Engineering Approaches in Plant Molecular Farming for Global Health
Abstract
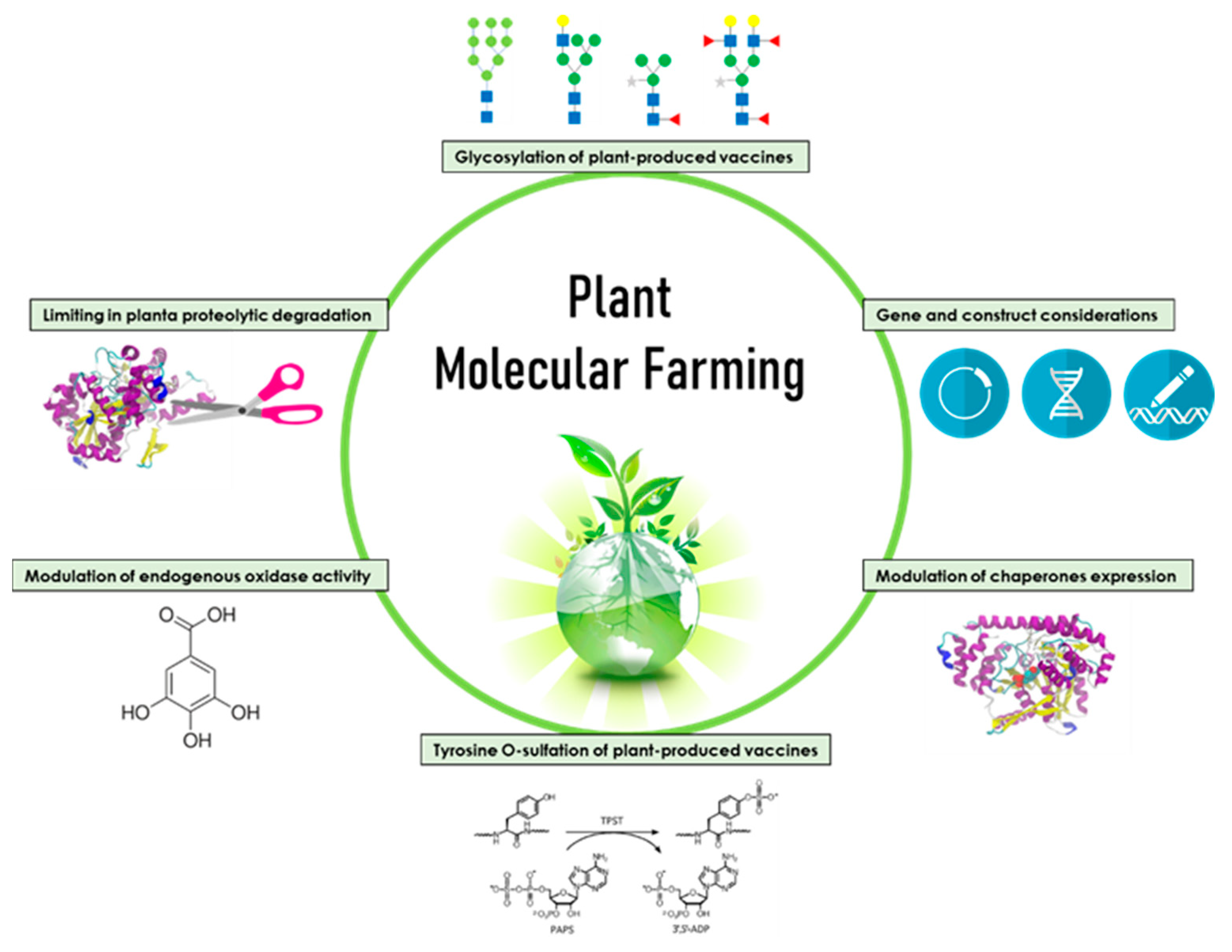
1. Introduction
2. Improving the Yields of Recombinant Protein Vaccines
2.1. Gene and Construct Considerations
2.2. Modulation of Chaperone Expression
2.3. Modulation of Endogenous Oxidase Activity
2.4. Limiting in Planta Proteolytic Degradation
3. Posttranslational Modifications of Plant-Produced Vaccines
3.1. Glycosylation of Plant-Produced Vaccines
3.2. Tyrosine O-Sulfation of Plant-Produced Vaccines
4. Conclusions and Future Impact of Engineering Strategies in Plant Molecular Farming
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
Abbreviations
References
- Pollard, A.J.; Bijker, E.M. A guide to vaccinology: From basic principles to new developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Swartz, J.R. Advances in Escherichia coli production of therapeutic proteins. Curr. Opin. Biotechnol. 2001, 12, 195–201. [Google Scholar] [CrossRef]
- Chu, L.; Robinson, D.K. Industrial choices for protein production by large-scale cell culture. Curr. Opin. Biotechnol. 2001, 12, 180–187. [Google Scholar] [CrossRef]
- Houdebine, L.M. Transgenic animal bioreactors. Transgenic Res. 2000, 9, 305–320. [Google Scholar] [CrossRef]
- Barta, A.; Sommergruber, K.; Thompson, D.; Hartmuth, K.; Matzke, M.A.; Matzke, A.J. The expression of a nopaline synthase—human growth hormone chimaeric gene in transformed tobacco and sunflower callus tissue. Plant Mol. Biol. 1986, 6, 347–357. [Google Scholar] [CrossRef]
- Hiatt, A.; Caffferkey, R.; Bowdish, K. Production of antibodies in transgenic plants. Nature 1989, 342, 76–78. [Google Scholar] [CrossRef]
- De Martinis, D.; Rybicki, E.P.; Fujiyama, K.; Franconi, R.; Benvenuto, E. Plant molecular farming: Fast, scalable, cheap, sustainable. Front. Plant Sci. 2016, 7, 1148. [Google Scholar] [CrossRef] [PubMed]
- Nandi, S.; Kwong, A.T.; Holtz, B.R.; Erwin, R.L.; Marcel, S.; McDonald, K.A. Techno-Economic Analysis of a Transient Plant-Based Platform for Monoclonal antibodyProduction. mAbs 2016, 8, 1456–1466. [Google Scholar] [CrossRef]
- Pillet, S.; Aubin, É.; Trépanier, S.; Bussière, D.; Dargis, M.; Poulin, J.-F.; Yassine-Diab, B.; Ward, B.J.; Landry, N. A plant-derived quadrivalent virus like particle influenza vaccine induces cross-reactive antibody and T cell response in healthy adults. Clin. Immunol. 2016, 168, 72–87. [Google Scholar] [CrossRef]
- De Muynck, B.; Navarre, C.; Boutry, M. Production of antibodies in plants: Status after twenty years. Plant Biotechnol. J. 2010, 8, 529–563. [Google Scholar] [CrossRef]
- Fischer, R.; Emans, N. Molecular farming of pharmaceutical proteins. Transgenic Res. 2000, 9, 279–299. [Google Scholar] [CrossRef] [PubMed]
- Giddings, G. Transgenic plants as protein factories. Curr. Opin. Biotechnol. 2001, 12, 450–454. [Google Scholar] [CrossRef]
- Spiegel, H.; Stöger, E.; Twyman, R.M.; Buyel, J.F. Current status and perspectives of the molecular farming landscape. In Molecular Pharming: Applications, Challenges and Emerging Areas; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 3–23. [Google Scholar]
