Development and Optimization of an Enzyme Immunoassay to Detect Serum Antibodies against the Hepatitis E Virus in Pigs, Using Plant-Derived ORF2 Recombinant Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Cloning, Plasmids Construction, and Agroinfiltration of Nicotiana Benthamiana
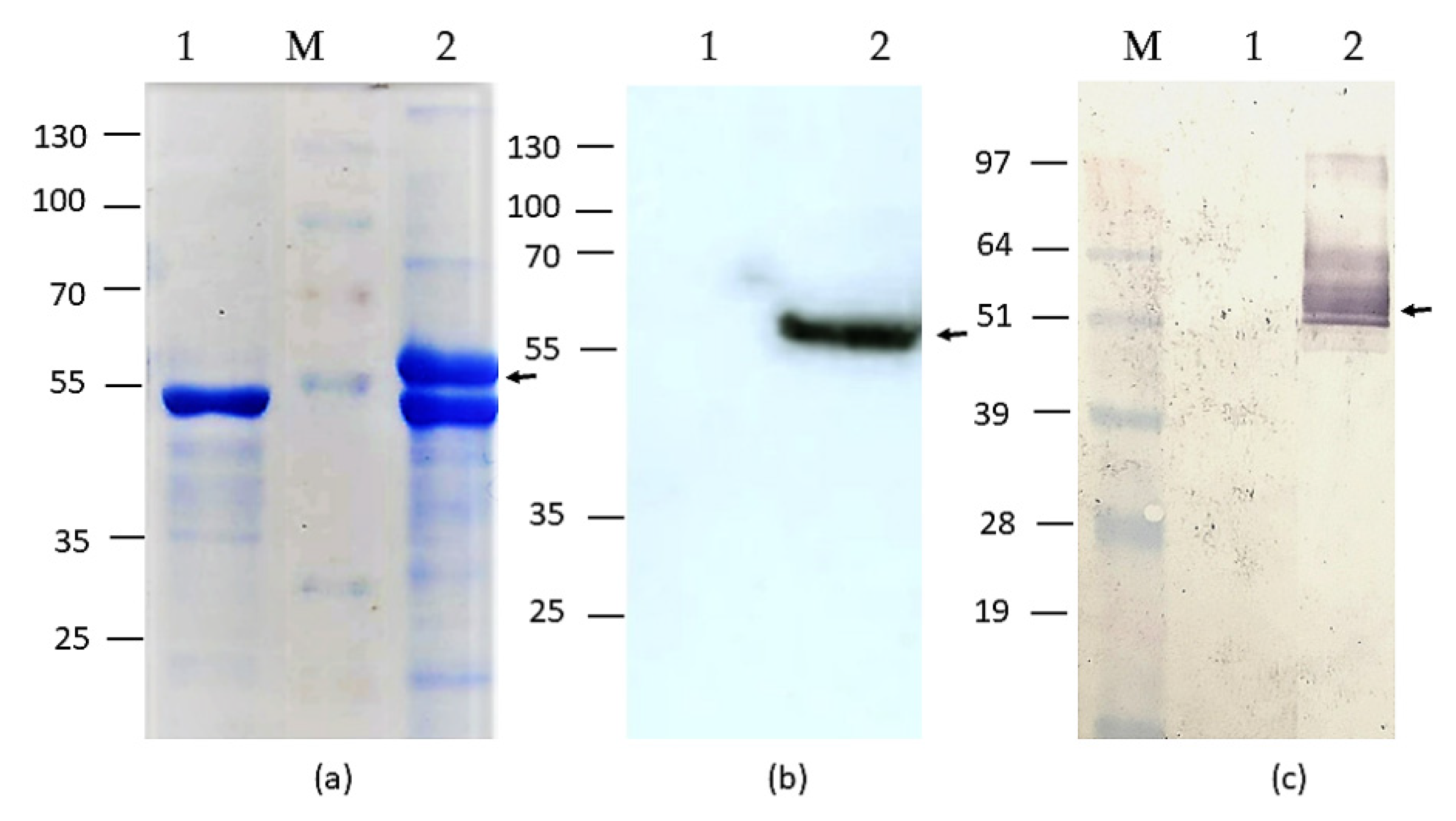
2.2. SDS-PAGE and Western Blot of HEV-3 OFR2 110-610_6his Protein Preparations
2.3. Large-Scale Isolation of HEV-3 ORF2 110-610_6his Recombinant Proteins from Plant Tissue
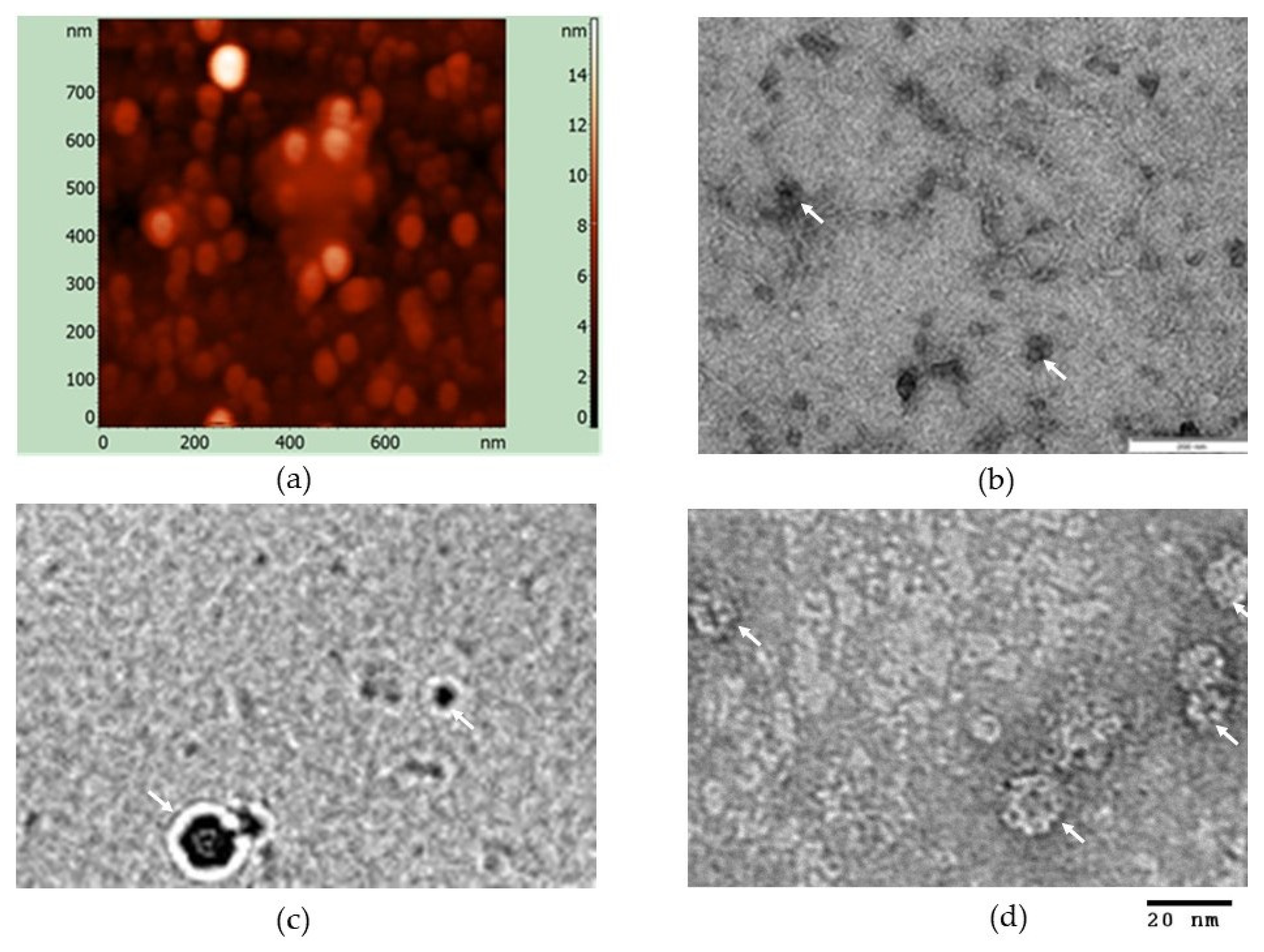
2.4. Transmission Electron Microscopy (TEM), Cryo-EM, Atomic Force Microscopy
2.5. Serum Samples
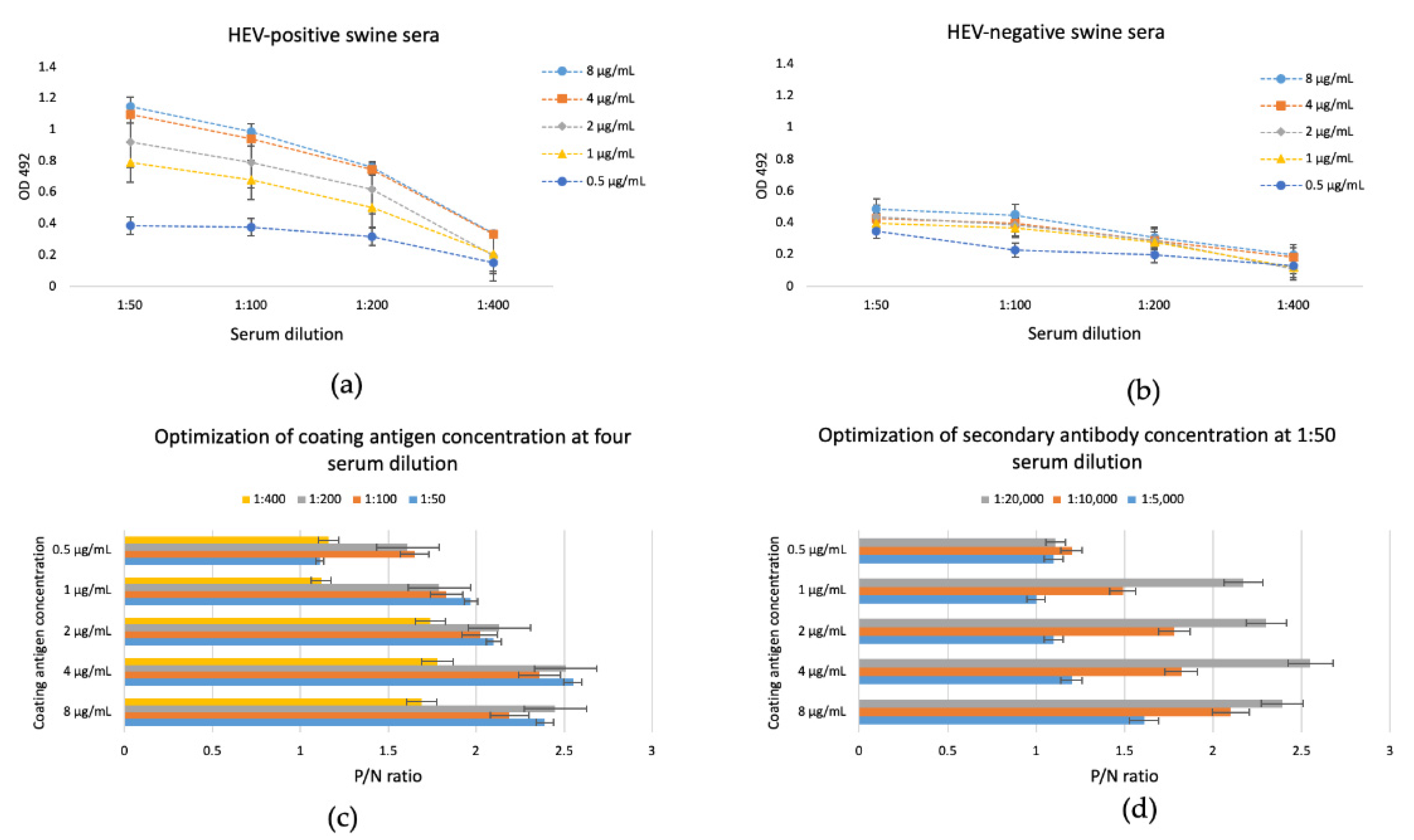
2.6. Optimization of iEIA Protocol Based on HEV-3 ORF2 110-610_6his Recombinant Protein
2.7. Validation, Statistical Analysis, and Cut-off Evaluation of the Developed iEIA Performance
- Specificity = no. of true negative/(no. of true negative + no. of false positive)
- Sensitivity = no. of true positive/(no. of true positive + no. of false negative)
- Efficiency (%) = ((no. of true positive + no. of true negative)/(no. of true positive + no. of false positive + no. of true negative + no. of false negative)) × 100
- Youden index = sensitivity + specificity − 1
3. Results
3.1. Production and Purification of HEV-3 ORF2 110-610_6his
3.2. In Vitro Particles Formation
3.3. In-House EIA Optimization Using the Plant-Derived HEV-3 ORF2 110-610_6his as a Coating Protein
3.4. Specificity and Sensitivity of the In-House Enzyme Immunosorbent Assay with the Plant-Derived HEV ORF2 110-610_6his Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Marion, O.; Abravanel, F.; Lhomme, S.; Izopet, J.; Kamar, N. Hepatitis E in Transplantation. Curr. Infect. Dis. Rep. 2016, 18, 8. [Google Scholar] [CrossRef]
- Wu, C.; Wu, X.; Xia, J. Hepatitis E Virus Infection during Pregnancy. Virol. J. 2020, 17, 73. [Google Scholar] [CrossRef]
- Montpellier, C.; Wychowski, C.; Sayed, I.M.; Meunier, J.-C.; Saliou, J.-M.; Ankavay, M.; Bull, A.; Pillez, A.; Abravanel, F.; Helle, F.; et al. Hepatitis E Virus Lifecycle and Identification of 3 Forms of the ORF2 Capsid Protein. Gastroenterology 2018, 154, 211–223.e8. [Google Scholar] [CrossRef] [PubMed]
- Purdy, M.A.; Harrison, T.J.; Jameel, S.; Meng, X.-J.; Okamoto, H.; Van der Poel, W.H.M.; Smith, D.B. Ictv Report Consortium, null ICTV Virus Taxonomy Profile: Hepeviridae. J. Gen. Virol. 2017, 98, 2645–2646. [Google Scholar] [CrossRef]
