat. Prod. Rep. 2010, 27, 768–789. [Google Scholar] [CrossRef]
- Xiao, Z.; Brose, J.; Schimo, S.; Ackland, S.M.; La Fontaine, S.; Wedd, A.G. Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: Detection probes and affinity standards. J. Biol. Chem. 2011, 286, 11047–11055. [Google Scholar] [CrossRef]
- Mercer, J.F. The molecular basis of copper-transport diseases. Trends Mol. Med. 2001, 7, 64–69. [Google Scholar] [CrossRef]
- Li, Y. Copper homeostasis: Emerging target for cancer treatment. IUBMB Life 2020. [Google Scholar] [CrossRef] [PubMed]
- Petruzzelli, R.; Polishchuk, R.S. Activity and Trafficking of Copper-Transporting ATPases in Tumor Development and Defense against Platinum-Based Drugs. Cells 2019, 8, 1080. [Google Scholar] [CrossRef] [PubMed]
- Tsang, T.; Posimo, J.M.; Gudiel, A.A.; Cicchini, M.; Feldser, D.M.; Brady, D.C. Copper is an essential regulator of the autophagic kinases ULK1/2 to drive lung adenocarcinoma. Nat. Cell Biol. 2020, 22, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Tsang, T.; Anderson, G.R.; Posimo, J.M.; Brady, D.C. Inhibition of BCL2 Family Members Increases the Efficacy of Copper