Zika Virus Pathogenesis: A Battle for Immune Evasion
Abstract
1. Introduction
2. Mechanisms of ZIKV Immune Evasion
2.1. Antiviral IFN and Associated Signals
2.2. ZIKV Structural E Protein Inhibits IFN Production
2.3. ZIKV NS Proteins as Modulators of Antiviral IFN and Associated Signals
2.4. Viral Genomic and Subgenomic Rnas Inhibit IFN Signaling
2.5. MicroRNA, ZIKV Infection, Immunity and Pathogenicity
2.6. ZIKV Infection and Adaptive Immune Responses Understanding Cross-Immune Protection and Vaccine Challenges
2.7. ZIKV Invades and Infects the Fetal Brain and Eye: Evading Immune Responses in Immuno-Privileged Organs
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- A World at Risk: Annual Report on Global Preparedness for Health Emergencies. Global Preparedness Monitoring Board. 2019. Available online: https://apps.who.int/gpmb/assets/annual_report/GPMB_annualreport_2019.pdf?utm_source=mandiner&utm_medium=link&utm_campaign=mandiner_202004 (accessed on September 2019).
- Coronavirus Disease (COVID-19) Weekly Epidemiological Update and Weekly Operational Update. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ (accessed on 29 December 2020).
- Statement on the 1st Meeting of the ihr Emergency Committee on the 2014 Ebola Outbreak in West Africa. 2014. Available online: https://www.who.int/mediacentre/news/statements/2014/ebola-20140808/en/ (accessed on 8 August 2014).
- Koenig, K.L.; Majestic, C.; Burns, M.J. Ebola virus disease: Essential public health principles for clinicians. West. J. Emerg. Med. 2014, 15, 728–731. [Google Scholar] [CrossRef][Green Version]
- McCoy, C.E.; Lotfipour, S.; Chakravarthy, B.; Schultz, C.; Barton, E. Emergency medical services public health implications and interim guidance for the ebola virus in the united states. West. J. Emerg. Med. 2014, 15, 723–727. [Google Scholar] [CrossRef]
- Safari, S.; Baratloo, A.; Rouhipour, A.; Ghelichkhani, P.; Yousefifard, M. Ebola hemorrhagic fever as a public health emergency of international concern: A review article. Emergency (TehranIran) 2015, 3, 3–7. [Google Scholar]
- Who Statement on the First Meeting of the International Health Regulations (2005) (ihr 2005) Emergency Committee on Zika Virus and Observed Increase in Neurological Disorders and Neonatal Malformations. 2016. Available online: https://www.who.int/news/item/01-02-2016-who-statement-on-the-first-meeting-of-the-international-health-regulations-(2005)-(ihr-2005)-emergency-committee-on-zika-virus-and-observed-increase-in-neurological-disorders-and-neonatal-malformations (accessed on 1 February 2016).
- Nicola, M.; Alsafi, Z.; Sohrabi, C.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, M.; Agha, R. The socio-economic implications of the coronavirus pandemic (covid-19): A review. Int. J. Surg. 2020, 78, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.; Wilson, M.E.; Touch, S.; McCloskey, B.; Mwaba, P.; Bates, M.; Dar, O.; Mattes, F.; Kidd, M.; Ippolito, G.; et al. Rapid spread of zika virus in the americas--implications for public health preparedness for mass gatherings at the 2016 brazil olympic games. Int. J. Infect. Dis. 2016, 44, 11–15. [Google Scholar] [CrossRef]
- Salvador, F.S.; Fujita, D.M. Entry routes for zika virus in brazil after 2014 world cup: New possibilities. Travel Med. Infect. Dis. 2016, 14, 49–51. [Google Scholar] [CrossRef]
- Miranda-Filho Dde, B.; Martelli, C.M.; Ximenes, R.A.; Araújo, T.V.; Rocha, M.A.; Ramos, R.C.; Dhalia, R.; França, R.F.; Marques Júnior, E.T.; Rodrigues, L.C. Initial description of the presumed congenital zika syndrome. Am. J. Public Health 2016, 106, 598–600. [Google Scholar] [CrossRef] [PubMed]
- Petersen, L.R.; Jamieson, D.J.; Powers, A.M.; Honein, M.A. Zika virus. N. Engl. J. Med. 2016, 374, 1552–1563. [Google Scholar] [CrossRef]
- Brasil, P.; Pereira, J.P., Jr.; Moreira, M.E.; Ribeiro Nogueira, R.M.; Damasceno, L.; Wakimoto, M.; Rabello, R.S.; Valderramos, S.G.; Halai, U.A.; Salles, T.S.; et al. Zika virus infection in pregnant women in rio de janeiro. N. Engl. J. Med. 2016, 375, 2321–2334. [Google Scholar] [CrossRef] [PubMed]
- Kleber de Oliveira, W.; Cortez-Escalante, J.; De Oliveira, W.T.; do Carmo, G.M.; Henriques, C.M.; Coelho, G.E.; Araújo de França, G.V. Increase in reported prevalence of microcephaly in infants born to women living in areas with confirmed zika virus transmission during the first trimester of pregnancy—Brazil, 2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 242–247. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; Jamieson, D.J.; Honein, M.A.; Petersen, L.R. Zika virus and birth defects—Reviewing the evidence for causality. N. Engl. J. Med. 2016, 374, 1981–1987. [Google Scholar] [CrossRef]
- Driggers, R.W.; Ho, C.Y.; Korhonen, E.M.; Kuivanen, S.; Jääskeläinen, A.J.; Smura, T.; Rosenberg, A.; Hill, D.A.; DeBiasi, R.L.; Vezina, G.; et al. Zika virus infection with prolonged maternal viremia and fetal brain abnormalities. N. Engl. J. Med. 2016, 374, 2142–2151. [Google Scholar] [CrossRef]
- Martines, R.B.; Bhatnagar, J.; Keating, M.K.; Silva-Flannery, L.; Muehlenbachs, A.; Gary, J.; Goldsmith, C.; Hale, G.; Ritter, J.; Rollin, D.; et al. Notes from the field: Evidence of zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses—Brazil, 2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 159–160. [Google Scholar] [CrossRef] [PubMed]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V.; et al. Zika virus associated with microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef]
- Schuler-Faccini, L.; Ribeiro, E.M.; Feitosa, I.M.; Horovitz, D.D.; Cavalcanti, D.P.; Pessoa, A.; Doriqui, M.J.; Neri, J.I.; Neto, J.M.; Wanderley, H.Y.; et al. Possible association between zika virus infection and microcephaly—Brazil, 2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 59–62. [Google Scholar] [CrossRef]
- Soares de Araújo, J.S.; Regis, C.T.; Gomes, R.G.; Tavares, T.R.; Rocha Dos Santos, C.; Assunção, P.M.; Nóbrega, R.V.; Pinto, D.F.; Bezerra, B.V.; Mattos, S.D. Microcephaly in north-east brazil: A retrospective study on neonates born between 2012 and 2015. Bull. World Health Organ. 2016, 94, 835–840. [Google Scholar] [CrossRef]
- Calvet, G.; Aguiar, R.S.; Melo, A.S.O.; Sampaio, S.A.; de Filippis, I.; Fabri, A.; Araujo, E.S.M.; de Sequeira, P.C.; de Mendonça, M.C.L.; de Oliveira, L.; et al. Detection and sequencing of zika virus from amniotic fluid of fetuses with microcephaly in brazil: A case study. Lancet Infect. Dis. 2016, 16, 653–660. [Google Scholar] [CrossRef]
- Dyer, O. Zika virus spreads across americas as concerns mount over birth defects. BMJ (Clin. Res. Ed.) 2015, 351, h6983. [Google Scholar] [CrossRef]
- WHO. WHO Situation Report. Zika Virus Microcephaly Guillain-Barré Syndrome. 2017. Available online: http://reliefweb.int/report/world/zika-virus-microcephaly-and-guillain-barr-syndrome-situation-report-10-march-2017 (accessed on 10 March 2017).