- Smith, D.B.; Simmonds, P. Classification and Genomic Diversity of Enterically Transmitted Hepatitis Viruses. Cold Spring Harb. Perspect. Med. 2018, 8, a031880. [Google Scholar] [CrossRef] [PubMed]
- Primadharsini, P.P.; Nagashima, S.; Okamoto, H. Genetic Variability and Evolution of Hepatitis E Virus. Viruses 2019, 11, 456. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Paddy, J.O.; Simmonds, P. The Use of Human Sewage Screening for Community Surveillance of Hepatitis E Virus in the UK. J. Med. Virol. 2016, 88, 915–918. [Google Scholar] [CrossRef]
- Izopet, J.; Dubois, M.; Bertagnoli, S.; Lhomme, S.; Marchandeau, S.; Boucher, S.; Kamar, N.; Abravanel, F.; Guérin, J.-L. Hepatitis E Virus Strains in Rabbits and Evidence of a Closely Related Strain in Humans, France. Emerg. Infect. Dis. 2012, 18, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Abravanel, F.; Lhomme, S.; El Costa, H.; Schvartz, B.; Peron, J.-M.; Kamar, N.; Izopet, J. Rabbit Hepatitis E Virus Infections in Humans, France. Emerg. Infect. Dis. 2017, 23, 1191–1193. [Google Scholar] [CrossRef]
- Oliveira-Filho, E.F.; Bank-Wolf, B.R.; Thiel, H.-J.; König, M. Phylogenetic Analysis of Hepatitis E Virus in Domestic Swine and Wild Boar in Germany. Vet. Microbiol. 2014, 174, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Debing, Y.; Moradpour, D.; Neyts, J.; Gouttenoire, J. Update on Hepatitis E Virology: Implications for Clinical Practice. J. Hepatol. 2016, 65, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Kenney, S.P.; Meng, X.-J. Hepatitis E Virus Genome Structure and Replication Strategy. Cold Spring Harb. Perspect. Med. 2019, 9, a031724. [Google Scholar] [CrossRef] [PubMed]
- Riddell, M.A.; Li, F.; Anderson, D.A. Identification of Immunodominant and Conformational Epitopes in the Capsid Protein of Hepatitis E Virus by Using Monoclonal Antibodies. J. Virol. 2000, 74, 8011–8017. [Google Scholar] [CrossRef]
- Ponterio, E.; Di Bartolo, I.; Orrù, G.; Liciardi, M.; Ostanello, F.; Ruggeri, F.M. Detection of Serum Antibodies to Hepatitis E Virus in Domestic Pigs in Italy Using a Recombinant Swine HEV Capsid Protein. BMC Vet. Res. 2014, 10, 133. [Google Scholar] [CrossRef] [PubMed]
- Mazalovska, M.; Varadinov, N.; Koynarski, T.; Minkov, I.; Teoharov, P.; Lomonossoff, G.P.; Zahmanova, G. Detection of Serum Antibodies to Hepatitis E Virus Based on HEV Genotype 3 ORF2 Capsid Protein Expressed in Nicotiana Benthamiana. Ann. Lab. Med. 2017, 37, 313–319. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arce, L.P.; Müller, M.F.; Martinez, A.; Baiker, A.; Marranzino, G.; Agote, F.; Vizoso-Pinto, M.G. A Novel In-House Enzyme-Linked Immunosorbent Assay for Genotype 3 Hepatitis E Virus Reveals High Seroprevalence in Blood Donors in Northern Argentina. Front. Microbiol. 2019, 10, 2481. [Google Scholar] [CrossRef]
