Strategies for Immunomonitoring after Vaccination and during Infection
Abstract
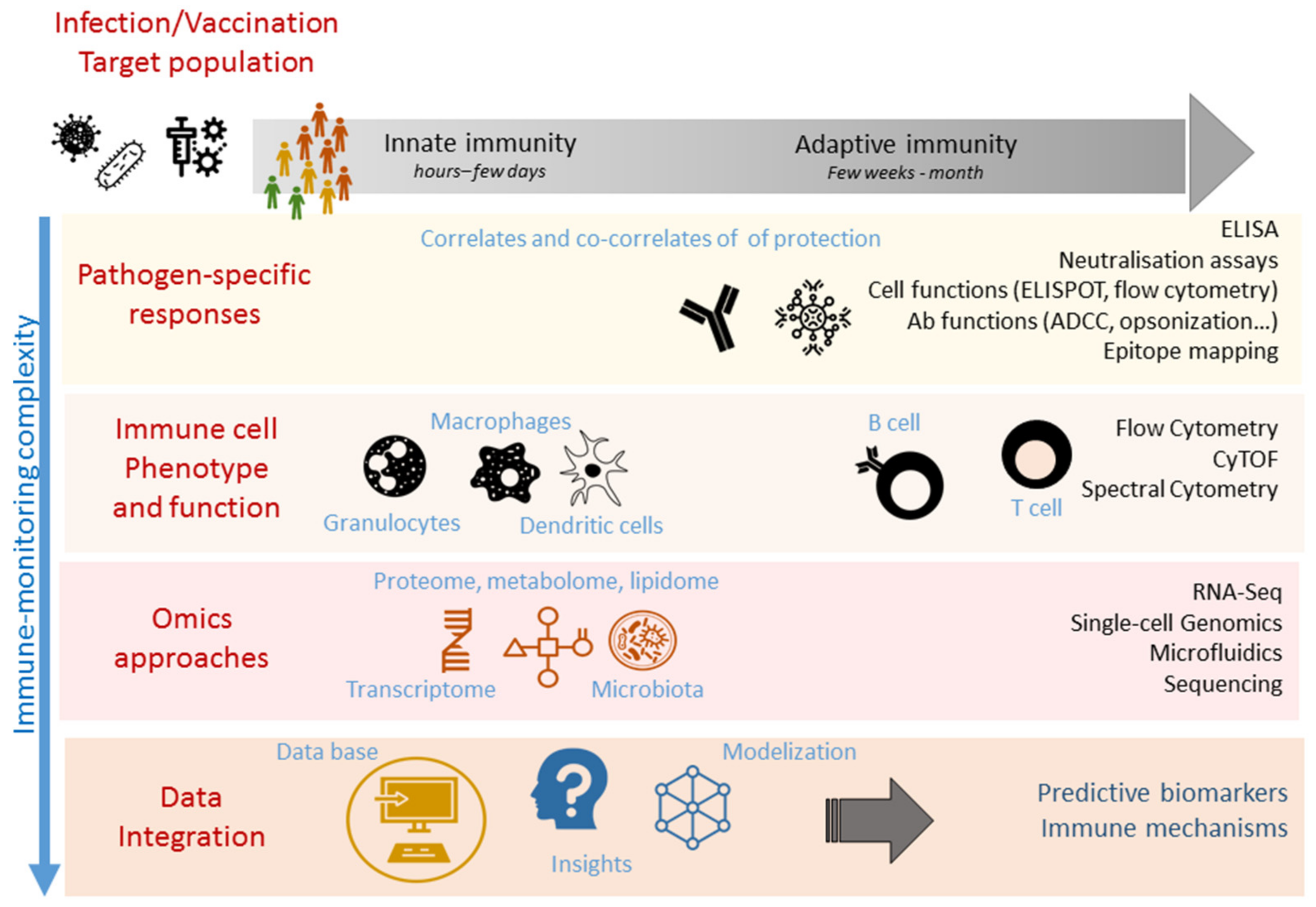
1. Introduction
2. Identification and Measurement of Correlates and Surrogates of Protection
3. Immunomonitoring of Correlates and Surrogates of Protection
3.1. Pathogen-Specific Easy-to-Use Assays
3.2. Immune Cell Phenotypic and Functional Analysis
4. High-Throughput Immunomonitoring Techniques
5. Systems Biology
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ELISA | enzyme-linked immunosorbent assay |
| ADCC | antibody-dependent cell cytotoxicity |
| CyTOF | cytometry by time-of-flight |
| t-SNE | t-distributed stochastic neighbor embedding |
| UMAP | Uniform Manifold Approximation and Projection |
| PCA | Principal component analysis |
| MDS | multidimensional scaling |
| LDA | Linear Discriminant analysis |
| LASSO | Least Absolute Shrinkage and Selection Operator |
References
- Bloom, D.E.; Cadarette, D. Infectious disease threats in the twenty-first century: Strengthening the global response. Front. Immunol. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rappuoli, R.; Santoni, A.; Mantovani, A. Vaccines: An achievement of civilization, a human right, our health insurance for the future. J. Exp. Med. 2019, 216, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Smith, K. A Edward jenner and the small pox vaccine. Front. Immunol. 2011, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.B.; Wu, J.J.; Tyring, S.K. A review of licensed viral vaccines, some of their safety concerns, and the advances in the development of investigational viral vaccines. J. Infect. 2004, 49, 179–209. [Google Scholar] [CrossRef]
- Gonçalves, E.; Bonduelle, O.; Soria, A.; Loulergue, P.; Rousseau, A.; Cachanado, M.; Bonnabau, H.; Thiebaut, R.; Tchitchek, N.; Behillil, S.; et al. Innate gene signature distinguishes humoral versus cytotoxic responses to influenza vaccination. J. Clin. Investig. 2019, 129, 1960–1971. [Google Scholar] [CrossRef]
- Rappuoli, R. MEDICINE: The Intangible Value of Vaccination. Science 2002, 297, 937–939. [Google Scholar] [CrossRef]
- Plotkin, S.A. Complex correlates of protection after vaccination. Clin. Infect. Dis. 2013, 56, 1458–1465. [Google Scholar] [CrossRef]
- Plotkin, S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010, 17, 1055–1065. [Google Scholar] [CrossRef]
- Rappuoli, R.; Aderem, A. A 2020 vision for vaccines against HIV, tuberculosis and malaria. Nature 2011, 473, 463–469. [Google Scholar] [CrossRef]
- Davis, M.M.; Tato, C.M.; Furman, D. Systems immunology: Just getting started. Nat. Immunol. 2017, 18, 725–732. [Google Scholar] [CrossRef]
- Sable, S.B.; Posey, J.E.; Scriba, T.J. Tuberculosis vaccine development: Progress in clinical evaluation. Clin. Microbiol. Rev. 2020, 33, 1–30. [Google Scholar] [CrossRef]
- Correlates of Vaccine-Induced Protection: Methods and Implications Immunization, Vaccines and Biologicals; WHO: Geneva, Switzerland, 2013.
- Zhong, Z.; Haltalli, M.; Holder, B.; Rice, T.; Donaldson, B.; O’Driscoll, M.; Le-Doare, K.; Kampmann, B.; Tregoning, J.S. The impact of timing of maternal influenza immunization on infant antibody levels at birth. Clin. Exp. Immunol. 2019, 195, 139–152. [Google Scholar] [CrossRef]
- Goodwin, K.; Viboud, C.; Simonsen, L. Antibody response to influenza vaccination in the elderly: A quantitative review. Vaccine 2006, 24, 1159–1169. [Google Scholar] [CrossRef]