- Carteaux, G.; Maquart, M.; Bedet, A.; Contou, D.; Brugières, P.; Fourati, S.; Cleret de Langavant, L.; de Broucker, T.; Brun-Buisson, C.; Leparc-Goffart, I.; et al. Zika virus associated with meningoencephalitis. N. Engl. J. Med. 2016, 374, 1595–1596. [Google Scholar] [CrossRef]
- Barbi, L.; Coelho, A.V.C.; Alencar, L.C.A.; Crovella, S. Prevalence of guillain-barré syndrome among zika virus infected cases: A systematic review and meta-analysis. Braz. J. Infect. Dis. 2018, 22, 137–141. [Google Scholar] [CrossRef]
- Costello, A.; Dua, T.; Duran, P.; Gülmezoglu, M.; Oladapo, O.T.; Perea, W.; Pires, J.; Ramon-Pardo, P.; Rollins, N.; Saxena, S. Defining the syndrome associated with congenital zika virus infection. Bull. World Health Organ. 2016, 94, 406–406a. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, T.; Rodriguez, A.; Almiron, M.; Sanhueza, A.; Ramon, P.; de Oliveira, W.K.; Coelho, G.E.; Badaró, R.; Cortez, J.; Ospina, M.; et al. Zika virus and the guillain-barré syndrome—Case series from seven countries. N. Engl. J. Med. 2016, 375, 1598–1601. [Google Scholar] [CrossRef]
- Hoen, B.; Schaub, B.; Funk, A.L.; Ardillon, V.; Boullard, M.; Cabié, A.; Callier, C.; Carles, G.; Cassadou, S.; Césaire, R.; et al. Pregnancy outcomes after zikv infection in french territories in the americas. N. Engl. J. Med. 2018, 378, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Zika Virus Disease Epidemic: Potential Association with Microcephaly and Guillain-Barré Syndrome (First Update). Eur. Cent. Dis. Prev. ControlStockh. Swed. 2016. Available online: http://ecdc.europa.eu/en/publications/Publications/rapid-risk-assessment-zika-virus-first-update-jan-2016.pdf (accessed on 21 January 2016).
- Pierson, T.C.; Diamond, M.S. The emergence of zika virus and its new clinical syndromes. Nature 2018, 560, 573–581. [Google Scholar] [CrossRef]
- Dudley, D.M.; Van Rompay, K.K.; Coffey, L.L.; Ardeshir, A.; Keesler, R.I.; Bliss-Moreau, E.; Grigsby, P.L.; Steinbach, R.J.; Hirsch, A.J.; MacAllister, R.P.; et al. Miscarriage and stillbirth following maternal zika virus infection in nonhuman primates. Nat. Med. 2018, 24, 1104–1107. [Google Scholar] [CrossRef] [PubMed]
- Van der Eijk, A.A.; van Genderen, P.J.; Verdijk, R.M.; Reusken, C.B.; Mögling, R.; van Kampen, J.J.; Widagdo, W.; Aron, G.I.; GeurtsvanKessel, C.H.; Pas, S.D.; et al. Miscarriage associated with zika virus infection. N. Engl. J. Med. 2016, 375, 1002–1004. [Google Scholar] [CrossRef]
- Sarno, M.; Sacramento, G.A.; Khouri, R.; do Rosário, M.S.; Costa, F.; Archanjo, G.; Santos, L.A.; Nery, N., Jr.; Vasilakis, N.; Ko, A.I.; et al. Zika virus infection and stillbirths: A case of hydrops fetalis, hydranencephaly and fetal demise. PLoS Negl. Trop. Dis. 2016, 10, e0004517. [Google Scholar] [CrossRef]
- Schaub, B.; Monthieux, A.; Najihoullah, F.; Harte, C.; Césaire, R.; Jolivet, E.; Voluménie, J.L. Late miscarriage: Another zika concern? Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 207, 240–241. [Google Scholar] [CrossRef] [PubMed]
- Leisher, S.H.; Balalian, A.A.; Reinebrant, H.; Shiau, S.; Flenady, V.; Kuhn, L.; Morse, S.S. Systematic review: Fetal death reporting and risk in zika-affected pregnancies. Trop. Med. Int. Health 2020, 26, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, I.R.F.; Frontera, J.A.; Bispo de Filippis, A.M.; Nascimento, O. Neurologic complications associated with the zika virus in brazilian adults. JAMA Neurol. 2017, 74, 1190–1198. [Google Scholar] [CrossRef]
- Rozé, B.; Najioullah, F.; Signate, A.; Apetse, K.; Brouste, Y.; Gourgoudou, S.; Fagour, L.; Abel, S.; Hochedez, P.; Cesaire, R.; et al. Zika virus detection in cerebrospinal fluid from two patients with encephalopathy, martinique, february 2016. Eur. Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2016, 21, 30205. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, R.S.; Araujo, M.T.; Martins Filho, A.J.; Oliveira, C.S.; Nunes, B.T.; Cruz, A.C.; Nascimento, A.G.; Medeiros, R.C.; Caldas, C.A.; Araujo, F.C.; et al. Zika virus epidemic in brazil. I. Fatal disease in adults: Clinical and laboratorial aspects. J. Clin. Virol. 2016, 85, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Alves-Leon, S.V.; Lima, M.D.R.; Nunes, P.C.G.; Chimelli, L.M.C.; Rabelo, K.; Nogueira, R.M.R.; de Bruycker-Nogueira, F.; de Azeredo, E.L.; Bahia, P.R.; Rueda Lopes, F.C.; et al. Zika virus found in brain tissue of a multiple sclerosis patient undergoing an acute disseminated encephalomyelitis-like episode. Mult. Scler. (Houndmills Basingstoke Engl.) 2019, 25, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Parra, B.; Lizarazo, J.; Jiménez-Arango, J.A.; Zea-Vera, A.F.; González-Manrique, G.; Vargas, J.; Angarita, J.A.; Zuñiga, G.; Lopez-Gonzalez, R.; Beltran, C.L.; et al. Guillain-barré syndrome associated with zika virus infection in colombia. N. Engl. J. Med. 2016, 375, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Brito Ferreira, M.L.; Antunes de Brito, C.A.; Moreira, Á.J.P.; de Morais Machado, M.; Henriques-Souza, A.; Cordeiro, M.T.; de Azevedo Marques, E.T.; Pena, L.J. Guillain-barré syndrome, acute disseminated encephalomyelitis and encephalitis associated with zika virus infection in brazil: Detection of viral rna and isolation of virus during late infection. Am. J. Trop. Med. Hyg. 2017, 97, 1405–1409. [Google Scholar] [CrossRef]
- Muñoz, L.S.; Barreras, P.; Pardo, C.A. Zika virus-associated neurological disease in the adult: Guillain-barré syndrome, encephalitis, and myelitis. Semin. Reprod. Med. 2016, 34, 273–279. [Google Scholar] [CrossRef]
- Soares, C.N.; Brasil, P.; Carrera, R.M.; Sequeira, P.; de Filippis, A.B.; Borges, V.A.; Theophilo, F.; Ellul, M.A.; Solomon, T. Fatal encephalitis associated with zika virus infection in an adult. J. Clin. Virol. 2016, 83, 63–65. [Google Scholar] [CrossRef]
- Schwartzmann, P.V.; Ramalho, L.N.; Neder, L.; Vilar, F.C.; Ayub-Ferreira, S.M.; Romeiro, M.F.; Takayanagui, O.M.; Dos Santos, A.C.; Schmidt, A.; Figueiredo, L.T.; et al. Zika virus meningoencephalitis in an immunocompromised patient. Mayo Clin. Proc. 2017, 92, 460–466. [Google Scholar] [CrossRef]
- Mécharles, S.; Herrmann, C.; Poullain, P.; Tran, T.H.; Deschamps, N.; Mathon, G.; Landais, A.; Breurec, S.; Lannuzel, A. Acute myelitis due to zika virus infection. Lancet 2016, 387, 1481. [Google Scholar] [CrossRef]
- Niemeyer, B.; Niemeyer, R.; Borges, R.; Marchiori, E. Acute disseminated encephalomyelitis following zika virus infection. Eur. Neurol. 2017, 77, 45–46. [Google Scholar] [CrossRef] [PubMed]
- Galliez, R.M.; Spitz, M.; Rafful, P.P.; Cagy, M.; Escosteguy, C.; Germano, C.S.; Sasse, E.; Gonçalves, A.L.; Silveira, P.P.; Pezzuto, P.; et al. Zika virus causing encephalomyelitis associated with immunoactivation. Open Forum Infect. Dis. 2016, 3, ofw203. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.T.; England, J.D.; Lorenzana, I.; Medina-Montoya, M.; Alvarado, D.; De Bastos, M.; Fontiveros, S.; Sierra, M.; Contreras, F. Zika virus associated with sensory polyneuropathy. J. Neurol. Sci. 2016, 369, 271–272. [Google Scholar] [CrossRef]
- Nicastri, E.; Castilletti, C.; Balestra, P.; Galgani, S.; Ippolito, G. Zika virus infection in the central nervous system and female genital tract. Emerg. Infect. Dis. 2016, 22, 2228–2230. [Google Scholar] [CrossRef] [PubMed]
- Bido-Medina, R.; Wirsich, J.; Rodríguez, M.; Oviedo, J.; Miches, I.; Bido, P.; Tusen, L.; Stoeter, P.; Sadaghiani, S. Impact of zika virus on adult human brain structure and functional organization. Ann. Clin. Transl. Neurol. 2018, 5, 752–762. [Google Scholar] [CrossRef]
- Musso, D.; Gubler, D.J. Zika virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef]
- Musso, D.; Baud, D.; Freedman, D.O. Should testing of donors be restricted to active zika virus areas? Lancet Infect. Dis. 2016, 16, 1108–1109. [Google Scholar] [CrossRef]
- McCarthy, M. Four in florida are infected with zika from local mosquitoes. BMJ (Clin. Res. Ed.) 2016, 354, i4235. [Google Scholar] [CrossRef]
- Wikan, N.; Smith, D.R. Zika virus: History of a newly emerging arbovirus. Lancet Infect. Dis. 2016, 16, e119–e126. [Google Scholar] [CrossRef]
- Baud, D.; Gubler, D.J.; Schaub, B.; Lanteri, M.C.; Musso, D. An update on zika virus infection. Lancet 2017, 390, 2099–2109. [Google Scholar] [CrossRef]
- Ferraris, P.; Yssel, H.; Missé, D. Zika virus infection: An update. Microbes Infect. 2019, 21, 353–360. [Google Scholar] [CrossRef]
- Fauci, A.S.; Morens, D.M. Zika virus in the americas—Yet another arbovirus threat. N. Engl. J. Med. 2016, 374, 601–604. [Google Scholar] [CrossRef]
- Meaney-Delman, D.; Hills, S.L.; Williams, C.; Galang, R.R.; Iyengar, P.; Hennenfent, A.K.; Rabe, I.B.; Panella, A.; Oduyebo, T.; Honein, M.A.; et al. Zika virus infection among u.S. Pregnant travelers—August 2015-february 2016. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 211–214. [Google Scholar] [CrossRef]
- Chan, M. Who Director-General Briefs Executive Board on Zika Situation. 2016. Available online: http://www.who.int/dg/speeches/2016/zika-situation/en (accessed on 28 January 2016).
