Cholera Toxin Subunit B for Sensitive and Rapid Determination of Exosomes by Gel Filtration
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Isolation of Exosomes
2.3. Preparation of Exosomal Liposomes
2.4. Gel Filtration Assay
2.5. Exosome Quantification
2.6. Liposome Quantification
3. Results
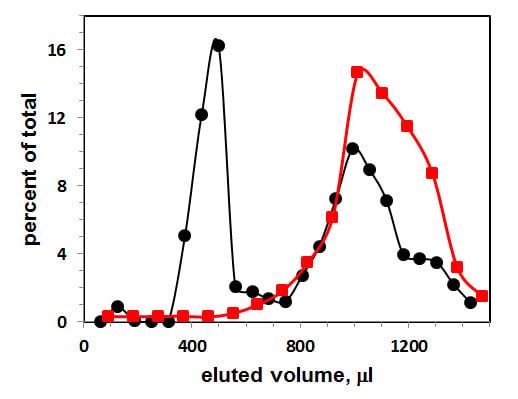
3.1. Gel Filtration of Exosomes
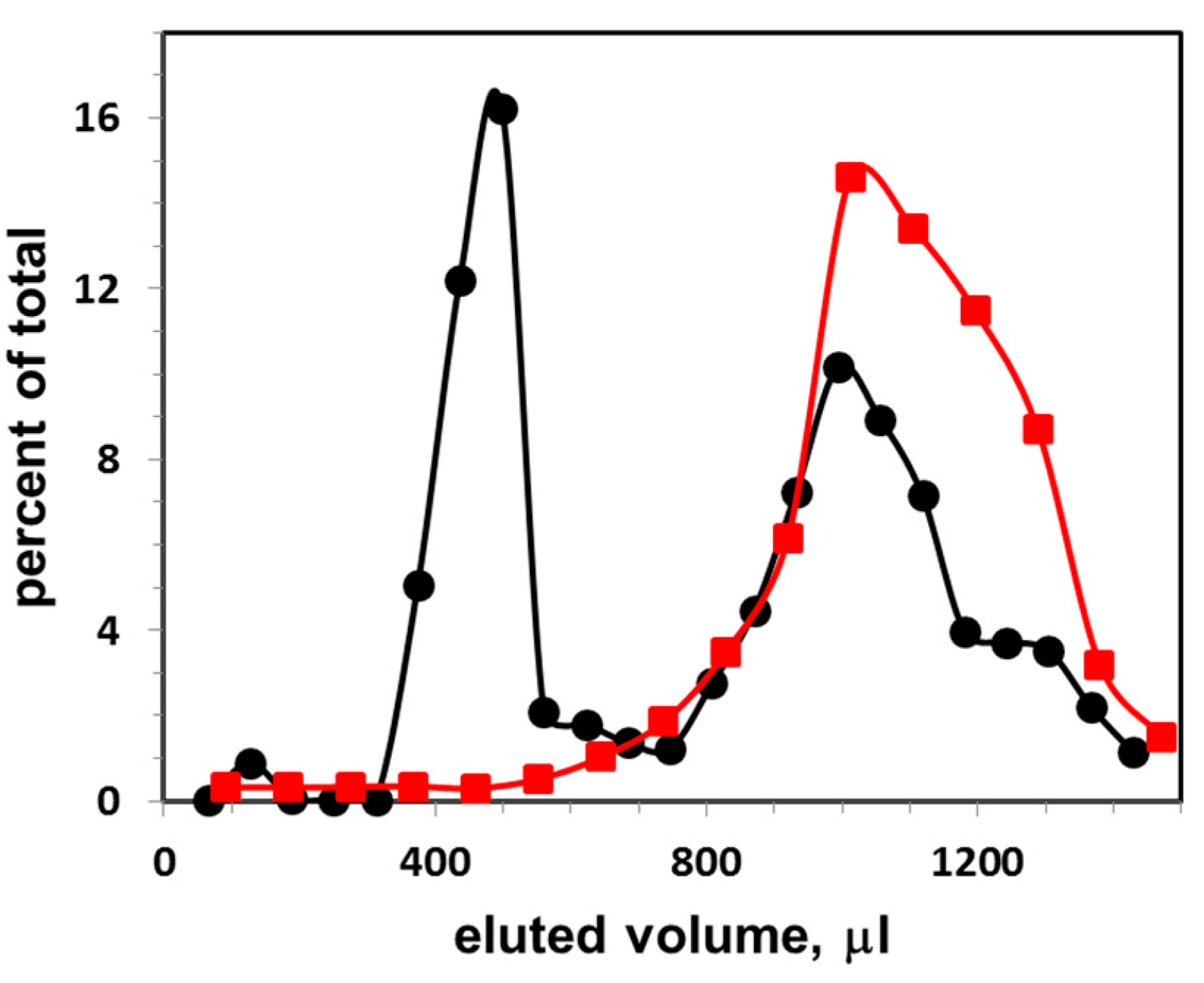
3.2. Calibration Curve for Exosome Quantification
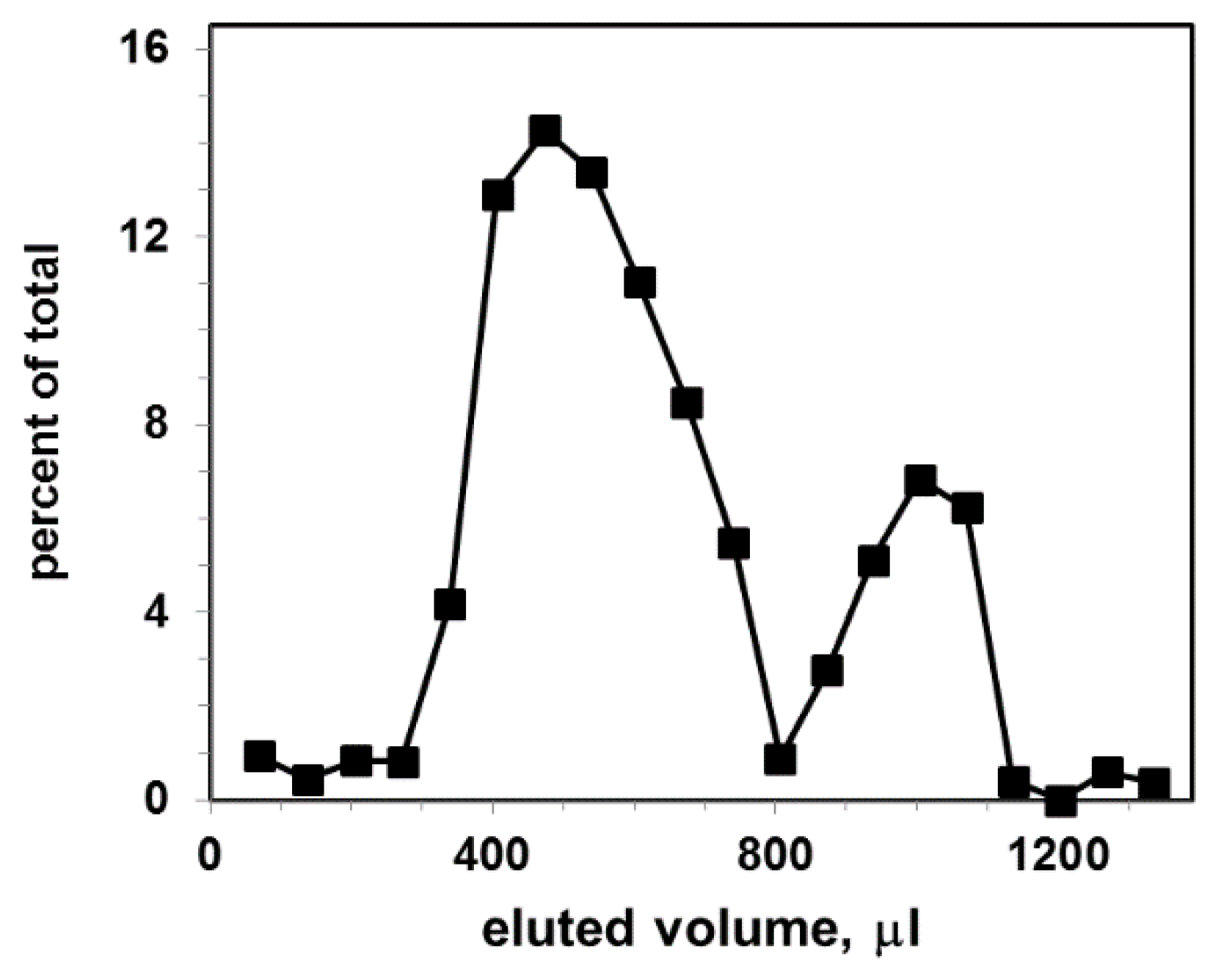
3.3. Gel Filtration of Exosomal Liposomes
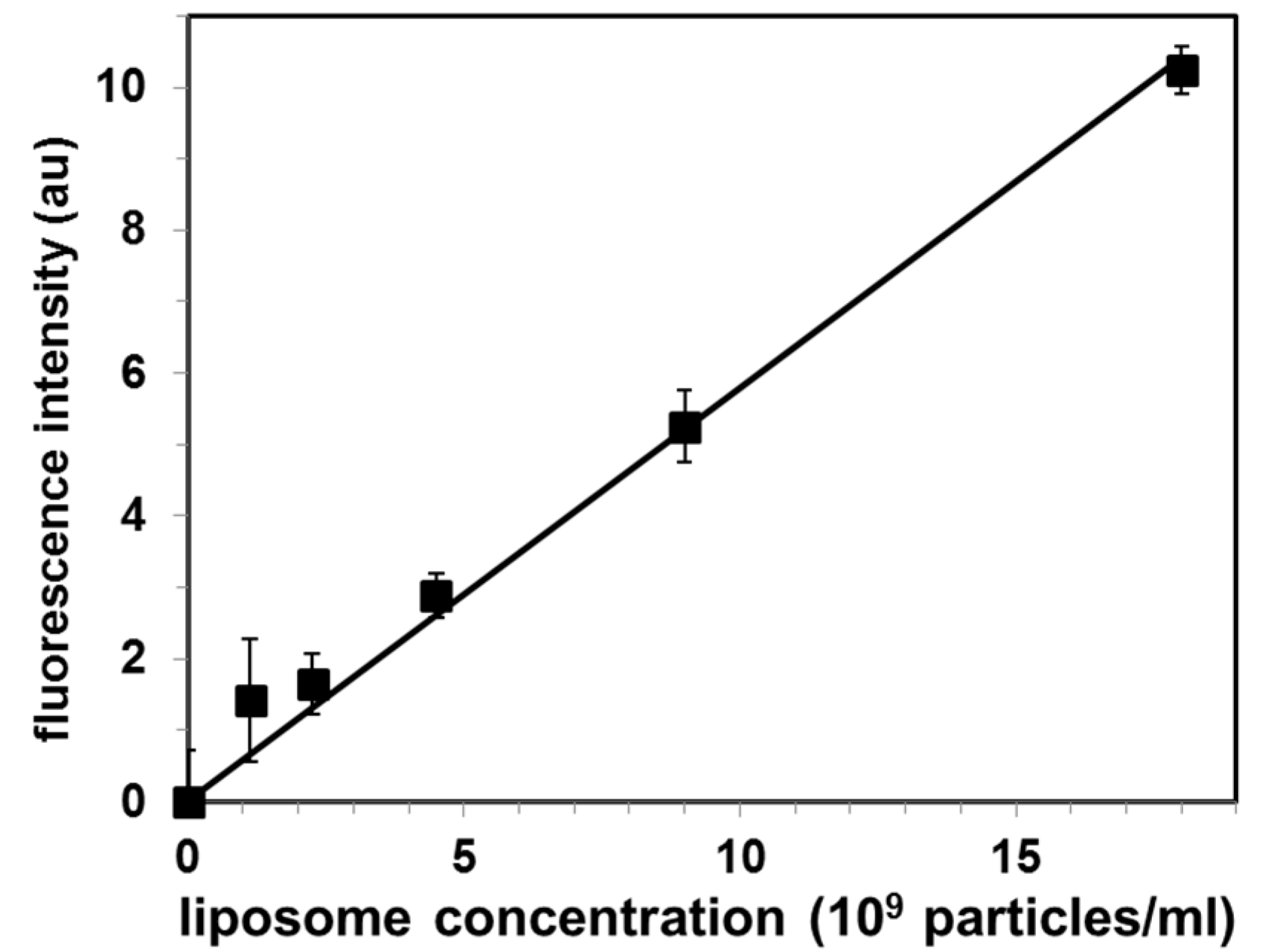
3.4. Calibration Curve for Exosomal Liposome Quantification
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef] [PubMed]
- Llorente, A.; Skotland, T.; Sylvanne, T.; Kauhanen, D.; Rog, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Osteikoetxea, X.; Balogh, A.; Szabó-Taylor, K.; Németh, A.; Szabó, T.G.; Pálóczi, K.; Sódar, B.; Kittel, A.; György, B.; Pállinger, E.; et al. Improved characterization of EV preparations based on protein to lipid ratio and lipid properties. PLoS ONE 2015, 10, e0121184. [Google Scholar] [CrossRef] [PubMed]
- Janas, A.M.; Sapoń, K.; Janas, T.; Stowell, M.H.; Janas, T. Exosomes and other extracellular vesicles in neural cells and neurodegenerative diseases. Biochim. Biophys. Acta 2016, 1858, 1139–1151. [Google Scholar] [CrossRef]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular vesicle heterogeneity: Subpopulations, isolation techniques, and diverse functions in cancer progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef]