- DiazGranados, C.A.; Dunning, A.J.; Kimmel, M.; Kirby, D.; Treanor, J.; Collins, A.; Pollak, R.; Christoff, J.; Earl, J.; Landolfi, V.; et al. Efficacy of High-Dose versus Standard-Dose Influenza Vaccine in Older Adults. N. Engl. J. Med. 2014, 371, 635–645. [Google Scholar] [CrossRef]
- Bruel, T.; Guivel-Benhassine, F.; Amraoui, S.; Malbec, M.; Richard, L.; Bourdic, K.; Donahue, D.A.; Lorin, V.; Casartelli, N.; Noël, N.; et al. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat. Commun. 2016, 7, 10844. [Google Scholar] [CrossRef]
- Glennie, M.J.; French, R.R.; Cragg, M.S.; Taylor, R.P. Mechanisms of killing by anti-CD20 monoclonal antibodies. Mol. Immunol. 2007, 44, 3823–3837. [Google Scholar] [CrossRef]
- Kohrt, H.; Rajasekaran, N.; Chester, C.; Yonezawa, A.; Zhao, X. Enhancement of antibody-dependent cell mediated cytotoxicity: A new era in cancer treatment. ImmunoTargets Ther. 2015, 4, 91. [Google Scholar] [CrossRef]
- Käyhty, H. Difficulties in establishing a serological correlate of protection after immunization with haemophilus influenzae conjugate vaccines. Biologicals 1994, 22, 397–402. [Google Scholar] [CrossRef]
- Siber, G.R.; Chang, I.; Baker, S.; Fernsten, P.; O’Brien, K.L.; Santosham, M.; Klugman, K.P.; Madhi, S.A.; Paradiso, P.; Kohberger, R. Estimating the protective concentration of anti-pneumococcal capsular polysaccharide antibodies. Vaccine 2007, 25, 3816–3826. [Google Scholar] [CrossRef]
- Maslanka, S.E.; Tappero, J.W.; Plikaytis, B.D.; Brumberg, R.S.; Dykes, J.K.; Gheesling, L.L.; Donaldson, K.B.J.; Schuchat, A.; Pullman, J.; Jones, M.; et al. Age-dependent Neisseria meningitidis serogroup C class-specific antibody concentrations and bactericidal titers in sera from young children from montana immunized with a licensed polysaccharide vaccine. Infect. Immun. 1998, 66, 2453–2459. [Google Scholar] [CrossRef]
- Danilova, E.; Jenum, P.A.; Skogen, V.; Pilnikov, V.F.; Sjursen, H. Antidiphtheria antibody responses in patients and carriers of Corynebacterium diphtheriae in the Arkhangelsk Region of Russia. Clin. Vaccine Immunol. 2006, 13, 627–632. [Google Scholar] [CrossRef][Green Version]
- Sonobe, M.H.; Trezena, A.G.; Guilhen, F.B.; Takano, V.L.; Fratelli, F.; Sakauchi, D.; Morais, J.F.; Prado, S.M.A.; Higashi, H.G. Determination of low tetanus or diphtheria antitoxin titers in sera by a toxin neutralization assay and a modified toxin-binding inhibition test. Brazilian J. Med. Biol. Res. 2007, 40, 69–76. [Google Scholar] [CrossRef][Green Version]
- Cherry, J.D.; Gornbein, J.; Heininger, U.; Stehr, K. A search for serologic correlates of immunity to Bordetella pertussis cough illnesses. Vaccine 1998, 16, 1901–1906. [Google Scholar] [CrossRef]
- Hobson, D.; Curry, R.L.; Beare, A.S.; Ward-Gardner, A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. Epidemiol. Infect. 1972, 70, 767–777. [Google Scholar] [CrossRef]
- Hannoun, C.; Megas, F.; Piercy, J. Immunogenicity and protective efficacy of influenza vaccination. Virus Res. 2004, 103, 133–138. [Google Scholar] [CrossRef]
- McMichael, A.J.; Gotch, F.M.; Noble, G.R.; Beare, P.A.S. Cytotoxic T-Cell Immunity to Influenza. N. Engl. J. Med. 1983, 309, 13–17. [Google Scholar] [CrossRef]
- Bhatt, K.; Verma, S.; Ellner, J.J.; Salgame, P. Quest for Correlates of Protection against Tuberculosis. Clin. Vaccine Immunol. 2015, 22, 258–266. [Google Scholar] [CrossRef]