- Rozé, B.; Najioullah, F.; Fergé, J.L.; Apetse, K.; Brouste, Y.; Cesaire, R.; Fagour, C.; Fagour, L.; Hochedez, P.; Jeannin, S.; et al. Zika virus detection in urine from patients with guillain-barré syndrome on martinique, january 2016. Euro Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2016, 21, 30154. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, E.M.; Huhtamo, E.; Smura, T.; Kallio-Kokko, H.; Raassina, M.; Vapalahti, O. Zika virus infection in a traveller returning from the maldives, june 2015. Eur. Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2016, 21, 30107. [Google Scholar] [CrossRef]
- Hennessey, M.; Fischer, M.; Staples, J.E. Zika virus spreads to new areas—Region of the americas, may 2015-january 2016. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Morales, A.J. Zika: The new arbovirus threat for latin america. J. Infect. Dev. Ctries. 2015, 9, 684–685. [Google Scholar] [CrossRef][Green Version]
- Martinez-Pulgarin, D.F.; Acevedo-Mendoza, W.F.; Cardona-Ospina, J.A.; Rodríguez-Morales, A.J.; Paniz-Mondolfi, A.E. A bibliometric analysis of global zika research. Travel Med. Infect. Dis. 2016, 14, 55–57. [Google Scholar] [CrossRef]
- Brown, C. Zika virus outbreaks in asia and south america. Cmaj Can. Med. Assoc. J. J. De L’association Med. Can. 2016, 188, E34. [Google Scholar] [CrossRef]
- Cauchemez, S.; Besnard, M.; Bompard, P.; Dub, T.; Guillemette-Artur, P.; Eyrolle-Guignot, D.; Salje, H.; Van Kerkhove, M.D.; Abadie, V.; Garel, C.; et al. Association between zika virus and microcephaly in french polynesia, 2013-2015: A retrospective study. Lancet 2016, 387, 2125–2132. [Google Scholar] [CrossRef]
- Cao-Lormeau, V.M.; Blake, A.; Mons, S.; Lastère, S.; Roche, C.; Vanhomwegen, J.; Dub, T.; Baudouin, L.; Teissier, A.; Larre, P.; et al. Guillain-barré syndrome outbreak associated with zika virus infection in french polynesia: A case-control study. Lancet 2016, 387, 1531–1539. [Google Scholar] [CrossRef]
- Oehler, E.; Watrin, L.; Larre, P.; Leparc-Goffart, I.; Lastere, S.; Valour, F.; Baudouin, L.; Mallet, H.; Musso, D.; Ghawche, F. Zika virus infection complicated by guillain-barre syndrome—Case report, french polynesia, december 2013. Eur. Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2014, 19, 20720. [Google Scholar] [CrossRef]
- Goncé, A.; Martínez, M.J.; Marbán-Castro, E.; Saco, A.; Soler, A.; Alvarez-Mora, M.I.; Peiro, A.; Gonzalo, V.; Hale, G.; Bhatnagar, J.; et al. Spontaneous abortion associated with zika virus infection and persistent viremia. Emerg. Infect. Dis. 2018, 24, 933–935. [Google Scholar] [CrossRef] [PubMed]
- Marques, V.M.; Santos, C.S.; Santiago, I.G.; Marques, S.M.; Nunes Brasil, M.D.G.; Lima, T.T.; Costa, P.S. Neurological complications of congenital zika virus infection. Pediatr. Neurol. 2019, 91, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Martines, R.B.; Bhatnagar, J.; de Oliveira Ramos, A.M.; Davi, H.P.; Iglezias, S.D.; Kanamura, C.T.; Keating, M.K.; Hale, G.; Silva-Flannery, L.; Muehlenbachs, A.; et al. Pathology of congenital zika syndrome in brazil: A case series. Lancet 2016, 388, 898–904. [Google Scholar] [CrossRef]
- Marchette, N.J.; Garcia, R.; Rudnick, A. Isolation of zika virus from aedes aegypti mosquitoes in malaysia. Am. J. Trop. Med. Hyg. 1969, 18, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G.; Chang, G.J. Full-length sequencing and genomic characterization of bagaza, kedougou, and zika viruses. Arch. Virol. 2007, 152, 687–696. [Google Scholar] [CrossRef]
- Mayer, S.V.; Tesh, R.B.; Vasilakis, N. The emergence of arthropod-borne viral diseases: A global prospective on dengue, chikungunya and zika fevers. Acta Trop. 2017, 166, 155–163. [Google Scholar] [CrossRef]
- Diallo, D.; Sall, A.A.; Diagne, C.T.; Faye, O.; Faye, O.; Ba, Y.; Hanley, K.A.; Buenemann, M.; Weaver, S.C.; Diallo, M. Zika virus emergence in mosquitoes in southeastern senegal, 2011. PLoS ONE 2014, 9, e109442. [Google Scholar] [CrossRef]
- Faye, O.; Faye, O.; Diallo, D.; Diallo, M.; Weidmann, M.; Sall, A.A. Quantitative real-time pcr detection of zika virus and evaluation with field-caught mosquitoes. Virol. J. 2013, 10, 311. [Google Scholar] [CrossRef]
- Weinbren, M.P.; Williams, M.C. Zika virus: Further isolations in the zika area, and some studies on the strains isolated. Trans. R. Soc. Trop. Med. Hyg. 1958, 52, 263–268. [Google Scholar] [CrossRef]
- Haddow, A.J.; Williams, M.C.; Woodall, J.P.; Simpson, D.I.; Goma, L.K. Twelve isolations of zika virus from aedes (stegomyia) africanus (theobald) taken in and above a uganda forest. Bull. World Health Organ. 1964, 31, 57–69. [Google Scholar] [PubMed]
- Berthet, N.; Nakouné, E.; Kamgang, B.; Selekon, B.; Descorps-Declère, S.; Gessain, A.; Manuguerra, J.C.; Kazanji, M. Molecular characterization of three zika flaviviruses obtained from sylvatic mosquitoes in the central african republic. Vector Borne Zoonotic Dis. 2014, 14, 862–865. [Google Scholar] [CrossRef] [PubMed]
- McCrae, A.W.; Kirya, B.G. Yellow fever and zika virus epizootics and enzootics in uganda. Trans. R. Soc. Trop. Med. Hyg. 1982, 76, 552–562. [Google Scholar] [CrossRef]
- Grard, G.; Caron, M.; Mombo, I.M.; Nkoghe, D.; Mboui Ondo, S.; Jiolle, D.; Fontenille, D.; Paupy, C.; Leroy, E.M. Zika virus in gabon (central africa)--2007: A new threat from aedes albopictus? PLoS Negl. Trop. Dis. 2014, 8, e2681. [Google Scholar] [CrossRef]
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Enfissi, A.; Codrington, J.; Roosblad, J.; Kazanji, M.; Rousset, D. Zika virus genome from the americas. Lancet 2016, 387, 227–228. [Google Scholar] [CrossRef]
- Haddow, A.D.; Woodall, J.P. Distinguishing between zika and spondweni viruses. Bull. World Health Organ. 2016, 94, 711. [Google Scholar] [CrossRef]
- Brès, P. Recent data from serological surveys on the prevalence of arbovirus infections in africa, with special reference to yellow fever. Bull. World Health Organ. 1970, 43, 223–267. [Google Scholar]
- Macnamara, F.N. Zika virus: A report on three cases of human infection during an epidemic of jaundice in nigeria. Trans. R. Soc. Trop. Med. Hyg. 1954, 48, 139–145. [Google Scholar] [CrossRef]
- Olson, J.G.; Ksiazek, T.G.; Suhandiman; Triwibowo. Zika virus, a cause of fever in central java, indonesia. Trans. R. Soc. Trop. Med. Hyg. 1981, 75, 389–393. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Lambert, A.J.; Holodniy, M.; Saavedra, S.; Signor Ldel, C. Phylogeny of zika virus in western hemisphere, 2015. Emerg. Infect. Dis. 2016, 22, 933–935. [Google Scholar] [CrossRef]
- Faye, O.; Freire, C.C.; Iamarino, A.; Faye, O.; de Oliveira, J.V.; Diallo, M.; Zanotto, P.M.; Sall, A.A. Molecular evolution of zika virus during its emergence in the 20(th) century. PLoS Negl. Trop. Dis. 2014, 8, e2636. [Google Scholar] [CrossRef]
- Collins, N.D.; Widen, S.G.; Li, L.; Swetnam, D.M.; Shi, P.Y.; Tesh, R.B.; Sarathy, V.V. Inter- and intra-lineage genetic diversity of wild-type zika viruses reveals both common and distinctive nucleotide variants and clusters of genomic diversity. Emerg. Microbes Infect. 2019, 8, 1126–1138. [Google Scholar] [CrossRef]