- Pandolfi, R.; de Almeida, D.R.; Pinto, M.A.; Kreutz, L.C.; Frandoloso, R. In House ELISA Based on Recombinant ORF2 Protein Underline High Prevalence of IgG Anti-Hepatitis E Virus amongst Blood Donors in South Brazil. PLoS ONE 2017, 12, e0176409. [Google Scholar] [CrossRef] [PubMed]
- Van Der Poel, W.H.M.; Berto, A. 19—Advances in understanding of hepatitis E virus as a food—And waterborne pathogen. In Viruses in Food and Water; Cook, N., Ed.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead Publishing: Cambridge, UK, 2013; pp. 401–441. ISBN 978-0-85709-430-8. [Google Scholar]
- Colson, P.; Borentain, P.; Queyriaux, B.; Kaba, M.; Moal, V.; Gallian, P.; Heyries, L.; Raoult, D.; Gerolami, R. Pig Liver Sausage as a Source of Hepatitis E Virus Transmission to Humans. J. Infect. Dis. 2010, 202, 825–834. [Google Scholar] [CrossRef]
- Tei, S.; Kitajima, N.; Takahashi, K.; Mishiro, S. Zoonotic Transmission of Hepatitis E Virus from Deer to Human Beings. Lancet 2003, 362, 371–373. [Google Scholar] [CrossRef]
- Haïm-Boukobza, S.; Ferey, M.-P.; Vétillard, A.-L.; Jeblaoui, A.; Pélissier, E.; Pelletier, G.; Teillet, L.; Roque-Afonso, A.-M. Transfusion-Transmitted Hepatitis E in a Misleading Context of Autoimmunity and Drug-Induced Toxicity. J. Hepatol. 2012, 57, 1374–1378. [Google Scholar] [CrossRef]
- Baymakova, M.; Terzieva, K.; Popov, R.; Grancharova, E.; Kundurzhiev, T.; Pepovich, R.; Tsachev, I. Seroprevalence of Hepatitis E Virus Infection among Blood Donors in Bulgaria. Viruses 2021, 13, 492. [Google Scholar] [CrossRef] [PubMed]
- Tsachev, I.; Baymakova, M.; Ciccozzi, M.; Pepovich, R.; Kundurzhiev, T.; Marutsov, P.; Dimitrov, K.K.; Gospodinova, K.; Pishmisheva, M.; Pekova, L. Seroprevalence of Hepatitis E Virus Infection in Pigs from Southern Bulgaria. Vector Borne Zoonotic Dis. 2019, 19, 767–772. [Google Scholar] [CrossRef]
- Pishmisheva, M.; Baymakova, M.; Golkocheva-Markova, E.; Kundurzhiev, T.; Pepovich, R.; Popov, G.T.; Tsachev, I. First Serological Study of Hepatitis E Virus Infection in Pigs in Bulgaria. Comptes Rendus L’Academie Bulg. Sci. 2018, 71, 1001–1008. [Google Scholar] [CrossRef]
- Takova, K.; Koynarski, T.; Minkov, I.; Ivanova, Z.; Toneva, V.; Zahmanova, G. Increasing Hepatitis E Virus Seroprevalence in Domestic Pigs and Wild Boar in Bulgaria. Animals 2020, 10, 1521. [Google Scholar] [CrossRef] [PubMed]