- Nölke, G.; Fischer, R.; Schillberg, S. Production of therapeutic antibodies in plants. Expert Opin. Biol. Ther. 2003, 3, 1153–1162. [Google Scholar] [CrossRef]
- Twyman, R.M.; Schillberg, S.; Fischer, R. Transgenic plants in the biopharmaceutical market. Expert Opin. Emerg. Drugs 2005, 10, 185–218. [Google Scholar] [CrossRef]
- Schillberg, S.; Raven, N.; Fischer, R.; Twyman, R.M.; Schiermeyer, A. Molecular farming of pharmaceutical proteins using plant suspension cell and tissue cultures. Curr. Pharm. Des. 2013, 19, 5531–5542. [Google Scholar] [CrossRef] [PubMed]
- Tsekoa, T.L.; Singh, A.A.; Buthelezi, S.G. Molecular farming for therapies and vaccines in Africa. Curr. Opin. Biotechnol. 2020, 61, 89–95. [Google Scholar] [CrossRef]
- Schillberg, S.; Finnern, R. Plant molecular farming for the production of valuable proteins–Critical evaluation of achievements and future challenges. J. Plant Physiol. 2021, 258, 153359. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.; Das, S.S.; Rakshit, R. Codon usage pattern and predicted gene expression in Arabidopsis thaliana. Gene X 2019, 2, 100012. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Gautier, C. Codon usage in bacteria: Correlation with gene expressivity. Nucleic Acids Res. 1982, 10, 7055–7074. [Google Scholar] [CrossRef]
- Webster, G.R.; Teh, A.Y.H.; Ma, J.K.C. Synthetic gene design—The rationale for codon optimization and implications for molecular pharming in plants. Biotechnol. Bioeng. 2017, 114, 492–502. [Google Scholar] [CrossRef]
- Mahalik, S.; Sharma, A.K.; Mukherjee, K.J. Genome engineering for improved recombinant protein expression in Escherichia coli. Microb. Cell Factories 2014, 13, 177. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beyer, A.; Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–353. [Google Scholar] [CrossRef]
- Buyel, J.; Stöger, E.; Bortesi, L. Targeted genome editing of plants and plant cells for biomanufacturing. Transgenic Res. 2021, 30, 101–426. [Google Scholar] [CrossRef]
- Salazar-González, J.A.; Bañuelos-Hernández, B.; Rosales-Mendoza, S. Current status of viral expression systems in plants and perspectives for oral vaccines development. Plant Mol. Biol. 2015, 87, 203–217. [Google Scholar] [CrossRef]
- Giritch, A.; Marillonnet, S.; Engler, C.; van Eldik, G.; Botterman, J.; Klimyuk, V.; Gleba, Y. Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc. Natl. Acad. Sci. USA 2006, 103, 14701–14706. [Google Scholar] [CrossRef]
- Peyret, H.; Brown, J.K.; Lomonossoff, G.P. Improving plant transient expression through the rational design of synthetic 5′ and 3′ untranslated regions. Plant Methods 2019, 15, 108. [Google Scholar] [CrossRef] [PubMed]
- Sainsbury, F.; Thuenemann, E.C.; Lomonossoff, G.P. pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 2009, 7, 682–693. [Google Scholar] [CrossRef]