- Flynn, J.L.; Chan, J. Immunology of tuberculosis. Annu. Rev. Immunol. 2001, 19, 93–129. [Google Scholar] [CrossRef]
- Tomaras, G.D.; Plotkin, S.A. Complex immune correlates of protection in HIV-1 vaccine efficacy trials. Immunol. Rev. 2017, 275, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Valmaseda, A.; Macete, E.; Nhabomba, A.; Guinovart, C.; Aide, P.; Bardají, A.; Bassat, Q.; Nhampossa, T.; Maculuve, S.; Casellas, A.; et al. Identifying immune correlates of protection against plasmodium falciparum through a novel approach to account for heterogeneity in malaria exposure. Clin. Infect. Dis. 2018, 66, 586–593. [Google Scholar] [CrossRef]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Addetia, A.; Crawford, K.H.D.; Dingens, A.; Zhu, H.; Roychoudhury, P.; Huang, M.-L.; Jerome, K.R.; Bloom, J.D.; Greninger, A.L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 2020, 58, 1–32. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Draft of the Landscape of COVID-19 Candidate Vaccines—13 August 2020; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Peeples, L. News Feature: Avoiding pitfalls in the pursuit of a COVID-19 vaccine. Proc. Natl. Acad. Sci. USA 2020, 117, 8218–8221. [Google Scholar] [CrossRef] [PubMed]
- Barouch, D.H.; Korber, B. HIV-1 vaccine development after STEP. Annu. Rev. Med. 2010, 61, 153–167. [Google Scholar] [CrossRef]
- Acosta, P.L.; Caballero, M.T.; Polack, F.P. Brief History and Characterization of Enhanced Respiratory Syncytial Virus Disease. Clin. Vaccine Immunol. 2015, 23, 189–195. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Dagan, N.; Barda, N.; Kepten, E.; Miron, O.; Perchik, S.; Katz, M.A.; Hernán, M.A.; Lipsitch, M.; Reis, B.; Balicer, R.D. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Combadiere, B.; Adam, L.; Quentric, P.; Rosenbaum, P.; Dorgham, K.; Bonduelle, O.; Parizot, C.; Sauce, D.; Mayaux, J.; Luyt, C.-E.; et al. LOX-1 + immature neutrophils predict severe COVID-19 patients at risk of thrombotic complications. bioRxiv 2020. [Google Scholar] [CrossRef]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.A.; Endeman, H.; van den Akker, J.P.C.; Molenkamp, R.; Koopmans, M.P.G.; van Gorp, E.C.M.; et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, 1–14. [Google Scholar] [CrossRef]
- Laing, A.G.; Lorenc, A.; del Molino del Barrio, I.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef]
- Wen, W.; Su, W.; Tang, H.; Le, W.; Zhang, X.; Zheng, Y.; Liu, X.; Xie, L.; Li, J.; Ye, J.; et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020, 6. [Google Scholar] [CrossRef]
- Silvin, A.; Chapuis, N.; Dunsmore, G.; Goubet, A.G.; Dubuisson, A.; Derosa, L.; Almire, C.; Hénon, C.; Kosmider, O.; Droin, N.; et al. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell 2020, 182, 1401–1418.e18. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1–2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatulin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G.; et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, G.; Pan, H.; Li, C.; Hu, Y.; Chu, K.; Han, W.; Chen, Z.; Tang, R.; Yin, W.; et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2020, 21, 181–192. [Google Scholar] [CrossRef]