- Ye, Q.; Liu, Z.Y.; Han, J.F.; Jiang, T.; Li, X.F.; Qin, C.F. Genomic characterization and phylogenetic analysis of zika virus circulating in the americas. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 43, 43–49. [Google Scholar] [CrossRef]
- Pettersson, J.H.; Eldholm, V.; Seligman, S.J.; Lundkvist, Å.; Falconar, A.K.; Gaunt, M.W.; Musso, D.; Nougairède, A.; Charrel, R.; Gould, E.A.; et al. How did zika virus emerge in the pacific islands and latin america? mBio 2016, 7. [Google Scholar] [CrossRef]
- Heinz, F.X.; Stiasny, K. The antigenic structure of zika virus and its relation to other flaviviruses: Implications for infection and immunoprophylaxis. Microbiol. Mol. Biol. Rev. Mmbr 2017, 81, e00055-16. [Google Scholar] [CrossRef]
- Gatherer, D.; Kohl, A. Zika virus: A previously slow pandemic spreads rapidly through the americas. J. Gen. Virol. 2016, 97, 269–273. [Google Scholar] [CrossRef]
- Duffy, M.R.; Chen, T.H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika virus outbreak on yap island, federated states of micronesia. N. Engl. J. Med. 2009, 360, 2536–2543. [Google Scholar] [CrossRef]
- Musso, D.; Nilles, E.J.; Cao-Lormeau, V.M. Rapid spread of emerging zika virus in the pacific area. Clin. Microbiol. Infect. 2014, 20, O595–O596. [Google Scholar] [CrossRef]
- Brito, C.A.; Cordeiro, M.T. One year after the zika virus outbreak in brazil: From hypotheses to evidence. Rev. Da Soc. Bras. De Med. Trop. 2016, 49, 537–543. [Google Scholar] [CrossRef]
- Brasil, P.; Calvet, G.A.; Siqueira, A.M.; Wakimoto, M.; de Sequeira, P.C.; Nobre, A.; Quintana Mde, S.; Mendonça, M.C.; Lupi, O.; de Souza, R.V.; et al. Zika virus outbreak in rio de janeiro, brazil: Clinical characterization, epidemiological and virological aspects. PLoS Negl. Trop. Dis. 2016, 10, e0004636. [Google Scholar] [CrossRef]
- Ikejezie, J.; Shapiro, C.N.; Kim, J.; Chiu, M.; Almiron, M.; Ugarte, C.; Espinal, M.A.; Aldighieri, S. Zika virus transmission—Region of the americas, 15 may 2015–15 december 2016. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 329–334. [Google Scholar] [CrossRef]
- White, M.K.; Wollebo, H.S.; David Beckham, J.; Tyler, K.L.; Khalili, K. Zika virus: An emergent neuropathological agent. Ann. Neurol. 2016, 80, 479–489. [Google Scholar] [CrossRef]
- Haddow, A.D.; Nasar, F.; Guzman, H.; Ponlawat, A.; Jarman, R.G.; Tesh, R.B.; Weaver, S.C. Genetic characterization of spondweni and zika viruses and susceptibility of geographically distinct strains of aedes aegypti, aedes albopictus and culex quinquefasciatus (diptera: Culicidae) to spondweni virus. PLoS Negl. Trop. Dis. 2016, 10, e0005083. [Google Scholar] [CrossRef] [PubMed]
- Boyer, S.; Calvez, E.; Chouin-Carneiro, T.; Diallo, D.; Failloux, A.B. An overview of mosquito vectors of zika virus. Microbes Infect. 2018, 20, 646–660. [Google Scholar] [CrossRef]
- Weaver, S.C.; Barrett, A.D. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2004, 2, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J. Dengue, urbanization and globalization: The unholy trinity of the 21(st) century. Trop. Med. Health 2011, 39, 3–11. [Google Scholar] [CrossRef]
- Imperato, P.J. The convergence of a virus, mosquitoes, and human travel in globalizing the zika epidemic. J. Community Health 2016, 41, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Cao-Lormeau, V.M.; Roche, C.; Teissier, A.; Robin, E.; Berry, A.L.; Mallet, H.P.; Sall, A.A.; Musso, D. Zika virus, french polynesia, south pacific, 2013. Emerg. Infect. Dis. 2014, 20, 1085–1086. [Google Scholar] [CrossRef]
- Musso, D. Zika virus transmission from french polynesia to brazil. Emerg. Infect. Dis. 2015, 21, 1887. [Google Scholar] [CrossRef]
- Zanluca, C.; Melo, V.C.; Mosimann, A.L.; Santos, G.I.; Santos, C.N.; Luz, K. First report of autochthonous transmission of zika virus in brazil. Mem. Do Inst. Oswaldo Cruz 2015, 110, 569–572. [Google Scholar] [CrossRef]
- Campos, G.S.; Bandeira, A.C.; Sardi, S.I. Zika virus outbreak, bahia, brazil. Emerg. Infect. Dis. 2015, 21, 1885–1886. [Google Scholar] [CrossRef] [PubMed]
- Paixão, E.S.; Teixeira, M.G.; Rodrigues, L.C. Zika, chikungunya and dengue: The causes and threats of new and re-emerging arboviral diseases. BMJ Glob. Health 2018, 3, e000530. [Google Scholar] [CrossRef] [PubMed]
- Eligio-García, L.; Crisóstomo-Vázquez, M.D.P.; Caballero-García, M.L.; Soria-Guerrero, M.; Méndez-Galván, J.F.; López-Cancino, S.A.; Jiménez-Cardoso, E. Co-infection of dengue, zika and chikungunya in a group of pregnant women from tuxtla gutiérrez, chiapas: Preliminary data. 2019. PLoS Negl. Trop. Dis. 2020, 14, e0008880. [Google Scholar] [CrossRef] [PubMed]
- Villamil-Gómez, W.E.; González-Camargo, O.; Rodriguez-Ayubi, J.; Zapata-Serpa, D.; Rodriguez-Morales, A.J. Dengue, chikungunya and zika co-infection in a patient from colombia. J. Infect. Public Health 2016, 9, 684–686. [Google Scholar] [CrossRef]
- Gautret, P.; Simon, F. Dengue, chikungunya and zika and mass gatherings: What happened in brazil, 2014. Travel Med. Infect. Dis. 2016, 14, 7–8. [Google Scholar] [CrossRef]
- Khandia, R.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Malik, Y.S.; Singh, R.K.; Chaicumpa, W. Modulation of dengue/zika virus pathogenicity by antibody-dependent enhancement and strategies to protect against enhancement in zika virus infection. Front. Immunol. 2018, 9, 597. [Google Scholar] [CrossRef]
- Willis, E.; Hensley, S.E. Characterization of zika virus binding and enhancement potential of a large panel of flavivirus murine monoclonal antibodies. Virology 2017, 508, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, K.; Göhner, C.; Kopp, A.; Schmidt, A.; Merz, W.M.; Markert, U.R.; Junglen, S.; Drosten, C. Zika virus infection in human placental tissue explants is enhanced in the presence of dengue virus antibodies in-vitro. Emerg. Microbes Infect. 2018, 7, 198. [Google Scholar] [CrossRef]
- Bardina, S.V.; Bunduc, P.; Tripathi, S.; Duehr, J.; Frere, J.J.; Brown, J.A.; Nachbagauer, R.; Foster, G.A.; Krysztof, D.; Tortorella, D.; et al. Enhancement of zika virus pathogenesis by preexisting antiflavivirus immunity. Science 2017, 356, 175–180. [Google Scholar] [CrossRef]
- Musso, D.; Despres, P. Serological diagnosis of flavivirus-associated human infections. Diagnostics 2020, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.P.S.; St John, A.L. Cross-reactive immunity among flaviviruses. Front. Immunol. 2020, 11, 334. [Google Scholar] [CrossRef] [PubMed]
- Rabe, I.B.; Staples, J.E.; Villanueva, J.; Hummel, K.B.; Johnson, J.A.; Rose, L.; Hills, S.; Wasley, A.; Fischer, M.; Powers, A.M. Interim guidance for interpretation of zika virus antibody test results. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 543–546. [Google Scholar] [CrossRef]