- Peyret, H.; Lomonossoff, G.P. The pEAQ vector series: The easy and quick way to produce recombinant proteins in plants. Plant Mol. Biol. 2013, 83, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Odell, J.T.; Nagy, F.; Chua, N.-H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985, 313, 810–812. [Google Scholar] [CrossRef]
- Lawton, M.A.; Tierney, M.A.; Nakamura, I.; Anderson, E.; Komeda, Y.; Dubé, P.; Hoffman, N.; Fraley, R.T.; Beachy, R.N. Expression of a soybean β-conclycinin gene under the control of the cauliflower mosaic virus 35S and 19S promoters in transformed petunia tissues. Plant Mol. Biol. 1987, 9, 315–324. [Google Scholar] [CrossRef]
- Kay, R.; Chan, A.; Daly, M.; McPherson, J. Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 1987, 236, 1299–1302. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.H.; Quail, P.H. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996, 5, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Voinnet, O. The diversity of RNA silencing pathways in plants. TRENDS Genet. 2006, 22, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Arzola, L.; Chen, J.; Rattanaporn, K.; Maclean, J.M.; McDonald, K.A. Transient co-expression of post-transcriptional gene silencing suppressors for increased in planta expression of a recombinant anthrax receptor fusion protein. Int. J. Mol. Sci. 2011, 12, 4975–4990. [Google Scholar] [CrossRef] [PubMed]
- Garabagi, F.; Gilbert, E.; Loos, A.; McLean, M.D.; Hall, J.C. Utility of the P 19 suppressor of gene-silencing protein for production of therapeutic antibodies in N icotiana expression hosts. Plant Biotechnol. J. 2012, 10, 1118–1128. [Google Scholar] [CrossRef]
- Strasser, R. Protein quality control in the endoplasmic reticulum of plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef]
- Wakasa, Y.; Yasuda, H.; Oono, Y.; Kawakatsu, T.; Hirose, S.; Takahashi, H.; Hayashi, S.; Yang, L.; Takaiwa, F. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J. 2011, 65, 675–689. [Google Scholar] [CrossRef]
- Margolin, E.; Oh, Y.J.; Verbeek, M.; Naude, J.; Ponndorf, D.; Meshcheriakova, Y.A.; Peyret, H.; van Diepen, M.T.; Chapman, R.; Meyers, A.E. Co-expression of human calreticulin significantly improves the production of HIV gp140 and other viral glycoproteins in plants. Plant Biotechnol. J. 2020, 18, 2109–2117. [Google Scholar] [CrossRef]
- Twyman, R.M.; Stoger, E.; Schillberg, S.; Christou, P.; Fischer, R. Molecular farming in plants: Host systems and expression technology. Trends Biotechnol. 2003, 21, 570–578. [Google Scholar] [CrossRef]
- Buyel, J.; Twyman, R.; Fischer, R. Extraction and downstream processing of plant-derived recombinant proteins. Biotechnol. Adv. 2015, 33, 902–913. [Google Scholar] [CrossRef]
- Nadakuduti, S.S.; Buell, C.R.; Voytas, D.F.; Starker, C.G.; Douches, D.S. Genome editing for crop improvement—Applications in clonally propagated polyploids with a focus on potato (Solanum tuberosum L.). Front. Plant Sci. 2018, 9, 1607. [Google Scholar] [CrossRef] [PubMed]
- González, M.N.; Massa, G.A.; Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Storani, L.; Décima Oneto, C.A.; Hofvander, P.; Feingold, S.E. Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant Sci. 2020, 10, 1649. [Google Scholar] [CrossRef]