- Sallusto, F.; Lanzavecchia, A.; Araki, K.; Ahmed, R. From Vaccines to Memory and Back. Immunity 2010, 33, 451–463. [Google Scholar] [CrossRef]
- Pulendran, B.; Ahmed, R. Immunological mechanisms of vaccination. Nat. Immunol. 2011, 12, 509–517. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Sallusto, F.; Geginat, J.; Lanzavecchia, A. C entral M emory and E ffector M emory T C ell S ubsets: Function, Generation, and Maintenance. Annu. Rev. Immunol. 2004, 22, 745–763. [Google Scholar] [CrossRef]
- Neumann, B.; Klippert, A.; Raue, K.; Sopper, S.; Stahl-Hennig, C. Characterization of B and plasma cells in blood, bone marrow, and secondary lymphoid organs of rhesus macaques by multicolor flow cytometry. J. Leukoc. Biol. 2014, 97, 19–30. [Google Scholar] [CrossRef]
- Maecker, H.T.; McCoy, J.P.; Nussenblatt, R. Standardizing immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol. 2012, 12, 191–200. [Google Scholar] [CrossRef]
- Coffman, R.L.; Sher, A.; Seder, R.A. Vaccine Adjuvants: Putting Innate Immunity to Work. Immunity 2010, 33, 492–503. [Google Scholar] [CrossRef]
- Zimmermann, P.; Curtis, N. Factors That Influence the Immune Response to Vaccination. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef]
- Cox, R.J.; Brokstad, K.A.; Ogra, P. Influenza Virus: Immunity and Vaccination Strategies. Comparison of the Immune Response to Inactivated and Live, Attenuated Influenza Vaccines. Scand. J. Immunol. 2004, 59, 1–15. [Google Scholar] [CrossRef]
- Zhaori, G.; Sun, M.; Ogra, P.L. Characteristics of the immune response to poliovirus virion polypeptides after immunization with live or inactivated polio vaccines. J. Infect. Dis. 1988, 158, 160–165. [Google Scholar] [CrossRef]
- Martinon, F.; Kaldma, K.; Sikut, R.; Çulina, S.; Romain, G.; Tuomela, M.; Adojaan, M.; Männik, A.; Toots, U.; Kivisild, T.; et al. Persistent immune responses induced by a human immunodeficiency virus dna vaccine delivered in association with electroporation in the skin of nonhuman primates. Hum. Gene Ther. 2009. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Blazevic, V.; Männik, A.; Malm, M.; Sikut, R.; Valtavaara, M.; Toots, U.; Ustav, M.; Krohn, K. Induction of human immunodeficiency virus type-1-specific immunity with a novel gene transport unit (GTU)-MultiHIV DNA vaccine. AIDS Res. Hum. Retrovir. 2006, 22, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.B.; Wilson, I.A. Innovations in structure-based antigen design and immune monitoring for next generation vaccines. Curr. Opin. Immunol. 2020, 65, 50–56. [Google Scholar] [CrossRef]
- Germain, R.N.; Meier-Schellersheim, M.; Nita-Lazar, A.; Fraser, I.D.C. Systems Biology in Immunology: A Computational Modeling Perspective. Annu. Rev. Immunol. 2011, 29, 527–585. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.G.; Harris, S.A.; Satti, I.; Bryan, D.; Walker, K.B.; Dockrell, H.M.; McShane, H.; Ho, M.M. Assay optimisation and technology transfer for multi-site immuno-monitoring in vaccine trials. PLoS ONE 2017, 12, e0184391. [Google Scholar] [CrossRef] [PubMed]
- Duffy, D. Standardized Immunomonitoring: Separating the Signals from the Noise. Trends Biotechnol. 2018, 36, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Muruato, A.E.; Fontes-Garfias, C.R.; Ren, P.; Garcia-Blanco, M.A.; Menachery, V.D.; Xie, X.; Shi, P.Y. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.H.; Frasch, C.E.; Falk, L.A.; Klein, D.L.; Deal, C.D. Correlates of immunity for pneumococcal conjugate vaccines. Vaccine 2003, 21, 2190–2196. [Google Scholar] [CrossRef]