- Pishmisheva, M.; Golkocheva-Markova, E.; Naseva, E.; Cachev, I. Distribution of Hepatitis E Virus Infection among Pigs in Pazardzik Region. Prev. Med. 2019, 7, 10. [Google Scholar]
- Tsachev, I.; Baymakova, M.; Pepovich, R.; Palova, N.; Marutsov, P.; Gospodinova, K.; Kundurzhiev, T.; Ciccozzi, M. High Seroprevalence of Hepatitis e Virus Infection among East Balkan Swine (Sus Scrofa) in Bulgaria: Preliminary Results. Pathogens 2020, 9, 911. [Google Scholar] [CrossRef] [PubMed]
- de Deus, N.; Peralta, B.; Pina, S.; Allepuz, A.; Mateu, E.; Vidal, D.; Ruiz-Fons, F.; Martín, M.; Gortázar, C.; Segalés, J. Epidemiological Study of Hepatitis E Virus Infection in European Wild Boars (Sus Scrofa) in Spain. Vet. Microbiol. 2008, 129, 163–170. [Google Scholar] [CrossRef]
- Rutjes, S.A.; Lodder-Verschoor, F.; Lodder, W.J.; van der Giessen, J.; Reesink, H.; Bouwknegt, M.; de Roda Husman, A.M. Seroprevalence and Molecular Detection of Hepatitis E Virus in Wild Boar and Red Deer in The Netherlands. J. Virol. Methods 2010, 168, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Zhang, J.; Huang, G.; Pang, S.; Zhou, K.; Xia, N. The immuno-protect study of a hepatitis E virus ORF2 peptide expressed in E. coli. Wei Sheng Wu Xue Bao 2003, 43, 35–42. [Google Scholar]
- Wei, M.; Zhang, X.; Yu, H.; Tang, Z.-M.; Wang, K.; Li, Z.; Zheng, Z.; Li, S.; Zhang, J.; Xia, N.; et al. Bacteria Expressed Hepatitis E Virus Capsid Proteins Maintain Virion-like Epitopes. Vaccine 2014, 32, 2859–2865. [Google Scholar] [CrossRef]
- Gupta, J.; Kaul, S.; Srivastava, A.; Kaushik, N.; Ghosh, S.; Sharma, C.; Batra, G.; Banerjee, M.; Shalimar; Nayak, B.; et al. Expression, Purification and Characterization of the Hepatitis E Virus Like-Particles in the Pichia Pastoris. Front. Microbiol. 2020, 11, 141. [Google Scholar] [CrossRef]
- Simanavicius, M.; Tamosiunas, P.L.; Petraityte-Burneikiene, R.; Johne, R.; Ulrich, R.G.; Zvirbliene, A.; Kucinskaite-Kodze, I. Generation in Yeast and Antigenic Characterization of Hepatitis E Virus Capsid Protein Virus-like Particles. Appl. Microbiol. Biotechnol. 2018, 102, 185–198. [Google Scholar] [CrossRef]
- Zhang, Y.; McAtee, P.; Yarbough, P.O.; Tam, A.W.; Fuerst, T. Expression, Characterization, and Immunoreactivities of a Soluble Hepatitis E Virus Putative Capsid Protein Species Expressed in Insect Cells. Clin. Diagn. Lab. Immunol. 1997, 4, 423–428. [Google Scholar] [CrossRef]