- van der Hoorn, R.A.L.; Jones, J.D.G. The plant proteolytic machinery and its role in defence. Curr. Opin. Plant Biol. 2004, 7, 400–407. [Google Scholar] [CrossRef] [PubMed]
- van der Hoorn, R.A.L. Plant Proteases: From Phenotypes to Molecular Mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223. [Google Scholar] [CrossRef]
- Grosse-Holz, F.; Kelly, S.; Blaskowski, S.; Kaschani, F.; Kaiser, M.; van der Hoorn, R.A. The transcriptome, extracellular proteome and active secretome of agroinfiltrated Nicotiana benthamiana uncover a large, diverse protease repertoire. Plant Biotechnol. J. 2018, 16, 1068–1084. [Google Scholar] [CrossRef]
- Schaller, A. A cut above the rest: The regulatory function of plant proteases. Planta 2004, 220, 183–197. [Google Scholar] [CrossRef]
- Zauner, F.B.; Dall, E.; Regl, C.; Grassi, L.; Huber, C.G.; Cabrele, C.; Brandstetter, H. Crystal structure of plant legumain reveals a unique two-chain state with pH-dependent activity regulation. Plant Cell 2018, 30, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Shabab, M.; Strasser, R.; Wolters, P.J.; Shindo, T.; Niemer, M.; Kaschani, F.; Mach, L.; van der Hoorn, R. Post-translational regulation and trafficking of the granulin-containing protease RD21 of Arabidopsis thaliana. PLoS ONE 2012, 7, e32422. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Doran, P.M. Foreign protein degradation and instability in plants and plant tissue cultures. Trends Biotechnol. 2006, 24, 426–432. [Google Scholar] [CrossRef]
- Benchabane, M.; Goulet, C.; Rivard, D.; Faye, L.; Gomord, V.; Michaud, D. Preventing unintended proteolysis in plant protein biofactories. Plant Biotechnol. J. 2008, 6, 633–648. [Google Scholar] [CrossRef]
- Rivard, D.; Anguenot, R.; Brunelle, F.; Le, V.Q.; Vézina, L.P.; Trépanier, S.; Michaud, D. An in-built proteinase inhibitor system for the protection of recombinant proteins recovered from transgenic plants. Plant Biotechnol. J. 2006, 4, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Hehle, V.K.; Lombardi, R.; Dolleweerd, C.J.; Paul, M.J.; Di Micco, P.; Morea, V.; Benvenuto, E.; Donini, M.; Ma, J.K.C. Site-specific proteolytic degradation of IgG monoclonal antibodies expressed in tobacco plants. Plant Biotechnol. J. 2015, 13, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.K.; Fischer, R.; Schillberg, S.; Schiermeyer, A. Inhibition of protease activity by antisense RNA improves recombinant protein production in Nicotiana tabacum cv. Bright Yellow 2 (BY-2) suspension cells. Biotechnol. J. 2014, 9, 1065–1073. [Google Scholar] [CrossRef]
- Niemer, M.; Mehofer, U.; Torres Acosta, J.A.; Verdianz, M.; Henkel, T.; Loos, A.; Strasser, R.; Maresch, D.; Rademacher, T.; Steinkellner, H. The human anti-HIV antibodies 2F5, 2G12, and PG9 differ in their susceptibility to proteolytic degradation: Down-regulation of endogenous serine and cysteine proteinase activities could improve antibody production in plant-based expression platforms. Biotechnol. J. 2014, 9, 493–500. [Google Scholar] [CrossRef]