- Boyle, M.J.; Reiling, L.; Feng, G.; Langer, C.; Osier, F.H.; Aspeling-Jones, H.; Cheng, Y.S.; Stubbs, J.; Tetteh, K.K.A.; Conway, D.J.; et al. Human antibodies fix complement to inhibit plasmodium falciparum invasion of erythrocytes andare associated with protection against malaria. Immunity 2015, 42, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, J.C.; Sébastien, I.; Germann, A.; Müller, S.C.; Meyerhans, A.; von Briesen, H.; Zimmermann, H. Towards standardized automated immunomonitoring: An automated ELISpot assay for safe and parallelized functionality analysis of immune cells. Cytotechnology 2017, 69, 57–73. [Google Scholar] [CrossRef] [PubMed]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Lo Tartaro, D.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020. [Google Scholar] [CrossRef]
- Mahnke, Y.D.; Brodie, T.M.; Sallusto, F.; Roederer, M.; Lugli, E. The who’s who of T-cell differentiation: Human memory T-cell subsets. Eur. J. Immunol. 2013, 43, 2797–2809. [Google Scholar] [CrossRef]
- Havenar-Daughton, C.; Reiss, S.M.; Carnathan, D.G.; Wu, J.E.; Kendric, K.; Torrents de la Peña, A.; Kasturi, S.P.; Dan, J.M.; Bothwell, M.; Sanders, R.W.; et al. Cytokine-Independent Detection of Antigen-Specific Germinal Center T Follicular Helper Cells in Immunized Nonhuman Primates Using a Live Cell Activation-Induced Marker Technique. J. Immunol. 2016, 197, 994–1002. [Google Scholar] [CrossRef]
- Herati, R.S.; Muselman, A.; Vella, L.; Bengsch, B.; Parkhouse, K.; Del Alcazar, D.; Kotzin, J.; Doyle, S.A.; Tebas, P.; Hensley, S.E.; et al. Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef]
- Morou, A.; Brunet-Ratnasingham, E.; Dubé, M.; Charlebois, R.; Mercier, E.; Darko, S.; Brassard, N.; Nganou-Makamdop, K.; Arumugam, S.; Gendron-Lepage, G.; et al. Altered differentiation is central to HIV-specific CD4+ T cell dysfunction in progressive disease. Nat. Immunol. 2019, 20, 1059–1070. [Google Scholar] [CrossRef]
- Dan, J.M.; Lindestam Arlehamn, C.S.; Weiskopf, D.; da Silva Antunes, R.; Havenar-Daughton, C.; Reiss, S.M.; Brigger, M.; Bothwell, M.; Sette, A.; Crotty, S. A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood. J. Immunol. 2016, 197, 983–993. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Amu, S.; Lavy-Shahaf, G.; Cagigi, A.; Hejdeman, B.; Nozza, S.; Lopalco, L.; Mehr, R.; Chiodi, F. Frequency and phenotype of B cell subpopulations in young and aged HIV-1 infected patients receiving ART. Retrovirology 2014, 11, 1–13. [Google Scholar] [CrossRef]
- Demberg, T.; Brocca-Cofano, E.; Xiao, P.; Venzon, D.; Vargas-Inchaustegui, D.; Lee, E.M.; Kalisz, I.; Kalyanaraman, V.S.; DiPasquale, J.; McKinnon, K.; et al. Dynamics of Memory B-Cell Populations in Blood, Lymph Nodes, and Bone Marrow during Antiretroviral Therapy and Envelope Boosting in Simian Immunodeficiency Virus SIVmac251-Infected Rhesus Macaques. J. Virol. 2012, 86, 12591–12604. [Google Scholar] [CrossRef]
- Le Lann, L.; Jouve, P.E.; Alarcón-Riquelme, M.; Jamin, C.; Pers, J.O.; Alvarez, M.; Alvarez-Errico, D.; Azevedo, N.; Barbarroja, N.; Buttgereit, A.; et al. Standardization procedure for flow cytometry data harmonization in prospective multicenter studies. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Palgen, J.-L.; Tchitchek, N.; Elhmouzi-Younes, J.; Delandre, S.; Namet, I.; Rosenbaum, P.; Dereuddre-Bosquet, N.; Martinon, F.; Cosma, A.; Lévy, Y.; et al. Prime and Boost Vaccination Elicit a Distinct Innate Myeloid Cell Immune Response. Sci. Rep. 2018, 8, 3087. [Google Scholar] [CrossRef]