- Terzian, A.C.B.; Schanoski, A.S.; Mota, M.T.O.; da Silva, R.A.; Estofolete, C.F.; Colombo, T.E.; Rahal, P.; Hanley, K.A.; Vasilakis, N.; Kalil, J.; et al. Viral load and cytokine response profile does not support antibody-dependent enhancement in dengue-primed zika virus-infected patients. Clin. Infect. Dis. 2017, 65, 1260–1265. [Google Scholar] [CrossRef] [PubMed]
- Rossati, A. Global warming and its health impact. Int. J. Occup. Environ. Med. 2017, 8, 7–20. [Google Scholar] [CrossRef]
- Bogoch, I.I.; Brady, O.J.; Kraemer, M.U.G.; German, M.; Creatore, M.I.; Kulkarni, M.A.; Brownstein, J.S.; Mekaru, S.R.; Hay, S.I.; Groot, E.; et al. Anticipating the international spread of zika virus from brazil. Lancet 2016, 387, 335–336. [Google Scholar] [CrossRef]
- Ali, S.; Gugliemini, O.; Harber, S.; Harrison, A.; Houle, L.; Ivory, J.; Kersten, S.; Khan, R.; Kim, J.; LeBoa, C.; et al. Environmental and social change drive the explosive emergence of zika virus in the americas. PLoS Negl. Trop. Dis. 2017, 11, e0005135. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Xie, X.; Shi, P.Y. Zika virus vaccine: Progress and challenges. Cell Host Microbe 2018, 24, 12–17. [Google Scholar] [CrossRef]
- Pierson, T.C.; Graham, B.S. Zika virus: Immunity and vaccine development. Cell 2016, 167, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Barouch, D.H.; Thomas, S.J.; Michael, N.L. Prospects for a zika virus vaccine. Immunity 2017, 46, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.H.; Yip, B.S.; Huang, L.M.; Wu, S.C. Zika virus structural biology and progress in vaccine development. Biotechnol. Adv. 2018, 36, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Morabito, K.M.; Graham, B.S. Zika virus vaccine development. J. Infect. Dis. 2017, 216, S957–S963. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Guida, J.P.; Costa Do Nascimento, M.L.; Mysorekar, I.U. Host and viral mechanisms of congenital zika syndrome. Virulence 2019, 10, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Nhan, T.; Robin, E.; Roche, C.; Bierlaire, D.; Zisou, K.; Shan Yan, A.; Cao-Lormeau, V.M.; Broult, J. Potential for zika virus transmission through blood transfusion demonstrated during an outbreak in french polynesia, november 2013 to february 2014. Eur. Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2014, 19, 20761. [Google Scholar] [CrossRef] [PubMed]
- Wiwanitkit, S.; Wiwanitkit, V. Afebrile, asymptomatic and non-thrombocytopenic zika virus infection: Don’t miss it! Asian Pac. J. Trop. Med. 2016, 9, 513. [Google Scholar] [CrossRef]
- Wikan, N.; Suputtamongkol, Y.; Yoksan, S.; Smith, D.R.; Auewarakul, P. Immunological evidence of zika virus transmission in thailand. Asian Pac. J. Trop. Med. 2016, 9, 141–144. [Google Scholar] [CrossRef]
- Oduyebo, T.; Petersen, E.E.; Rasmussen, S.A.; Mead, P.S.; Meaney-Delman, D.; Renquist, C.M.; Ellington, S.R.; Fischer, M.; Staples, J.E.; Powers, A.M.; et al. Update: Interim guidelines for health care providers caring for pregnant women and women of reproductive age with possible zika virus exposure—United states, 2016. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Haby, M.M.; Pinart, M.; Elias, V.; Reveiz, L. Prevalence of asymptomatic zika virus infection: A systematic review. Bull. World Health Organ. 2018, 96, 402–413. [Google Scholar] [CrossRef]
- Grossi-Soyster, E.N.; LaBeaud, A.D. Clinical aspects of zika virus. Curr. Opin. Pediatr. 2017, 29, 102–106. [Google Scholar] [CrossRef]
- Plourde, A.R.; Bloch, E.M. A literature review of zika virus. Emerg. Infect. Dis. 2016, 22, 1185–1192. [Google Scholar] [CrossRef]
- Mercado-Reyes, M.; Acosta-Reyes, J.; Navarro-Lechuga, E.; Corchuelo, S.; Rico, A.; Parra, E.; Tolosa, N.; Pardo, L.; González, M.; Martìn-Rodriguez-Hernández, J.; et al. Dengue, chikungunya and zika virus coinfection: Results of the national surveillance during the zika epidemic in colombia. Epidemiol. Infect. 2019, 147, e77. [Google Scholar] [CrossRef]
- Chaves, B.A.; Orfano, A.S.; Nogueira, P.M.; Rodrigues, N.B.; Campolina, T.B.; Nacif-Pimenta, R.; Pires, A.; Júnior, A.B.V.; Paz, A.D.C.; Vaz, E.; et al. Coinfection with zika virus (zikv) and dengue virus results in preferential zikv transmission by vector bite to vertebrate host. J. Infect. Dis. 2018, 218, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.; Guimarães, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The brazilian zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef]
- Halai, U.A.; Nielsen-Saines, K.; Moreira, M.L.; de Sequeira, P.C.; Junior, J.P.P.; de Araujo Zin, A.; Cherry, J.; Gabaglia, C.R.; Gaw, S.L.; Adachi, K.; et al. Maternal zika virus disease severity, virus load, prior dengue antibodies, and their relationship to birth outcomes. Clin. Infect. Dis. 2017, 65, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Herrlinger, S.; Zhu, Y.N.; Yang, M.; Goodfellow, F.; Stice, S.L.; Qi, X.P.; Brindley, M.A.; Chen, J.F. The african zika virus mr-766 is more virulent and causes more severe brain damage than current asian lineage and dengue virus. Development 2017, 144, 4114–4124. [Google Scholar] [CrossRef]
- Esser-Nobis, K.; Aarreberg, L.D.; Roby, J.A.; Fairgrieve, M.R.; Green, R.; Gale, M., Jr. Comparative analysis of african and asian lineage-derived zika virus strains reveals differences in activation of and sensitivity to antiviral innate immunity. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Dowall, S.D.; Graham, V.A.; Hewson, R. Lineage-dependent differences of zika virus infection in a susceptible mouse model are associated with different profiles of cytokines, chemokines, growth factors and acute phase proteins. Cytokine 2020, 125, 154864. [Google Scholar] [CrossRef] [PubMed]
- Avilés-Vergara, P.A.; Trujillo-Correa, A.; Gómez-Suárez, L.A.; Ricardo-Caldera, D.; Soto-De León, S.C.; Brango, H.; Tovar Acero, C. Denv and zikv detection in patients with acute febrile syndrome in córdoba, colombia. Int. J. Infect. Dis. 2020, 99, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Bourdon, M.; Manet, C.; Montagutelli, X. Host genetic susceptibility to viral infections: The role of type i interferon induction. Genes Immun. 2020, 21, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Sultan, N.; Bukhari, S.A.; Ali, I.; Asif, M.; Umar, Z.; Akash, M.S.H. Zika virus: A critical analysis and pharmaceutical perspectives. Crit. Rev. Eukaryot. Gene Expr. 2018, 28, 357–371. [Google Scholar] [CrossRef]
- Osuna, C.E.; Lim, S.Y.; Deleage, C.; Griffin, B.D.; Stein, D.; Schroeder, L.T.; Omange, R.W.; Best, K.; Luo, M.; Hraber, P.T.; et al. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat. Med. 2016, 22, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, M.C.; Ribeiro, I.P.; Lima, N.S.; Dos Santos, A.A.; Menezes, L.S.; da Cruz, S.O.; de Mello, I.S.; Furtado, N.D.; de Moura, E.E.; Damasceno, L.; et al. Isolation of infective zika virus from urine and saliva of patients in brazil. PLoS Negl. Trop. Dis. 2016, 10, e0004816. [Google Scholar] [CrossRef] [PubMed]
- Sotelo, J.R.; Sotelo, A.B.; Sotelo, F.J.B.; Doi, A.M.; Pinho, J.R.R.; Oliveira, R.C.; Bezerra, A.; Deutsch, A.D.; Villas-Boas, L.S.; Felix, A.C.; et al. Persistence of zika virus in breast milk after infection in late stage of pregnancy. Emerg. Infect. Dis. 2017, 23, 856–857. [Google Scholar] [CrossRef]