Chelation in BRAF(V600E)-Driven Melanoma. Cancer Res. 2020, 80, 1387–1400. [Google Scholar] [CrossRef] [PubMed]
- Blockhuys, S.; Brady, D.C.; Wittung-Stafshede, P. Evaluation of copper chaperone ATOX1 as prognostic biomarker in breast cancer. Breast Cancer 2020, 27, 505–509. [Google Scholar] [CrossRef] [PubMed]
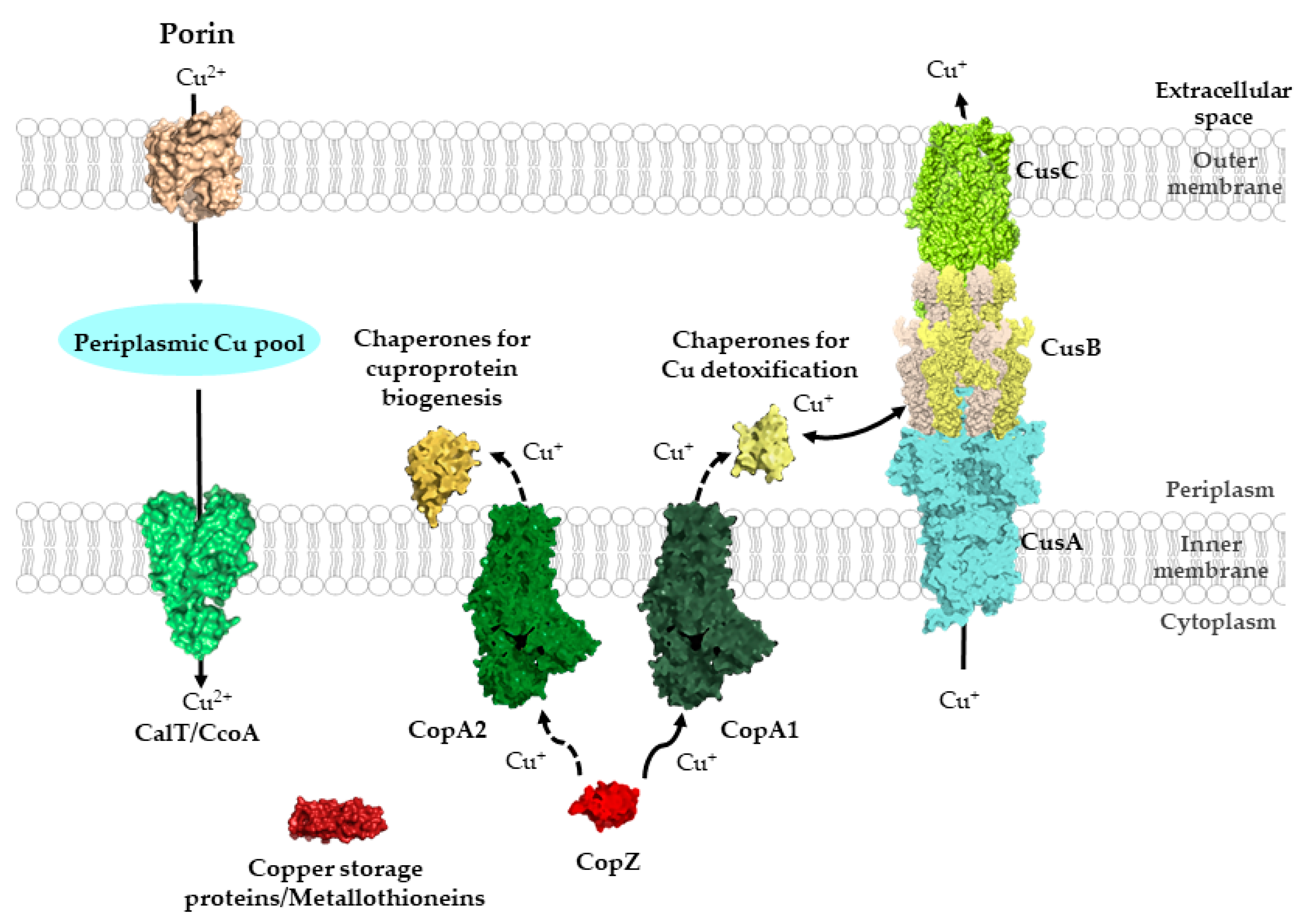
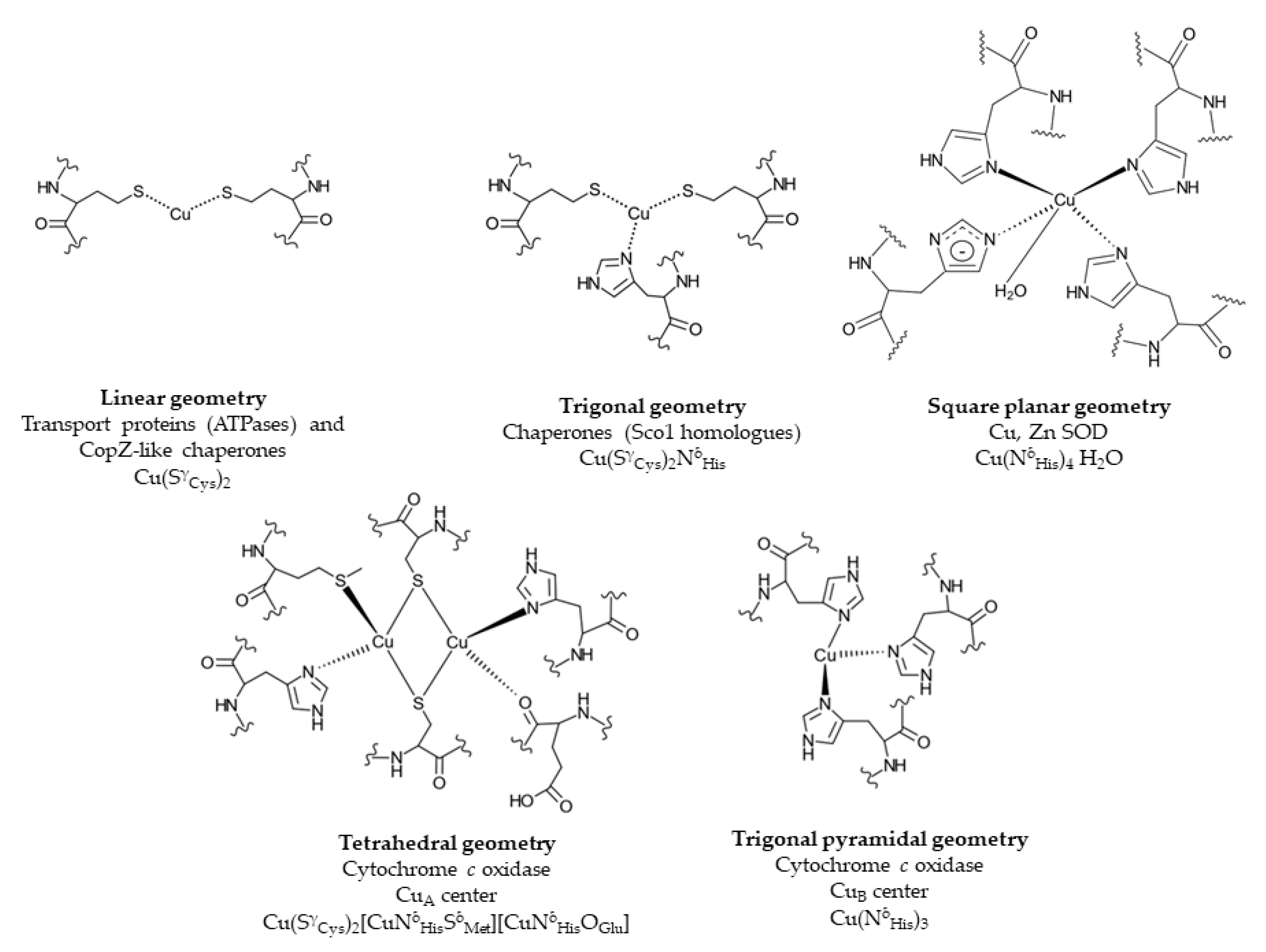

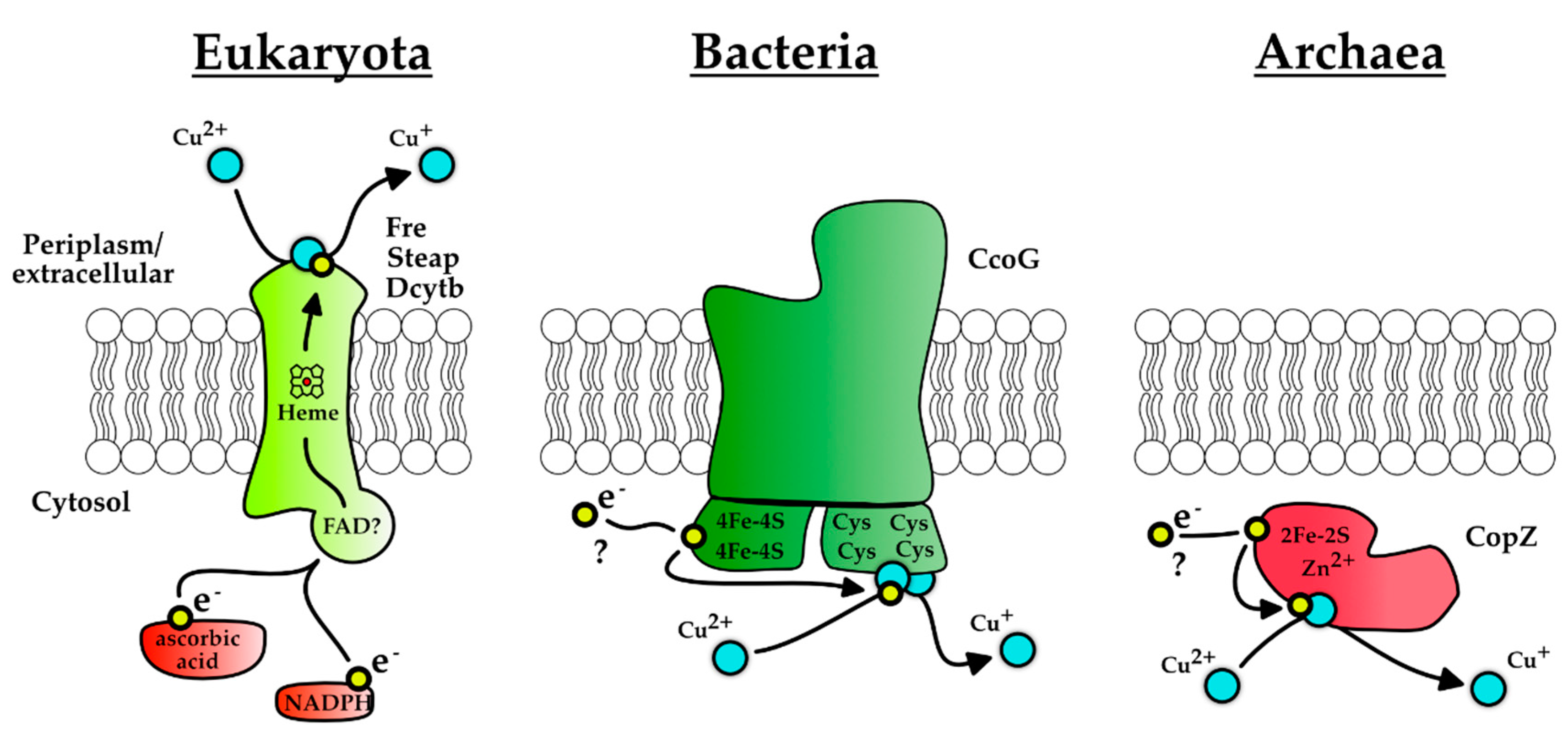
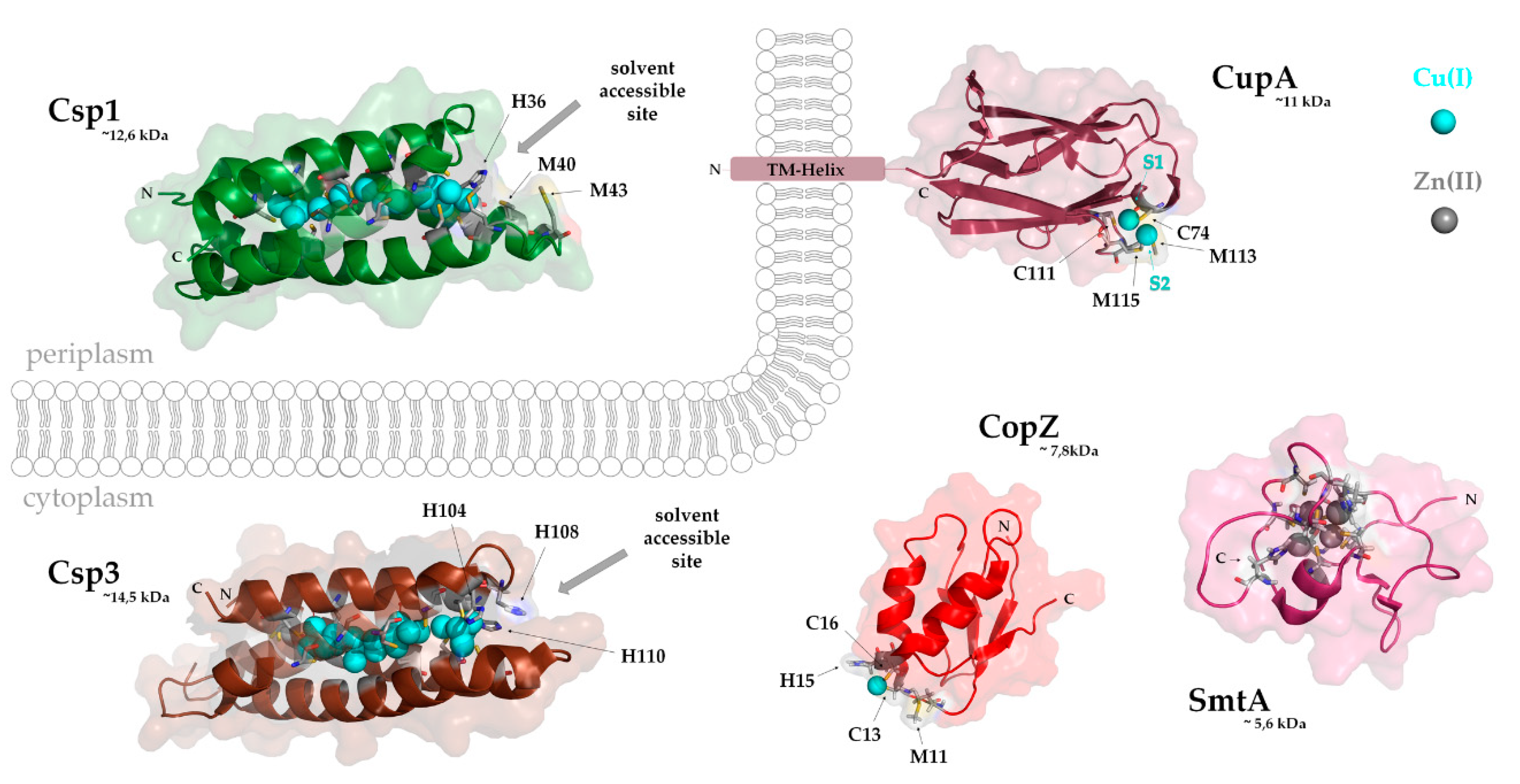
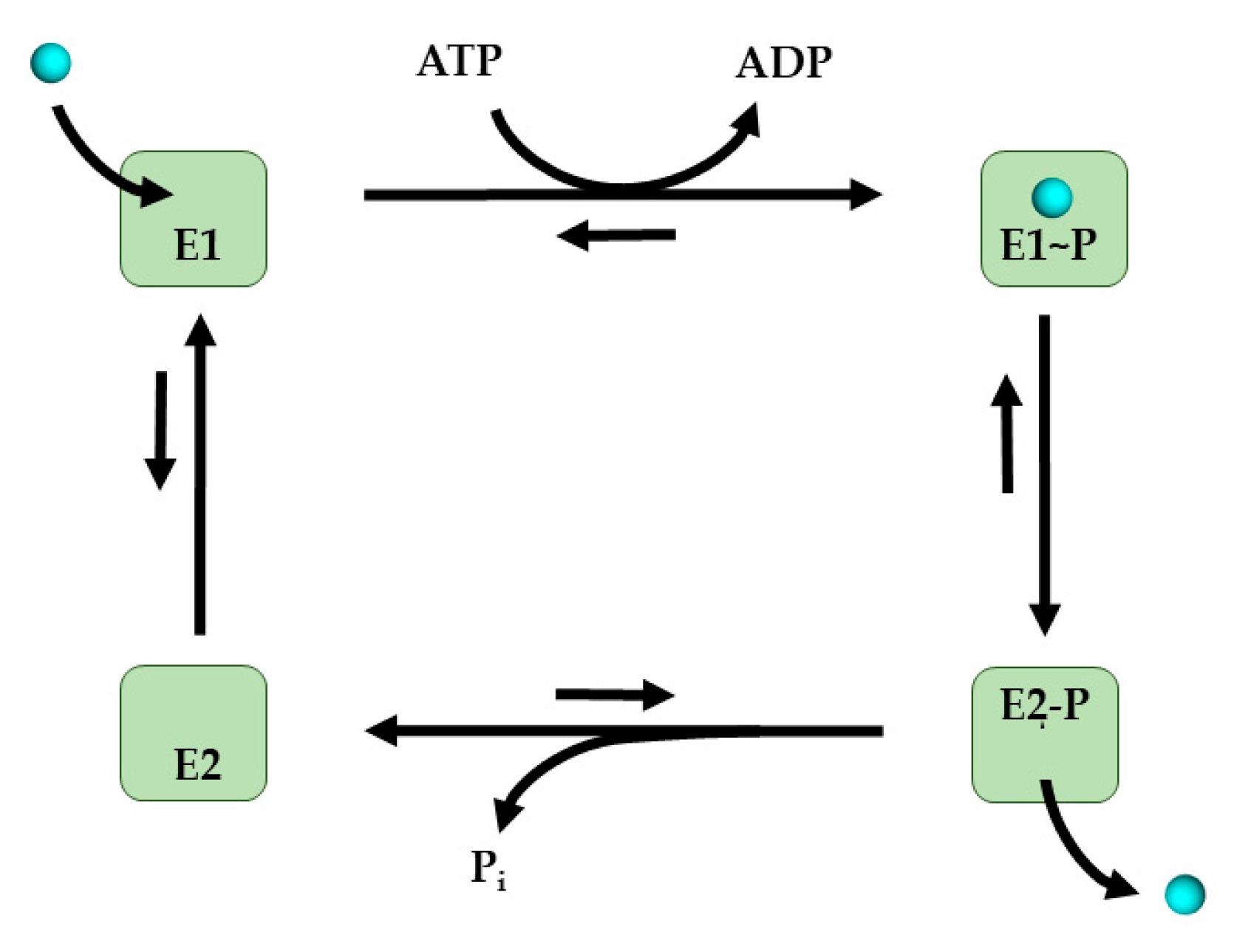
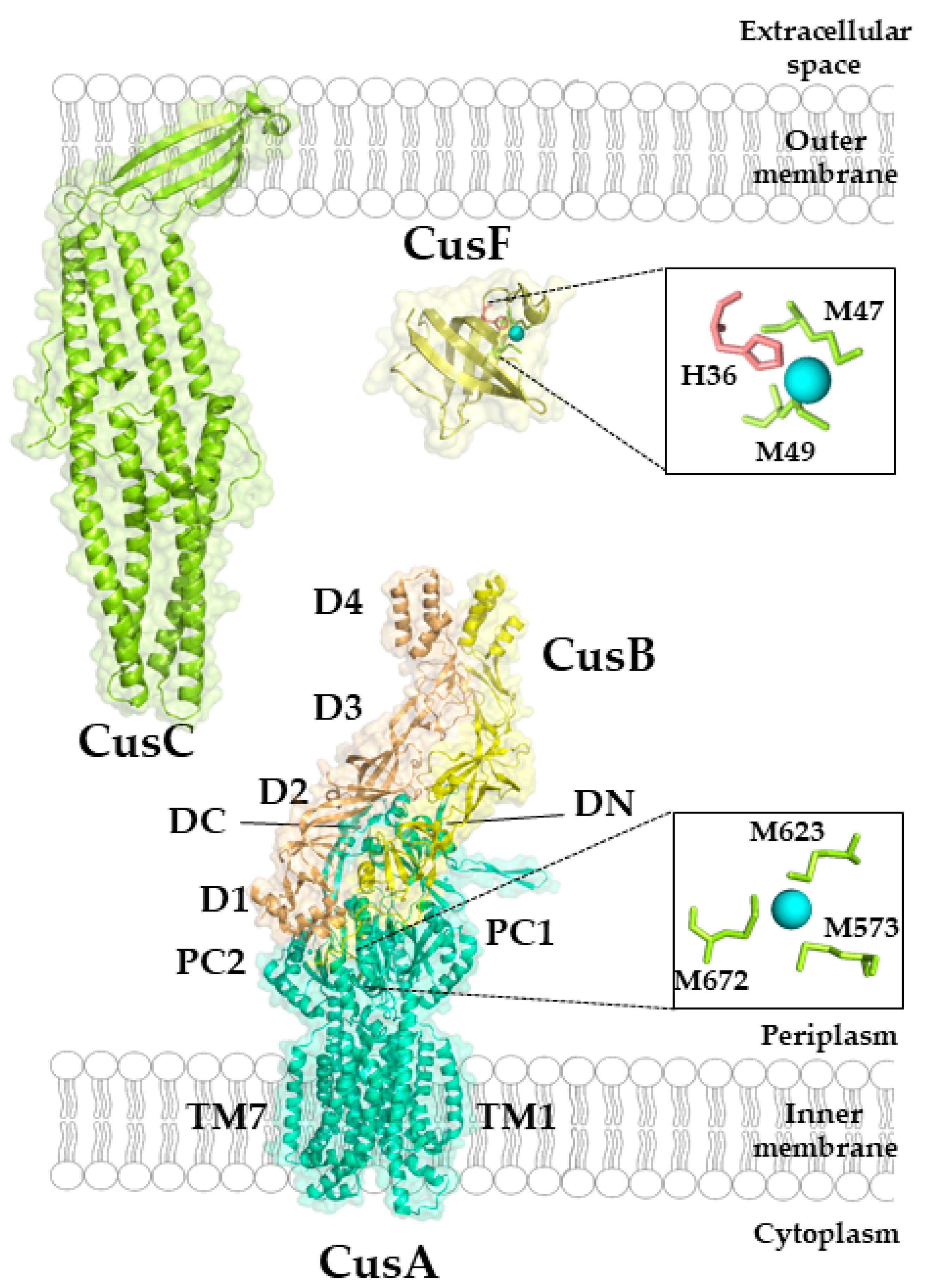