- Nolan, J.P.; Condello, D. Spectral Flow Cytometry. Curr. Protoc. Cytom. 2013. [Google Scholar] [CrossRef]
- Niewold, P.; Ashhurst, T.M.; Smith, A.L.; King, N.J.C. Evaluating spectral cytometry for immune profiling in viral disease. Cytom. Part A 2020, 97, 1165–1179. [Google Scholar] [CrossRef]
- Mair, F.; Hartmann, F.J.; Mrdjen, D.; Tosevski, V.; Krieg, C.; Becher, B. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur. J. Immunol. 2016, 46, 34–43. [Google Scholar] [CrossRef]
- Reeves, P.M.; Raju Paul, S.; Baeten, L.; Korek, S.E.; Yi, Y.; Hess, J.; Sobell, D.; Scholzen, A.; Garritsen, A.; De Groot, A.S.; et al. Novel multiparameter correlates of Coxiella burnetii infection and vaccination identified by longitudinal deep immune profiling. Sci. Rep. 2020, 10, 1–20. [Google Scholar] [CrossRef]
- Pejoski, D.; Tchitchek, N.; Pozo, A.R.; Elhmouzi-Younes, J.; Yousfi-Bogniaho, R.; Rogez-Kreuz, C.; Clayette, P.; Dereuddre-Bosquet, N.; Lévy, Y.; Cosma, A.; et al. Identification of vaccine-altered circulating B cell phenotypes using mass cytometry and a two-step clustering analysis. J. Immunol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Mathian, A.; Mouries-Martin, S.; Dorgham, K.; Devilliers, H.; Barnabei, L.; Ben Salah, E.; Cohen-Aubart, F.; Garrido Castillo, L.; Haroche, J.; Hie, M.; et al. Monitoring Disease Activity in Systemic Lupus Erythematosus With Single-Molecule Array Digital Enzyme-Linked Immunosorbent Assay Quantification of Serum Interferon-α. Arthritis Rheumatol. 2019, 71, 756–765. [Google Scholar] [CrossRef]
- Furman, D.; Davis, M.M. New approaches to understanding the immune response to vaccination and infection. Vaccine 2015, 33, 5271–5281. [Google Scholar] [CrossRef]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Svensson, V.; Vento-Tormo, R.; Teichmann, S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018, 13, 599–604. [Google Scholar] [CrossRef]
- Stuart, T.; Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 2019, 20, 257–272. [Google Scholar] [CrossRef]
- Benichou, J.; Ben-Hamo, R.; Louzoun, Y.; Efroni, S. Rep-Seq: Uncovering the immunological repertoire through next-generation sequencing. Immunology 2012, 135, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Dalloul, I.; Boyer, F.; Dalloul, Z.; Pignarre, A.; Lacombe, G.; Fest, T.; Chatonnet, F.; Delaloy, C.; Durandy, A.; Jeannet, R.; et al. Locus Suicide Recombination actively occurs on the functionally rearranged IgH allele in B-cells from inflamed human lymphoid tissues. bioRxiv 2018, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Bender, S.; Javaugue, V.; Saintamand, A.; Ayala, M.V.; Alizadeh, M.; Filloux, M.; Pascal, V.; Gachard, N.; Lavergne, D.; Auroy, F.; et al. Immunoglobulin variable domain high-throughput sequencing reveals specific novel mutational patterns in POEMS syndrome. Blood 2020, 135, 1750–1758. [Google Scholar] [CrossRef] [PubMed]
- Kreer, C.; Gruell, H.; Mora, T.; Walczak, A.M.; Klein, F. Exploiting B cell receptor analyses to inform on HIV-1 vaccination strategies. Vaccines 2020, 8, 13. [Google Scholar] [CrossRef]