- Stratmann, T. Cholera toxin subunit B as adjuvant—An accelerator in protective immunity and a break in autoimmunity. Vaccines 2015, 3, 579–596. [Google Scholar] [CrossRef]
- Skocaj, M.; Bakrac, B.; Krizaj, I.; Macek, P.; Anderluh, G.; Sepcic, K. The sensing of membrane microdomains based on pore-forming toxins. Curr. Med. Chem. 2013, 20, 491–501. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 8, 1535750. [Google Scholar] [CrossRef]
- Xu, R.; Fitts, A.; Li, X.; Fernandes, J.; Pochampally, R.; Mao, J.; Liu, Y.M. Quantification of small extracellular vesicles by size exclusion chromatography with fluorescence detection. Anal. Chem. 2016, 88, 10390–10394. [Google Scholar] [CrossRef]
- Kitka, D.; Mihaly, J.; Fraikin, J.-L.; Beke-Somfai, T.Z.; Varga, Z. Detection and phenotyping of extracellular vesicles by size exclusion chromatography coupled with on-line fluorescence detection. Sci. Rep. 2019, 9, 19868. [Google Scholar] [CrossRef] [PubMed]
- Sapoń, K.; Gawrońska, I.; Janas, T.; Sikorski, A.F.; Janas, T. Exosome-associated polysialic acid modulates membrane potentials, membrane thermotropic properties, and raft-dependent interactions between vesicles. FEBS Lett. 2020, 594, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Sapoń, K.; Janas, T.; Sikorski, A.F.; Janas, T. Polysialic acid chains exhibit enhanced affinity for ordered regions of membranes. Biochim. Biophys. Acta-Biomembr. 