- Aid, M.; Abbink, P.; Larocca, R.A.; Boyd, M.; Nityanandam, R.; Nanayakkara, O.; Martinot, A.J.; Moseley, E.T.; Blass, E.; Borducchi, E.N.; et al. Zika virus persistence in the central nervous system and lymph nodes of rhesus monkeys. Cell 2017, 169, 610–620.e614. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Armstrong, N.; Obwolo, L.A.; Thomas, M.; Pang, X.; Jones, K.S.; Tang, Q. Determination of the cell permissiveness spectrum, mode of rna replication, and rna-protein interaction of zika virus. BMC Infect. Dis. 2017, 17, 239. [Google Scholar] [CrossRef]
- Retallack, H.; Di Lullo, E.; Arias, C.; Knopp, K.A.; Laurie, M.T.; Sandoval-Espinosa, C.; Mancia Leon, W.R.; Krencik, R.; Ullian, E.M.; Spatazza, J.; et al. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. USA 2016, 113, 14408–14413. [Google Scholar] [CrossRef] [PubMed]
- Meertens, L.; Labeau, A.; Dejarnac, O.; Cipriani, S.; Sinigaglia, L.; Bonnet-Madin, L.; Le Charpentier, T.; Hafirassou, M.L.; Zamborlini, A.; Cao-Lormeau, V.M.; et al. Axl mediates zika virus entry in human glial cells and modulates innate immune responses. Cell Rep. 2017, 18, 324–333. [Google Scholar] [CrossRef]
- Aagaard, K.M.; Lahon, A.; Suter, M.A.; Arya, R.P.; Seferovic, M.D.; Vogt, M.B.; Hu, M.; Stossi, F.; Mancini, M.A.; Harris, R.A.; et al. Primary human placental trophoblasts are permissive for zika virus (zikv) replication. Sci. Rep. 2017, 7, 41389. [Google Scholar] [CrossRef]
- Tabata, T.; Petitt, M.; Puerta-Guardo, H.; Michlmayr, D.; Wang, C.; Fang-Hoover, J.; Harris, E.; Pereira, L. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe 2016, 20, 155–166. [Google Scholar] [CrossRef]
- Chen, J.C.; Wang, Z.; Huang, H.; Weitz, S.H.; Wang, A.; Qiu, X.; Baumeister, M.A.; Uzgiris, A. Infection of human uterine fibroblasts by zika virus in vitro: Implications for viral transmission in women. Int. J. Infect. Dis. 2016, 51, 139–140. [Google Scholar] [CrossRef]
- Miner, J.J.; Diamond, M.S. Zika virus pathogenesis and tissue tropism. Cell Host Microbe 2017, 21, 134–142. [Google Scholar] [CrossRef]
- Ngono, A.E.; Shresta, S. Immune response to dengue and zika. Annu. Rev. Immunol. 2018, 36, 279–308. [Google Scholar] [CrossRef] [PubMed]
- Wiwanitkit, V. Eye problem in zika virus infection. Eur. J. Intern. Med. 2018, 47, e17. [Google Scholar] [CrossRef]
- Carson, M.J.; Doose, J.M.; Melchior, B.; Schmid, C.D.; Ploix, C.C. Cns immune privilege: Hiding in plain sight. Immunol. Rev. 2006, 213, 48–65. [Google Scholar] [CrossRef] [PubMed]
- Russo, F.B.; Jungmann, P.; Beltrão-Braga, P.C.B. Zika infection and the development of neurological defects. Cell. Microbiol. 2017, 19, e12744. [Google Scholar] [CrossRef]
- Serman, T.M.; Gack, M.U. Evasion of innate and intrinsic antiviral pathways by the zika virus. Viruses 2019, 11, 970. [Google Scholar] [CrossRef]
- Tonnerre, P.; Melgaço, J.G.; Torres-Cornejo, A.; Pinto, M.A.; Yue, C.; Blümel, J.; de Sousa, P.S.F.; de Mello, V.D.M.; Moran, J.; de Filippis, A.M.B.; et al. Evolution of the innate and adaptive immune response in women with acute zika virus infection. Nat. Microbiol. 2020, 5, 76–83. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Meylan, E.; Tschopp, J.; Karin, M. Intracellular pattern recognition receptors in the host response. Nature 2006, 442, 39–44. [Google Scholar] [CrossRef]
- Suthar, M.S.; Aguirre, S.; Fernandez-Sesma, A. Innate immune sensing of flaviviruses. PLoS Pathog. 2013, 9, e1003541. [Google Scholar] [CrossRef]
- Sirohi, D.; Kuhn, R.J. Zika virus structure, maturation, and receptors. J. Infect. Dis. 2017, 216, S935–s944. [Google Scholar] [CrossRef] [PubMed]
- Valente, A.P.; Moraes, A.H. Zika virus proteins at an atomic scale: How does structural biology help us to understand and develop vaccines and drugs against zika virus infection? J. Venom. Anim. Toxins Incl. Trop. Dis. 2019, 25, e20190013. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Kuhn, R.J.; Rossmann, M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005, 3, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.L.; Jones, C.T.; Rice, C.M. Architects of assembly: Roles of flaviviridae non-structural proteins in virion morphogenesis. Nat. Rev. Microbiol. 2008, 6, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Agrelli, A.; de Moura, R.R.; Crovella, S.; Brandão, L.A.C. Zika virus entry mechanisms in human cells. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 69, 22–29. [Google Scholar] [CrossRef]
- Best, S.M. The many faces of the flavivirus ns5 protein in antagonism of type i interferon signaling. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus ns1: A multifaceted enigmatic viral protein. Virol. J. 2016, 13, 131. [Google Scholar] [CrossRef]
- Nunes, B.T.D.; Fontes-Garfias, C.R.; Shan, C.; Muruato, A.E.; Nunes, J.G.C.; Burbano, R.M.R.; Vasconcelos, P.F.C.; Shi, P.Y.; Medeiros, D.B.A. Zika structural genes determine the virulence of african and asian lineages. Emerg. Microbes Infect. 2020, 9, 1023–1033. [Google Scholar] [CrossRef]
- Xu, H.; Cheng, M.; Chi, X.; Liu, X.; Zhou, J.; Lin, T.; Yang, W. High-throughput screening identifies mixed-lineage kinase 3 as a key host regulatory factor in zika virus infection. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Shan, C.; Xie, X.; Muruato, A.E.; Rossi, S.L.; Roundy, C.M.; Azar, S.R.; Yang, Y.; Tesh, R.B.; Bourne, N.; Barrett, A.D.; et al. An infectious cdna clone of zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe 2016, 19, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liao, H.M.; Li, B.; Tsai, S.; Hung, G.C.; Lo, S.C. Comparative genomics, infectivity and cytopathogenicity of american isolates of zika virus that developed persistent infections in human embryonic kidney (hek293) cells. Int. J. Mol. Sci. 2019, 20, 3035. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of mda5 and rig-i helicases in the recognition of rna viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Hamel, R.; Dejarnac, O.; Wichit, S.; Ekchariyawat, P.; Neyret, A.; Luplertlop, N.; Perera-Lecoin, M.; Surasombatpattana, P.; Talignani, L.; Thomas, F.; et al. Biology of zika virus infection in human skin cells. J. Virol. 2015, 89, 8880–8896. [Google Scholar] [CrossRef] [PubMed]
- Hiscott, J.; Grandvaux, N.; Sharma, S.; Tenoever, B.R.; Servant, M.J.; Lin, R. Convergence of the nf-kappab and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 2003, 1010, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Chevaliez, S.; Pawlotsky, J.M. Interferons and their use in persistent viral infections. Handb. Exp. Pharmacol. 2009, 189, 203–241. [Google Scholar]
- De Veer, M.J.; Holko, M.; Frevel, M.; Walker, E.; Der, S.; Paranjape, J.M.; Silverman, R.H.; Williams, B.R. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001, 69, 912–920. [Google Scholar]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type i interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Singh, P.K.; Singh, S.; Farr, D.; Kumar, A. Interferon-stimulated gene 15 (isg15) restricts zika virus replication in primary human corneal epithelial cells. Ocul. Surf. 2019, 17, 551–559. [Google Scholar] [CrossRef]