- Noé, A.; Cargill, T.N.; Nielsen, C.M.; Russell, A.J.C.; Barnes, E. The Application of Single-Cell RNA Sequencing in Vaccinology. J. Immunol. Res. 2020, 2020. [Google Scholar] [CrossRef]
- Querec, T.; Bennouna, S.; Alkan, S.; Laouar, Y.; Gorden, K.; Flavell, R.; Akira, S.; Ahmed, R.; Pulendran, B. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J. Exp. Med. 2006, 203, 413–424. [Google Scholar] [CrossRef]
- Gaucher, D.; Therrien, R.; Kettaf, N.; Angermann, B.R.; Boucher, G.; Filali-Mouhim, A.; Moser, J.M.; Mehta, R.S.; Drake, D.R.; Castro, E.; et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 2008, 205, 3119–3131. [Google Scholar] [CrossRef]
- Kasturi, S.P.; Skountzou, I.; Albrecht, R.A.; Koutsonanos, D.; Hua, T.; Nakaya, H.I.; Ravindran, R.; Stewart, S.; Alam, M.; Kwissa, M.; et al. Programming the magnitude and persistence of antibody responses with innate immunity. Nature 2011, 470, 543–547. [Google Scholar] [CrossRef]
- Querec, T.D.; Akondy, R.S.; Lee, E.K.; Cao, W.; Nakaya, H.I.; Teuwen, D.; Pirani, A.; Gernert, K.; Deng, J.; Marzolf, B.; et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009, 10, 116–125. [Google Scholar] [CrossRef]
- Ravindran, R.; Khan, N.; Nakaya, H.I.; Li, S.; Loebbermann, J.; Maddur, M.S.; Park, Y.; Jones, D.P.; Chappert, P.; Davoust, J.; et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science 2014, 343, 313–317. [Google Scholar] [CrossRef]
- Nakaya, H.I.; Wrammert, J.; Lee, E.K.; Racioppi, L.; Marie-Kunze, S.; Haining, W.N.; Means, A.R.; Kasturi, S.P.; Khan, N.; Li, G.-M.; et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011, 12, 786–795. [Google Scholar] [CrossRef]
- Furman, D.; Hejblum, B.P.; Simon, N.; Jojic, V.; Dekker, C.L.; Thiebaut, R.; Tibshirani, R.J.; Davis, M.M. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc. Natl. Acad. Sci. USA 2014, 111, 869–874. [Google Scholar] [CrossRef]
- Lynn, D.J.; Pulendran, B. The potential of the microbiota to influence vaccine responses. J. Leukoc. Biol. 2017, 103. [Google Scholar] [CrossRef]
- Oh, J.Z.; Ravindran, R.; Chassaing, B.; Carvalho, F.A.; Maddur, M.S.; Bower, M.; Hakimpour, P.; Gill, K.P.; Nakaya, H.I.; Yarovinsky, F.; et al. TLR5-Mediated Sensing of Gut Microbiota Is Necessary for Antibody Responses to Seasonal Influenza Vaccination. Immunity 2014, 41, 478–492. [Google Scholar] [CrossRef]
- De Groot, A.S.; Moise, L.; Terry, F.; Gutierrez, A.H.; Hindocha, P.; Richard, G.; Hoft, D.F.; Ross, T.M.; Noe, A.R.; Takahashi, Y.; et al. Better epitope discovery, precision immune engineering, and accelerated vaccine design using Immunoinformatics tools. Front. Immunol. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Poland, G.A.; Ovsyannikova, I.G.; Jacobson, R.M. Personalized vaccines: The emerging field of vaccinomics. Expert Opin. Biol. Ther. 2008, 8, 1659. [Google Scholar] [CrossRef]
| References | Patient Cohort | Findings | Immunitoring Techniques |
|---|---|---|---|
| Hadjaj et al. [40] | 18 Healthy Donors 15 Mild 17 Severe 18 Critical | Type I IFN impairment Exacerbated inflammatory response | Mass Cytometry mRNA Expression Analysis Multiplex Cytokine Detection |
| Combadiere et al. [41] | 38 Critical | Myelemia with overabundance of CD123+ and LOX-1+ neutrophils | Flow Cytometry Ultra-sensitive Digital Immunoassay (Quanterix) |