2019, 1861, 245–255. [Google Scholar] [CrossRef]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef]
- Janas, T.; Sapon, K.; Stowell, M.H.B.; Janas, T. Selection of membrane RNA aptamers to amyloid beta peptide: Implications for exosome-based antioxidant strategies. Int. J. Mol. Sci. 2019, 20, 299. [Google Scholar] [CrossRef]
- Takamori, S.; Holt, M.; Stenius, K.; Lemke, E.A.; Gronborg, M.; Riedel, D.; Urlaub, H.; Schenck, S.; Brügger, B.; Ringler, P.; et al. Molecular anatomy of a trafficking organelle. Cell 2006, 127, 831–846. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, A.D.; Engelman, D.M. Protein area occupancy at the center of the red blood cell membrane. Proc. Natl. Acad. Sci. USA 2008, 105, 2848–2852. [Google Scholar] [CrossRef]
- Houser, J.R.; Busch, D.J.; Bell, D.R.; Li, B.; Ren, P.; Stachowiak, J.C. The impact of physiological Crowding on the Diffusivity of Membrane Bound Proteins. Soft Matter 2016, 12, 2127–2134. [Google Scholar] [CrossRef]
- Pandit, S.A.; Vasudevan, S.; Chiu, S.W.; Mashl, R.J.; Jakobsson, E.; Scott, H.L. Sphingomyelin-cholesterol domains in phospholipid membranes: Atomistic simulation. Biophys. J. 2004, 87, 1092–1100. [Google Scholar] [CrossRef]