- Mandal, M.K.; Ahvari, H.; Schillberg, S.; Schiermeyer, A. Tackling Unwanted Proteolysis in Plant Production Hosts Used for Molecular Farming. Front. Plant Sci. 2016, 7, 267. [Google Scholar] [CrossRef]
- Castilho, A.; Windwarder, M.; Gattinger, P.; Mach, L.; Strasser, R.; Altmann, F.; Steinkellner, H. Proteolytic and N-Glycan Processing of Human α1-Antitrypsin Expressed in Nicotiana benthamiana. Plant Physiol. 2014, 166, 1839–1851. [Google Scholar] [CrossRef] [PubMed]
- Faye, L.; Boulaflous, A.; Benchabane, M.; Gomord, V.; Michaud, D. Protein modifications in the plant secretory pathway: Current status and practical implications in molecular pharming. Vaccine 2005, 23, 1770–1778. [Google Scholar] [CrossRef]
- Goulet, C.; Goulet, C.; Goulet, M.-C.; Michaud, D. 2-DE proteome maps for the leaf apoplast of Nicotiana benthamiana. Proteomics 2010, 10, 2536–2544. [Google Scholar] [CrossRef]
- Zhou, Y.; Cox, A.M.; Kearney, C.M. Pathogenesis-related proteins induced by agroinoculation-associated cell wall weakening can be obviated by spray-on inoculation or mannitol ex vivo culture. Plant Biotechnol. Rep. 2017, 11, 161–169. [Google Scholar] [CrossRef]
- Pitzschke, A. Agrobacterium infection and plant defense—transformation success hangs by a thread. Front. Plant Sci. 2015, 4, 519. [Google Scholar] [CrossRef]
- Li, W.; Cao, J.Y.; Xu, Y.P.; Cai, X.Z. Artificial Agrobacterium tumefaciens strains exhibit diverse mechanisms to repress Xanthomonas oryzae pv. oryzae-induced hypersensitive response and non-host resistance in Nicotiana benthamiana. Mol. Plant Pathol. 2017, 18, 489–502. [Google Scholar] [CrossRef]
- Rico, A.; Bennett, M.H.; Forcat, S.; Huang, W.E.; Preston, G.M. Agroinfiltration reduces ABA levels and suppresses Pseudomonas syringae-elicited salicylic acid production in Nicotiana tabacum. PLoS ONE 2010, 5, e8977. [Google Scholar] [CrossRef]
- Robinette, D.; Matthysse, A. Inhibition by Agrobacterium tumefaciens and Pseudomonas savastanoi of development of the hypersensitive response elicited by Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 1990, 172, 5742–5749. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.H.; Raghuram, B.; Eschen-Lippold, L.; Scheel, D.; Lee, J.; Sinha, A.K. Agroinfiltration by Cytokinin-Producing Agrobacterium sp. Strain GV3101 Primes Defense Responses in Nicotiana tabacum. Mol. Plant-Microbe Interact. 2014, 27, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Hörtensteiner, S.; Feller, U. Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 2002, 53, 927–937. [Google Scholar] [CrossRef]
- Duwadi, K.; Chen, L.; Menassa, R.; Dhaubhadel, S. Identification, characterization and down-Regulation of cysteine Protease genes in tobacco for use in recombinant protein production. PLoS ONE 2015, 10, e0130556. [Google Scholar] [CrossRef]