- Dukhovny, A.; Lamkiewicz, K.; Chen, Q.; Fricke, M.; Jabrane-Ferrat, N.; Marz, M.; Jung, J.U.; Sklan, E.H. A crispr activation screen identifies genes that protect against zika virus infection. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Wichit, S.; Hamel, R.; Yainoy, S.; Gumpangseth, N.; Panich, S.; Phuadraksa, T.; Saetear, P.; Monteil, A.; Morales Vargas, R.; Missé, D. Interferon-inducible protein (ifi) 16 regulates chikungunya and zika virus infection in human skin fibroblasts. EXCLI J. 2019, 18, 467–476. [Google Scholar] [PubMed]
- Zhang, D.; Zhang, D.E. Interferon-stimulated gene 15 and the protein isgylation system. J. Interferon Cytokine Res. 2011, 31, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ren, K.; Li, S.; Yang, C.; Chen, L. Interferon stimulated gene 15 promotes zika virus replication through regulating jak/stat and isgylation pathways. Virus Res. 2020, 287, 198087. [Google Scholar] [CrossRef]
- Ojha, C.R.; Rodriguez, M.; Karuppan, M.K.M.; Lapierre, J.; Kashanchi, F.; El-Hage, N. Toll-like receptor 3 regulates zika virus infection and associated host inflammatory response in primary human astrocytes. PLoS ONE 2019, 14, e0208543. [Google Scholar] [CrossRef] [PubMed]
- Vanwalscappel, B.; Tada, T.; Landau, N.R. Toll-like receptor agonist r848 blocks zika virus replication by inducing the antiviral protein viperin. Virology 2018, 522, 199–208. [Google Scholar] [CrossRef]
- Khan, S.; Lew, I.; Wu, F.; Fritts, L.; Fontaine, K.A.; Tomar, S.; Trapecar, M.; Shehata, H.M.; Ott, M.; Miller, C.J.; et al. Low expression of rna sensors impacts zika virus infection in the lower female reproductive tract. Nat. Commun. 2019, 10, 4344. [Google Scholar] [CrossRef] [PubMed]
- Ledur, P.F.; Karmirian, K.; Pedrosa, C.; Souza, L.R.Q.; Assis-de-Lemos, G.; Martins, T.M.; Ferreira, J.; de Azevedo Reis, G.F.; Silva, E.S.; Silva, D.; et al. Zika virus infection leads to mitochondrial failure, oxidative stress and DNA damage in human ipsc-derived astrocytes. Sci. Rep. 2020, 10, 1218. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, Q.; Wu, Y.; Ma, L.; Zhang, Z.; Liu, T.; Jin, S.; She, Y.; Li, Y.P.; Cui, J. Zika virus elicits inflammation to evade antiviral response by cleaving cgas via ns1-caspase-1 axis. Embo J. 2018, 37, e99347. [Google Scholar] [CrossRef]
- Coldbeck-Shackley, R.C.; Eyre, N.S.; Beard, M.R. The molecular interactions of zikv and denv with the type-i ifn response. Vaccines 2020, 8, 530. [Google Scholar] [CrossRef]
- Bowen, J.R.; Zimmerman, M.G.; Suthar, M.S. Taking the defensive: Immune control of zika virus infection. Virus Res. 2018, 254, 21–26. [Google Scholar] [CrossRef] [PubMed]
- De Weerd, N.A.; Nguyen, T. The interferons and their receptors--distribution and regulation. Immunol. Cell Biol. 2012, 90, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type i interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Darnell, J.E., Jr. The jak-stat pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef]
- MacMicking, J.D. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 2012, 12, 367–382. [Google Scholar] [CrossRef]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Bieniasz, P.; Rice, C.M. A diverse range of gene products are effectors of the type i interferon antiviral response. Nature 2011, 472, 481–485. [Google Scholar] [CrossRef]
- Rusinova, I.; Forster, S.; Yu, S.; Kannan, A.; Masse, M.; Cumming, H.; Chapman, R.; Hertzog, P.J. Interferome v2.0: An updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013, 41, D1040–D1046. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Govero, J.; Smith, A.M.; Platt, D.J.; Fernandez, E.; Miner, J.J.; Diamond, M.S. A mouse model of zika virus pathogenesis. Cell Host Microbe 2016, 19, 720–730. [Google Scholar] [CrossRef]
- Frumence, E.; Roche, M.; Krejbich-Trotot, P.; El-Kalamouni, C.; Nativel, B.; Rondeau, P.; Missé, D.; Gadea, G.; Viranaicken, W.; Desprès, P. The south pacific epidemic strain of zika virus replicates efficiently in human epithelial a549 cells leading to ifn-β production and apoptosis induction. Virology 2016, 493, 217–226. [Google Scholar] [CrossRef]
- Bayer, A.; Lennemann, N.J.; Ouyang, Y.; Bramley, J.C.; Morosky, S.; Marques, E.T., Jr.; Cherry, S.; Sadovsky, Y.; Coyne, C.B. Type iii interferons produced by human placental trophoblasts confer protection against zika virus infection. Cell Host Microbe 2016, 19, 705–712. [Google Scholar] [CrossRef]
- Savidis, G.; Perreira, J.M.; Portmann, J.M.; Meraner, P.; Guo, Z.; Green, S.; Brass, A.L. The ifitms inhibit zika virus replication. Cell Rep. 2016, 15, 2323–2330. [Google Scholar] [CrossRef]
- Goubau, D.; Deddouche, S.; Reis e Sousa, C. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef]
- Bowen, J.R.; Quicke, K.M.; Maddur, M.S.; O’Neal, J.T.; McDonald, C.E.; Fedorova, N.B.; Puri, V.; Shabman, R.S.; Pulendran, B.; Suthar, M.S. Zika virus antagonizes type i interferon responses during infection of human dendritic cells. PLoS Pathog. 2017, 13, e1006164. [Google Scholar] [CrossRef]
- Marzi, A.; Emanuel, J.; Callison, J.; McNally, K.L.; Arndt, N.; Chadinha, S.; Martellaro, C.; Rosenke, R.; Scott, D.P.; Safronetz, D.; et al. Lethal zika virus disease models in young and older interferon α/β receptor knock out mice. Front. Cell. Infect. Microbiol. 2018, 8, 117. [Google Scholar] [CrossRef]
- Metz, S.W.; Thomas, A.; Brackbill, A.; Forsberg, J.; Miley, M.J.; Lopez, C.A.; Lazear, H.M.; Tian, S.; de Silva, A.M. Oligomeric state of the zikv e protein defines protective immune responses. Nat. Commun. 2019, 10, 4606. [Google Scholar] [CrossRef]
- Kim, S.Y.; Zhao, J.; Liu, X.; Fraser, K.; Lin, L.; Zhang, X.; Zhang, F.; Dordick, J.S.; Linhardt, R.J. Interaction of zika virus envelope protein with glycosaminoglycans. Biochemistry 2017, 56, 1151–1162. [Google Scholar] [CrossRef]
- Strange, D.P.; Jiyarom, B.; Pourhabibi Zarandi, N.; Xie, X.; Baker, C.; Sadri-Ardekani, H.; Shi, P.Y.; Verma, S. Axl promotes zika virus entry and modulates the antiviral state of human sertoli cells. mBio 2019, 10. [Google Scholar] [CrossRef]
- Di Scipio, R.G.; Hermodson, M.A.; Yates, S.G.; Davie, E.W. A comparison of human prothrombin, factor ix (christmas factor), factor x (stuart factor), and protein s. Biochemistry 1977, 16, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Stitt, T.N.; Conn, G.; Gore, M.; Lai, C.; Bruno, J.; Radziejewski, C.; Mattsson, K.; Fisher, J.; Gies, D.R.; Jones, P.F.; et al. The anticoagulation factor protein s and its relative, gas6, are ligands for the tyro 3/axl family of receptor tyrosine kinases. Cell 1995, 80, 661–670. [Google Scholar] [CrossRef]