- Edholm, O.; Nagle, J.F. Areas of molecules in membranes consisting of mixtures. Biophys. J. 2005, 89, 1827–1832. [Google Scholar] [CrossRef]
- Zrimi, J.; Ling, A.N.; Arifin, E.G.-R.; Feverati, G.; Lesieur, C. Cholera toxin B subunits assemble into pentamers—Proposition of a fly-casting mechanism. PLoS ONE 2010, 5, e15347. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, X.; Gong, X.; Wang, G. Human umbilical cord mesenchymal stem cells derived exosomes exert antiapoptosis effect via activating PI3K/Akt/mTOR pathway on H9C2 cells. J. Cell. Biochem. 2019, 120, 14455–14464. [Google Scholar] [CrossRef] [PubMed]
- Margheri, G.; Agostino, R.D’.; Trigari, S.; Sottiniet, S.; Rosso, M.D. The β-subunit of cholera toxin has a high affinity for ganglioside GM1 embedded into solidsupported lipid membranes with a lipid raft-like composition. Lipids 2014, 49, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Wands, A.M.; Fujita, A.; McCombs, J.E.; Cervin, J.; Dedic, B.; Rodriguez, A.C.; Nischan, N.; Bond, M.R.; Mettlen, M.; Trudgian, D.C.; et al. Fucosylation and protein glycosylation create functional receptors for cholera toxin. Elife 2015, 4, e09545. [Google Scholar] [CrossRef]
- Harada, Y.; Kizuka, Y.; Tokoro, Y.; Kondo, K.; Yagi, H.; Kato, K.; Inoue, H.; Taniguchi, N.; Maruyama, I. N-glycome inheritance from cells to extracellular vesicles in B16 melanomas. FEBS Lett. 2019, 593, 942–951. [Google Scholar] [CrossRef]