- Jameel, S.; Zafrullah, M.; Ozdener, M.H.; Panda, S.K. Expression in Animal Cells and Characterization of the Hepatitis E Virus Structural Proteins. J. Virol. 1996, 70, 207–216. [Google Scholar] [CrossRef]
- Maloney, B.J.; Takeda, N.; Suzaki, Y.; Ami, Y.; Li, T.C.; Miyamura, T.; Arntzen, C.J.; Mason, H.S. Challenges in Creating a Vaccine to Prevent Hepatitis, E. Vaccine 2005, 23, 1870–1874. [Google Scholar] [CrossRef]
- Zahmanova, G.G.; Mazalovska, M.; Takova, K.H.; Toneva, V.T.; Minkov, I.N.; Mardanova, E.S.; Ravin, N.V.; Lomonossoff, G.P. Rapid High-Yield Transient Expression of Swine Hepatitis E ORF2 Capsid Proteins in Nicotiana Benthamiana Plants and Production of Chimeric Hepatitis E Virus-Like Particles Bearing the M2e Influenza Epitope. Plants 2020, 9, 29. [Google Scholar] [CrossRef]
- Lomonossoff, G.P.; D’Aoust, M.-A. Plant-Produced Biopharmaceuticals: A Case of Technical Developments Driving Clinical Deployment. Science 2016, 353, 1237–1240. [Google Scholar] [CrossRef]
- Tschofen, M.; Knopp, D.; Hood, E.; Stöger, E. Plant Molecular Farming: Much More than Medicines. Annu. Rev. Anal. Chem. 2016, 9, 271–294. [Google Scholar] [CrossRef] [PubMed]
- Giritch, A.; Marillonnet, S.; Engler, C.; van Eldik, G.; Botterman, J.; Klimyuk, V.; Gleba, Y. Rapid High-Yield Expression of Full-Size IgG Antibodies in Plants Coinfected with Noncompeting Viral Vectors. Proc. Natl. Acad. Sci. USA 2006, 103, 14701–14706. [Google Scholar] [CrossRef] [PubMed]
- Mardanova, E.S.; Blokhina, E.A.; Tsybalova, L.M.; Peyret, H.; Lomonossoff, G.P.; Ravin, N.V. Efficient Transient Expression of Recombinant Proteins in Plants by the Novel PEff Vector Based on the Genome of Potato Virus X. Front. Plant. Sci. 2017, 8, 247. [Google Scholar] [CrossRef] [PubMed]
- Sainsbury, F.; Lomonossoff, G.P. Extremely High-Level and Rapid Transient Protein Production in Plants without the Use of Viral Replication. Plant. Physiol. 2008, 148, 1212–1218. [Google Scholar] [CrossRef]
- Sainsbury, F.; Liu, L.; Lomonossoff, G.P. Cowpea Mosaic Virus-Based Systems for the Expression of Antigens and Antibodies in Plants. Methods Mol. Biol. 2009, 483, 25–39. [Google Scholar] [CrossRef]
- Siew, Q.Y.; Pang, E.L.; Loh, H.-S.; Tan, M.T.T. Highly Sensitive and Specific Graphene/TiO2 Impedimetric Immunosensor Based on Plant-Derived Tetravalent Envelope Glycoprotein Domain III (EDIII) Probe Antigen for Dengue Diagnosis. Biosens. Bioelectron. 2021, 176, 112895. [Google Scholar] [CrossRef]