| Protein | Localization | Function | Reference |
|---|---|---|---|
| Plastocyanin | Thylakoid lumen | Photosynthetic electron transfer | [26] |
| Cytochrome c Oxidase | Membrane | O2-reduction | [34] |
| Particulate Methane monooxygenase | Membrane- associated | Methane hydroxylation | [35] |
| Multi-Copper oxidases | Periplasm | Cu detoxification | [36] |
| Nitrite reductase | Periplasm | Nitrite reduction | [37] |
| Azurin | Periplasm | Respiratory electron transfer | [38] |
| Cu-Zn Superoxide dismutase | Periplasm | Superoxide detoxification | [39] |
| Nitrous oxide reductase | Periplasm | Denitrification | [40] |
| Amine oxidases | Periplasm | Amine oxidation | [41] |
| MccA-type Sulfite reductase | Periplasm | Sulfite reduction | [42] |
| Tyrosinase | Extracellular | Monooxygenase | [43] |
| Protein | Organisms | Function | Reference |
|---|---|---|---|
| CcoG | Rhodobacter capsulatus | Specific Cu(II) reduction via two tetranuclear iron–sulfur clusters | [157] |
| NDH-2 | Escherichia coli | NADH-Dehydrogenase 2, unspecific quinol-dependent Cu(II) reduction | [162] |
| ? | Lactococcus lactis | Unspecific quinol-mediated Cu(II) reduction | [163] |