- Paireder, M.; Tholen, S.; Porodko, A.; Biniossek, M.L.; Mayer, B.; Novinec, M.; Schilling, O.; Mach, L. The papain-like cysteine proteinases NbCysP6 and NbCysP7 are highly processive enzymes with substrate specificities complementary to Nicotiana benthamiana cathepsin B. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2017, 1865, 444–452. [Google Scholar] [CrossRef]
- Pillay, P.; Kibido, T.; Plessis, M.; Vyver, C.; Beyene, G.; Vorster, B.J.; Kunert, K.J.; Schlüter, U. Use of Transgenic Oryzacystatin-I-Expressing Plants Enhances Recombinant Protein Production. Appl. Biochem Biotechnol. 2012, 168, 1608–1620. [Google Scholar] [CrossRef]
- Girard, C.; Rivard, D.; Kiggundu, A.; Kunert, K.; Gleddie, S.C.; Cloutier, C.; Michaud, D. A multicomponent, elicitor-inducible cystatin complex in tomato, Solanum lycopersicum. New Phytol. 2007, 173, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Van der Vyver, C.; Schneidereit, J.; Driscoll, S.; Turner, J.; Kunert, K.; Foyer, C.H. Oryzacystatin I expression in transformed tobacco produces a conditional growth phenotype and enhances chilling tolerance. Plant Biotechnol. J. 2003, 1, 101–112. [Google Scholar] [CrossRef]
- Jutras, P.V.; D’Aoust, M.A.; Couture, M.M.J.; Vézina, L.P.; Goulet, M.C.; Michaud, D.; Sainsbury, F. Modulating secretory pathway pH by proton channel co-expression can increase recombinant protein stability in plants. Biotechnol. J. 2015, 10, 1478–1486. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Du, J.; Jiang, H. Post-Translation Modifications to Regulate Protein Function; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008; pp. 1–31. [Google Scholar]
- Lewis, G.K. Qualitative and quantitative variables that affect the potency of Fc-mediated effector function in vitro and in vivo: Considerations for passive immunization using non-neutralizing antibodies. Curr. HIV Res. 2013, 11, 354–364. [Google Scholar] [CrossRef]
- Baum, L.L.; Cassutt, K.J.; Knigge, K.; Khattri, R.; Margolick, J.; Rinaldo, C.; Kleeberger, C.A.; Nishanian, P.; Henrard, D.R.; Phair, J. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J. Immunol. 1996, 157, 2168–2173. [Google Scholar] [PubMed]
- Shields, R.L.; Lai, J.; Keck, R.; O’Connell, L.Y.; Hong, K.; Meng, Y.G.; Weikert, S.H.; Presta, L.G. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002, 277, 26733–26740. [Google Scholar] [CrossRef]
- Whaley, K.J.; Hiatt, A.; Zeitlin, L. Emerging antibody products and Nicotiana manufacturing. Hum. Vaccines 2011, 7, 349–356. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Strasser, R.; Stadlmann, J.; Schähs, M.; Stiegler, G.; Quendler, H.; Mach, L.; Glössl, J.; Weterings, K.; Pabst, M.; Steinkellner, H. Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol. J. 2008, 6, 392–402. [Google Scholar] [CrossRef]
- Strasser, R.; Castilho, A.; Stadlmann, J.; Kunert, R.; Quendler, H.; Gattinger, P.; Jez, J.; Rademacher, T.; Altmann, F.; Mach, L. Improved virus neutralization by plant-produced anti-HIV antibodies with a homogeneous β1, 4-galactosylated N-glycan profile. J. Biol. Chem. 2009, 284, 20479–20485. [Google Scholar] [CrossRef]