| Weiskopf et al. [42] | 10 Severe/Critical | Immunomodulation of T-cell responses depending on severity | Flow Cytometry ELISA Multiplex Cytokine Detection |
| Laing et al. [43] | 55 Health Donors 56 Severe/Critical | CXCL10, IL-10, IL-6; B and T, and monocyte subset signatures related with severity | Flow Cytometry mRNA Expression Analysis Multiplex Cytokine |
| Wen et al. [44] | 10 Recovering COVID-19 Patients | SARS-CoV2-specific IGHV3-23-IGHJ4 pairing TNFSF13, IL-18, IL-2, and IL-4 genes may benefit from COVID-19 recovery | TCR and BCR Sequencing Single-Cell RNA-Seq |
| Silvin et al. [45] | 72 Healthy Donors 27 Mild 16 Moderate 43 Severe | Non-classical monocytes and calprotectin-producing immature neutrophils increase in severe cases | Spectral Cytometry Mass Cytometry Flow Cytometry Single-Cell RNA-Seq Multiplex Cytokine Detection |
| References Clinical Trial ID | Phase Patient Cohort | Vaccine | Immunitoring Techniques |
|---|---|---|---|
| Jackon et al. [46] NCT04283461 | Phase I 45 healthy adults Age: 18–55 | Moderna vaccine RNA-based vaccine mRNA-1273 → Spike Dose escalation (25 μg, 100 μg, 250 μg) Homologous prime boost | ELISA Neutralization assay ICS—flow cytometry |
| Keech et al. [47] NCT04368988 | Phase I–II 132 healthy adults Age: 18–59 | Novavax vaccine Protein-based vaccine NVX-CoV2373 → Spike with/without Matrix-M1 adjuvant dose escalation (5–25 ug) Homologous prime boost | ELISA microneutralization assay ICS—flow cytometry |
| Logunov et al. [48] NCT04436471 NCT04437875 | Phase I–II 120 healthy adults Age 18–60 | Sputnik V vaccine Viral vector-based vaccine rAd26 and rAd5 → spike Heterologous prime boost Prime rAd26-S Boost rAd5 | ELISA Neutralization assay Proliferation assay |
| Mulligan et al. [49] NCT04368728 | Phase I–II 45 healthy adults Age 18–55 | Pfizer-BioNTech vaccine RNA-based vaccine BNT162b1→ Lipid nanoparticle-formulated nucleoside-modified mRNA vaccine Trimerized SARS-CoV-2 RBD Dose escalation: 10 μg–30 μg–100 μg Homologous prime boost | IgG binding assay Neutralization assay |
| Folegatti et al. [50] NCT04324606 | Phase I–II 1077 adults Age: 18-55 | Astrazeneca vaccine Viral vector-based vaccine ChAdOx1 nCoV-19 → spike Homologous prime boost | ELISA Neutralization assay ELISpot |
| Zhang et al. [51] NCT04352608 | Phase I 143 healthy adults Phase II 600 healthy adults Age: 18–59 | Sinovac vaccine Inactivated virus-based vaccine CoronaVac → inactivated SARS-CoV-2 Dose escalation | ELISA Microcytophathogenic effect assay ELISpot |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adam, L.; Rosenbaum, P.; Bonduelle, O.; Combadière, B. Strategies for Immunomonitoring after Vaccination and during Infection. Vaccines 2021, 9, 365. https://doi.org/10.3390/vaccines9040365
Adam L, Rosenbaum P, Bonduelle O, Combadière B. Strategies for Immunomonitoring after Vaccination and during Infection. Vaccines. 2021; 9(4):365. https://doi.org/10.3390/vaccines9040365
Chicago/Turabian StyleAdam, Lucille, Pierre Rosenbaum, Olivia Bonduelle, and Behazine Combadière. 2021. "Strategies for Immunomonitoring after Vaccination and during Infection" Vaccines 9, no. 4: 365. https://doi.org/10.3390/vaccines9040365
APA StyleAdam, L., Rosenbaum, P., Bonduelle, O., & Combadière, B. (2021). Strategies for Immunomonitoring after Vaccination and during Infection. Vaccines, 9(4), 365. https://doi.org/10.3390/vaccines9040365