| ? | Pseudomonas sp. | Cell-free Cu(II) reduction, not further characterized | [164] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrei, A.; Öztürk, Y.; Khalfaoui-Hassani, B.; Rauch, J.; Marckmann, D.; Trasnea, P.-I.; Daldal, F.; Koch, H.-G. Cu Homeostasis in Bacteria: The Ins and Outs. Membranes 2020, 10, 242. https://doi.org/10.3390/membranes10090242
Andrei A, Öztürk Y, Khalfaoui-Hassani B, Rauch J, Marckmann D, Trasnea P-I, Daldal F, Koch H-G. Cu Homeostasis in Bacteria: The Ins and Outs. Membranes. 2020; 10(9):242. https://doi.org/10.3390/membranes10090242
Chicago/Turabian StyleAndrei, Andreea, Yavuz Öztürk, Bahia Khalfaoui-Hassani, Juna Rauch, Dorian Marckmann, Petru-Iulian Trasnea, Fevzi Daldal, and Hans-Georg Koch. 2020. "Cu Homeostasis in Bacteria: The Ins and Outs" Membranes 10, no. 9: 242. https://doi.org/10.3390/membranes10090242
APA StyleAndrei, A., Öztürk, Y., Khalfaoui-Hassani, B., Rauch, J., Marckmann, D., Trasnea, P.-I., Daldal, F., & Koch, H.-G. (2020). Cu Homeostasis in Bacteria: The Ins and Outs. Membranes, 10(9), 242. https://doi.org/10.3390/membranes10090242