- Jansing, J.; Sack, M.; Augustine, S.M.; Fischer, R.; Bortesi, L. CRISPR/Cas9-mediated knockout of six glycosyltransferase genes in Nicotiana benthamiana for the production of recombinant proteins lacking β-1, 2-xylose and core α-1, 3-fucose. Plant Biotechnol. J. 2019, 17, 350–361. [Google Scholar] [CrossRef]
- Montero-Morales, L.; Steinkellner, H. Advanced Plant-Based Glycan Engineering. Front. Bioeng. Biotechnol. 2018. [Google Scholar] [CrossRef]
- Montero-Morales, L.; Maresch, D.; Crescioli, S.; Castilho, A.; Ilieva, K.M.; Mele, S.; Karagiannis, S.N.; Altmann, F.; Steinkellner, H. In planta glycan engineering and functional activities of IgE antibodies. Front. Bioeng. Biotechnol. 2019, 7, 242. [Google Scholar] [CrossRef] [PubMed]
- Göritzer, K.; Turupcu, A.; Maresch, D.; Novak, J.; Altmann, F.; Oostenbrink, C.; Obinger, C.; Strasser, R. Distinct Fcα receptor N-glycans modulate the binding affinity to immunoglobulin A (IgA) antibodies. J. Biol. Chem. 2019, 294, 13995–14008. [Google Scholar] [CrossRef]
- Kallolimath, S.; Castilho, A.; Strasser, R.; Grünwald-Gruber, C.; Altmann, F.; Strubl, S.; Galuska, C.E.; Zlatina, K.; Galuska, S.P.; Werner, S. Engineering of complex protein sialylation in plants. Proc. Natl. Acad. Sci. USA 2016, 113, 9498–9503. [Google Scholar] [CrossRef]
- Castilho, A.; Beihammer, G.; Pfeiffer, C.; Göritzer, K.; Montero-Morales, L.; Vavra, U.; Maresch, D.; Grünwald-Gruber, C.; Altmann, F.; Steinkellner, H. An oligosaccharyltransferase from Leishmania major increases the N-glycan occupancy on recombinant glycoproteins produced in Nicotiana benthamiana. Plant Biotechnol. J. 2018, 16, 1700–1709. [Google Scholar] [CrossRef]
- Singh, A.A.; Pooe, O.; Kwezi, L.; Lotter-Stark, T.; Stoychev, S.H.; Alexandra, K.; Gerber, I.; Bhiman, J.N.; Vorster, J.; Pauly, M. Plant-based production of highly potent anti-HiV antibodies with engineered posttranslational modifications. Sci. Rep. 2020, 10, 6201. [Google Scholar] [CrossRef] [PubMed]
- Bendandi, M.; Marillonnet, S.; Kandzia, R.; Thieme, F.; Nickstadt, A.; Herz, S.; Fröde, R.; Inoges, S.; de Cerio, A.L.-D.; Soria, E. Rapid, high-yield production in plants of individualized idiotype vaccines for non-Hodgkin’s lymphoma. Ann. Oncol. 2010, 21, 2420–2427. [Google Scholar] [CrossRef] [PubMed]
- Loos, A.; Gach, J.S.; Hackl, T.; Maresch, D.; Henkel, T.; Porodko, A.; Bui-Minh, D.; Sommeregger, W.; Wozniak-Knopp, G.; Forthal, D.N. Glycan modulation and sulfoengineering of anti–HIV-1 monoclonal antibody PG9 in plants. Proc. Natl. Acad. Sci. USA 2015, 112, 12675–12680. [Google Scholar] [CrossRef]
- Stelter, S.; Paul, M.; Teh, A.; Grandits, M.; Altmann, F.; Vanier, J.; Bardor, M.; Castilho, A.; Allen, R.; Ma, J. Engineering the interactions between a plant-produced HIV antibody and human Fc receptors. Plant Biotechnol. J. 2020, 18, 402–414. [Google Scholar] [CrossRef]
- Strasser, R. Plant protein glycosylation. Glycobiology 2016, 26, 926–939. [Google Scholar] [CrossRef]