- Marques, L.É.C.; Silva, B.B.; Dutra, R.F.; Florean, E.O.P.T.; Menassa, R.; Guedes, M.I.F. Transient Expression of Dengue Virus NS1 Antigen in Nicotiana Benthamiana for Use as a Diagnostic Antigen. Front. Plant. Sci. 2020, 10, 1674. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Lai, H.; Brock, C.; Chen, Q. A Novel System for Rapid and Cost-Effective Production of Detection and Diagnostic Reagents of West Nile Virus in Plants. J. Biomed. Biotechnol. 2011, 2012, e106783. [Google Scholar] [CrossRef] [PubMed]
- Mbewana, S.; Meyers, A.E.; Weber, B.; Mareledwane, V.; Ferreira, M.L.; Majiwa, P.A.O.; Rybicki, E.P. Expression of Rift Valley Fever Virus N-Protein in Nicotiana Benthamiana for Use as a Diagnostic Antigen. BMC Biotechnol. 2018, 18, 77. [Google Scholar] [CrossRef]
- Mardanova, E.S.; Takova, K.H.; Toneva, V.T.; Zahmanova, G.G.; Tsybalova, L.M.; Ravin, N.V. A Plant-Based Transient Expression System for the Rapid Production of Highly Immunogenic Hepatitis E Virus-like Particles. Biotechnol. Lett. 2020, 42, 2441–2446. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.J.; Steele, J.F.C.; Hesketh, E.L.; Walden, M.; Thompson, R.F.; Lomonossoff, G.P.; Ranson, N.A. Combining Transient Expression and Cryo-EM to Obtain High-Resolution Structures of Luteovirid Particles. Structure 2019, 27, 1761–1770.e3. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Shin, M.K.; Cha, S.B.; Yoo, H.S. Development of a Novel Enzyme-Linked Immunosorbent Assay to Detect Anti-IgG against Swine Hepatitis E Virus. J. Vet. Sci. 2013, 14, 467–472. [Google Scholar] [CrossRef]
- Dichtl, K.; Zimmermann, J.; Koeppel, M.B.; Böhm, S.; Osterman, A. Evaluation of a Novel CLIA Monotest Assay for the Detection of Anti-Hepatitis E Virus-IgG and IgM: A Retrospective Comparison with a Line Blot and an ELISA. Pathogens 2021, 10, 689. [Google Scholar] [CrossRef]
- Peyret, H.; Steele, J.F.C.; Jung, J.-W.; Thuenemann, E.C.; Meshcheriakova, Y.; Lomonossoff, G.P. Producing Vaccines against Enveloped Viruses in Plants: Making the Impossible, Difficult. Vaccines 2021, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Takeyama, N.; Kiyono, H.; Yuki, Y. Plant-Based Vaccines for Animals and Humans: Recent Advances in Technology and Clinical Trials. Ther. Adv. Vaccines 2015, 3, 139–154. [Google Scholar] [CrossRef]
- Kumar, M.; Kumari, N.; Thakur, N.; Bhatia, S.K.; Saratale, G.D.; Ghodake, G.; Mistry, B.M.; Alavilli, H.; Kishor, D.S.; Du, X.; et al. A Comprehensive Overview on the Production of Vaccines in Plant-Based Expression Systems and the Scope of Plant Biotechnology to Combat against SARS-CoV-2 Virus Pandemics. Plants 2021, 10, 1213. [Google Scholar] [CrossRef] [PubMed]