- Vermassen, T.; Speeckaert, M.M.; Lumen, N.; Rottey, S.; Delanghe, J.R. Glycosylation of prostate specific antigen and its potential diagnostic applications. Clin. Chim. Acta 2012, 413, 1500–1505. [Google Scholar] [CrossRef]
- Costa, J.; Gatermann, M.; Nimtz, M.; Kandzia, S.; Glatzel, M.; Conradt, H.S. N-glycosylation of extracellular vesicles from HEK-293 and glioma cell lines. Anal. Chem. 2018, 90, 7871–7879. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapoń, K.; Maziarz, D.; Janas, T.; Sikorski, A.F.; Janas, T. Cholera Toxin Subunit B for Sensitive and Rapid Determination of Exosomes by Gel Filtration. Membranes 2020, 10, 172. https://doi.org/10.3390/membranes10080172
Sapoń K, Maziarz D, Janas T, Sikorski AF, Janas T. Cholera Toxin Subunit B for Sensitive and Rapid Determination of Exosomes by Gel Filtration. Membranes. 2020; 10(8):172. https://doi.org/10.3390/membranes10080172
Chicago/Turabian StyleSapoń, Karolina, Dominika Maziarz, Teresa Janas, Aleksander F. Sikorski, and Tadeusz Janas. 2020. "Cholera Toxin Subunit B for Sensitive and Rapid Determination of Exosomes by Gel Filtration" Membranes 10, no. 8: 172. https://doi.org/10.3390/membranes10080172
APA StyleSapoń, K., Maziarz, D., Janas, T., Sikorski, A. F., & Janas, T. (2020). Cholera Toxin Subunit B for Sensitive and Rapid Determination of Exosomes by Gel Filtration. Membranes, 10(8), 172. https://doi.org/10.3390/membranes10080172