- Jeong, I.S.; Lee, S.; Bonkhofer, F.; Tolley, J.; Fukudome, A.; Nagashima, Y.; May, K.; Rips, S.; Lee, S.Y.; Gallois, P. Purification and characterization of Arabidopsis thaliana oligosaccharyltransferase complexes from the native host: A protein super-expression system for structural studies. Plant J. 2018, 94, 131–145. [Google Scholar] [CrossRef]
- Wohlschlager, T.; Scheffler, K.; Forstenlehner, I.C.; Skala, W.; Senn, S.; Damoc, E.; Holzmann, J.; Huber, C.G. Native mass spectrometry combined with enzymatic dissection unravels glycoform heterogeneity of biopharmaceuticals. Nat. Commun. 2018, 9, 1713. [Google Scholar] [CrossRef]
- Bagdonaite, I.; Wandall, H.H. Global aspects of viral glycosylation. Glycobiology 2018, 28, 443–467. [Google Scholar] [CrossRef]
- Castilho, A.; Steinkellner, H. Glyco-engineering in plants to produce human-like N-glycan structures. Biotechnol. J. 2012, 7, 1088–1098. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.J.; Chuang, S.; Hou, X.; Shoham, M.; Zhu, J.Z. Tyrosine sulfation: An increasingly recognised post-translational modification of secreted proteins. New Biotechnol. 2009, 25, 299–317. [Google Scholar] [CrossRef]
- Doria-Rose, N.A.; Schramm, C.A.; Gorman, J.; Moore, P.L.; Bhiman, J.N.; DeKosky, B.J.; Ernandes, M.J.; Georgiev, I.S.; Kim, H.J.; Pancera, M. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 2014, 509, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Choe, H.; Li, W.; Wright, P.L.; Vasilieva, N.; Venturi, M.; Huang, C.-C.; Grundner, C.; Dorfman, T.; Zwick, M.B.; Wang, L. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 2003, 114, 161–170. [Google Scholar] [CrossRef]
- Komori, R.; Amano, Y.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 15067–15072. [Google Scholar] [CrossRef]
- Moore, K.L. Protein tyrosine sulfation: A critical posttranslation modification in plants and animals. Proc. Natl. Acad. Sci. USA 2009, 106, 14741–14742. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, Y.; Sack, M.; Montefiori, D.; Labranche, C.; Lewis, M.; Urban, L. Pharmacokinetics and Immunogenicity of Broadly Neutralizing HIV Monoclonal Antibodies in Macaques. PLoS ONE 2015, 10, e0120451. [Google Scholar] [CrossRef]
- Bombarely, A.; Rosli, H.G.; Vrebalov, J.; Moffett, P.; Mueller, L.A.; Martin, G.B. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 2012, 25, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.A.; Pillay, P.; Tsekoa, T.L. Engineering Approaches in Plant Molecular Farming for Global Health. Vaccines 2021, 9, 1270. https://doi.org/10.3390/vaccines9111270
Singh AA, Pillay P, Tsekoa TL. Engineering Approaches in Plant Molecular Farming for Global Health. Vaccines. 2021; 9(11):1270. https://doi.org/10.3390/vaccines9111270
Chicago/Turabian StyleSingh, Advaita Acarya, Priyen Pillay, and Tsepo Lebiletsa Tsekoa. 2021. "Engineering Approaches in Plant Molecular Farming for Global Health" Vaccines 9, no. 11: 1270. https://doi.org/10.3390/vaccines9111270
APA StyleSingh, A. A., Pillay, P., & Tsekoa, T. L. (2021). Engineering Approaches in Plant Molecular Farming for Global Health. Vaccines, 9(11), 1270. https://doi.org/10.3390/vaccines9111270