- Shanmugaraj, B.; I. Bulaon, C.J.; Phoolcharoen, W. Plant Molecular Farming: A Viable Platform for Recombinant Biopharmaceutical Production. Plants 2020, 9, 842. [Google Scholar] [CrossRef] [PubMed]
- Myint, K.S.A.; Endy, T.P.; Gibbons, R.V.; Laras, K.; Mammen, M.P.; Sedyaningsih, E.R.; Seriwatana, J.; Glass, J.S.; Narupiti, S.; Corwin, A.L. Evaluation of Diagnostic Assays for Hepatitis E Virus in Outbreak Settings. J. Clin. Microbiol. 2006, 44, 1581–1583. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.C.; Flower, R.L.P.; Seed, C.R.; Keller, A.J.; Harley, R.; Chan, H.-T.; Hoad, V.; Warrilow, D.; Northill, J.; Holmberg, J.A.; et al. Hepatitis E Virus RNA in Australian Blood Donations. Transfusion 2016, 56, 3086–3093. [Google Scholar] [CrossRef] [PubMed]
- Engle, R.E.; Yu, C.; Emerson, S.U.; Meng, X.-J.; Purcell, R.H. Hepatitis E Virus (HEV) Capsid Antigens Derived from Viruses of Human and Swine Origin Are Equally Efficient for Detecting Anti-HEV by Enzyme Immunoassay. J. Clin. Microbiol. 2002, 40, 4576–4580. [Google Scholar] [CrossRef]
- Schnegg, A.; Bürgisser, P.; André, C.; Kenfak-Foguena, A.; Canellini, G.; Moradpour, D.; Abravanel, F.; Izopet, J.; Cavassini, M.; Darling, K.E.A. An Analysis of the Benefit of Using HEV Genotype 3 Antigens in Detecting Anti-HEV IgG in a European Population. PLoS ONE 2013, 8, e62980. [Google Scholar] [CrossRef]
- Tang, X.; Yang, C.; Gu, Y.; Song, C.; Zhang, X.; Wang, Y.; Zhang, J.; Hew, C.L.; Li, S.; Xia, N.; et al. Structural Basis for the Neutralization and Genotype Specificity of Hepatitis E Virus. Proc. Natl. Acad. Sci. USA 2011, 108, 10266–10271. [Google Scholar] [CrossRef]
- Li, S.; Tang, X.; Seetharaman, J.; Yang, C.; Gu, Y.; Zhang, J.; Du, H.; Shih, J.W.K.; Hew, C.-L.; Sivaraman, J.; et al. Dimerization of Hepatitis E Virus Capsid Protein E2s Domain Is Essential for Virus–Host Interaction. PLoS Pathog. 2009, 5, e1000537. [Google Scholar] [CrossRef]
- Folgado, A.; Abranches, R. Plant Aspartic Proteases for Industrial Applications: Thistle Get Better. Plants 2020, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Ding, X.; Li, Z.; Wang, S. Co-Expression with Replicating Vector Overcoming Competitive Effects Derived by a Companion Protease Inhibitor in Plants. Front. Plant. Sci. 2021, 12, 1168. [Google Scholar] [CrossRef]
- Castilho, A.; Schwestka, J.; Kienzl, N.F.; Vavra, U.; Grünwald-Gruber, C.; Izadi, S.; Hiremath, C.; Niederhöfer, J.; Laurent, E.; Monteil, V.; et al. Generation of Enzymatically Competent SARS-CoV-2 Decoy Receptor ACE2-Fc in Glycoengineered Nicotiana Benthamiana. Biotechnol. J. 2021, 16, 2000566. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, A.A.P.; Maresch, D.; Grill, A.; Bakalarz, J.; Acosta, J.A.T.; Castilho, A.; Steinkellner, H.; Mach, L. Identification of Two Subtilisin-like Serine Proteases Engaged in the Degradation of Recombinant Proteins in Nicotiana Benthamiana. FEBS Lett. 2021, 595, 379–388. [Google Scholar] [CrossRef]
- Sainsbury, F.; Varennes-Jutras, P.; Goulet, M.-C.; D’Aoust, M.-A.; Michaud, D. Tomato Cystatin SlCYS8 as a Stabilizing Fusion Partner for Human Serpin Expression in Plants. Plant. Biotechnol. J. 2013, 11, 1058–1068. [Google Scholar] [CrossRef] [PubMed]


| IN-HOUSE EIA | ||||
|---|---|---|---|---|
| Commercial ELISA | Positive | Negative | Total | |
| Positive | 67 | 2 | 69 | |
| Negative | 1 | 71 | 72 | |
| Total | 68 | 73 | 141 | |
| Kappa index Sensitivity Specificity | 0.9399 97.1% (95% Cl: 89.9–99.65) 98.6% (95% Cl: 92.5-99.96) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takova, K.; Koynarski, T.; Minkov, G.; Toneva, V.; Mardanova, E.; Ravin, N.; Lukov, G.L.; Zahmanova, G. Development and Optimization of an Enzyme Immunoassay to Detect Serum Antibodies against the Hepatitis E Virus in Pigs, Using Plant-Derived ORF2 Recombinant Protein. Vaccines 2021, 9, 991. https://doi.org/10.3390/vaccines9090991
Takova K, Koynarski T, Minkov G, Toneva V, Mardanova E, Ravin N, Lukov GL, Zahmanova G. Development and Optimization of an Enzyme Immunoassay to Detect Serum Antibodies against the Hepatitis E Virus in Pigs, Using Plant-Derived ORF2 Recombinant Protein. Vaccines. 2021; 9(9):991. https://doi.org/10.3390/vaccines9090991
Chicago/Turabian StyleTakova, Katerina, Tsvetoslav Koynarski, George Minkov, Valentina Toneva, Eugenia Mardanova, Nikolai Ravin, Georgi L. Lukov, and Gergana Zahmanova. 2021. "Development and Optimization of an Enzyme Immunoassay to Detect Serum Antibodies against the Hepatitis E Virus in Pigs, Using Plant-Derived ORF2 Recombinant Protein" Vaccines 9, no. 9: 991. https://doi.org/10.3390/vaccines9090991
APA StyleTakova, K., Koynarski, T., Minkov, G., Toneva, V., Mardanova, E., Ravin, N., Lukov, G. L., & Zahmanova, G. (2021). Development and Optimization of an Enzyme Immunoassay to Detect Serum Antibodies against the Hepatitis E Virus in Pigs, Using Plant-Derived ORF2 Recombinant Protein. Vaccines, 9(9), 991. https://doi.org/10.3390/vaccines9090991