- Amara, A.; Mercer, J. Viral apoptotic mimicry. Nat. Rev. Microbiol. 2015, 13, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Richard, A.S.; Shim, B.S.; Kwon, Y.C.; Zhang, R.; Otsuka, Y.; Schmitt, K.; Berri, F.; Diamond, M.S.; Choe, H. Axl-dependent infection of human fetal endothelial cells distinguishes zika virus from other pathogenic flaviviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 2024–2029. [Google Scholar] [CrossRef] [PubMed]
- Lemke, G. Biology of the tam receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a009076. [Google Scholar] [CrossRef]
- In, H.J.; Lee, Y.H.; Jang, S.; Lim, H.J.; Kim, M.Y.; Kim, J.A.; Yoo, J.S.; Chung, G.T.; Kim, Y.J. Enhanced effect of modified zika virus e antigen on the immunogenicity of DNA vaccine. Virology 2020, 549, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Larocca, R.A.; Abbink, P.; Peron, J.P.; Zanotto, P.M.; Iampietro, M.J.; Badamchi-Zadeh, A.; Boyd, M.; Ng’ang’a, D.; Kirilova, M.; Nityanandam, R.; et al. Vaccine protection against zika virus from brazil. Nature 2016, 536, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Abbink, P.; Larocca, R.A.; De La Barrera, R.A.; Bricault, C.A.; Moseley, E.T.; Boyd, M.; Kirilova, M.; Li, Z.; Ng’ang’a, D.; Nanayakkara, O.; et al. Protective efficacy of multiple vaccine platforms against zika virus challenge in rhesus monkeys. Science 2016, 353, 1129–1132. [Google Scholar] [CrossRef]
- López-Camacho, C.; Abbink, P.; Larocca, R.A.; Dejnirattisai, W.; Boyd, M.; Badamchi-Zadeh, A.; Wallace, Z.R.; Doig, J.; Velazquez, R.S.; Neto, R.D.L.; et al. Rational zika vaccine design via the modulation of antigen membrane anchors in chimpanzee adenoviral vectors. Nat. Commun. 2018, 9, 2441. [Google Scholar] [CrossRef] [PubMed]
- Sager, G.; Gabaglio, S.; Sztul, E.; Belov, G.A. Role of host cell secretory machinery in zika virus life cycle. Viruses 2018, 10, 559. [Google Scholar] [CrossRef]
- Xia, H.; Luo, H.; Shan, C.; Muruato, A.E.; Nunes, B.T.D.; Medeiros, D.B.A.; Zou, J.; Xie, X.; Giraldo, M.I.; Vasconcelos, P.F.C.; et al. An evolutionary ns1 mutation enhances zika virus evasion of host interferon induction. Nat. Commun. 2018, 9, 414. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, Q.; Zhou, J.; Xie, W.; Chen, C.; Wang, Z.; Yang, H.; Cui, J. Zika virus evades interferon-mediated antiviral response through the co-operation of multiple nonstructural proteins in vitro. Cell Discov. 2017, 3, 17006. [Google Scholar] [CrossRef]
- Ngueyen, T.T.N.; Kim, S.J.; Lee, J.Y.; Myoung, J. Zika virus proteins ns2a and ns4a are major antagonists that reduce ifn-β promoter activity induced by the mda5/rig-i signaling pathway. J. Microbiol. Biotechnol. 2019, 29, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Gale, M., Jr. Immune signaling by rig-i-like receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef]
- Ding, Q.; Gaska, J.M.; Douam, F.; Wei, L.; Kim, D.; Balev, M.; Heller, B.; Ploss, A. Species-specific disruption of sting-dependent antiviral cellular defenses by the zika virus ns2b3 protease. Proc. Natl. Acad. Sci. USA 2018, 115, E6310–e6318. [Google Scholar] [CrossRef] [PubMed]
- Riedl, W.; Acharya, D.; Lee, J.H.; Liu, G.; Serman, T.; Chiang, C.; Chan, Y.K.; Diamond, M.S.; Gack, M.U. Zika virus ns3 mimics a cellular 14-3-3-binding motif to antagonize rig-i- and mda5-mediated innate immunity. Cell Host Microbe 2019, 26, 493–503.e496. [Google Scholar] [CrossRef]
- Lin, J.P.; Fan, Y.K.; Liu, H.M. The 14-3-3η chaperone protein promotes antiviral innate immunity via facilitating mda5 oligomerization and intracellular redistribution. PLoS Pathog. 2019, 15, e1007582. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.M.; Loo, Y.M.; Horner, S.M.; Zornetzer, G.A.; Katze, M.G.; Gale, M., Jr. The mitochondrial targeting chaperone 14-3-3ε regulates a rig-i translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe 2012, 11, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Salazar, M.; Gutiérrez-Escolano, A.L.; Reyes-Ruiz, J.M.; del Angel, R.M. The nonstructural proteins 3 and 5 from flavivirus modulate nuclear-cytoplasmic transport and innate immune response targeting nuclear proteins. bioRxiv 2018, 375899. [Google Scholar] [CrossRef]
- Ma, J.; Ketkar, H.; Geng, T.; Lo, E.; Wang, L.; Xi, J.; Sun, Q.; Zhu, Z.; Cui, Y.; Yang, L.; et al. Zika virus non-structural protein 4a blocks the rlr-mavs signaling. Front. Microbiol. 2018, 9, 1350. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Hou, S.; Airo, A.M.; Limonta, D.; Mancinelli, V.; Branton, W.; Power, C.; Hobman, T.C. Zika virus inhibits type-i interferon production and downstream signaling. Embo Rep. 2016, 17, 1766–1775. [Google Scholar] [CrossRef]
- Hu, Y.; Dong, X.; He, Z.; Wu, Y.; Zhang, S.; Lin, J.; Yang, Y.; Chen, J.; An, S.; Yin, Y.; et al. Zika virus antagonizes interferon response in patients and disrupts rig-i-mavs interaction through its card-tm domains. Cell Biosci. 2019, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Chan, J.F.; Tee, K.M.; Choi, G.K.; Lau, S.K.; Woo, P.C.; Tse, H.; Yuen, K.Y. Comparative genomic analysis of pre-epidemic and epidemic zika virus strains for virological factors potentially associated with the rapidly expanding epidemic. Emerg. Microbes Infect. 2016, 5, e22. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Thurmond, S.; Hai, R.; Song, J. Structure and function of zika virus ns5 protein: Perspectives for drug design. Cell. Mol. Life Sci. 2018, 75, 1723–1736. [Google Scholar] [CrossRef]
- Elshahawi, H.; Syed Hassan, S.; Balasubramaniam, V. Importance of zika virus ns5 protein for viral replication. Pathogens 2019, 8, 169. [Google Scholar] [CrossRef]
- Ng, I.H.W.; Chan, K.W.; Tan, M.J.A.; Gwee, C.P.; Smith, K.M.; Jeffress, S.J.; Saw, W.G.; Swarbrick, C.M.D.; Watanabe, S.; Jans, D.A.; et al. Zika virus ns5 forms supramolecular nuclear bodies that sequester importin-α and modulate the host immune and pro-inflammatory response in neuronal cells. Acs Infect. Dis. 2019, 5, 932–948. [Google Scholar] [CrossRef]
- Tan, M.J.A.; Chan, K.W.K.; Ng, I.H.W.; Kong, S.Y.Z.; Gwee, C.P.; Watanabe, S.; Vasudevan, S.G. The potential role of the zikv ns5 nuclear spherical-shell structures in cell type-specific host immune modulation during zikv infection. Cells 2019, 8, 1519. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Wang, W.; Wang, Y.; Chen, K.; Xiao, F.; Hu, D.; Hui, L.; Liu, W.; Feng, Y.; Li, G.; et al. Ns5 conservative site is required for zika virus to restrict the rig-i signaling. Front. Immunol. 2020, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, R.; Melén, K.; Westenius, V.; Jiang, M.; Österlund, P.; Khan, H.; Vapalahti, O.; Julkunen, I.; Kakkola, L. Zika virus non-structural protein ns5 inhibits the rig-i pathway and interferon lambda 1 promoter activation by targeting ikk epsilon. Viruses 2019, 11, 1024. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yang, S.; He, J.; Guest, J.D.; Ma, Z.; Yang, L.; Pierce, B.G.; Tang, Q.; Zhang, Y.J. Zika virus ns5 protein antagonizes type i interferon production via blocking tbk1 activation. Virology 2019, 527, 180–187. [Google Scholar] [CrossRef]
- Grant, A.; Ponia, S.S.; Tripathi, S.; Balasubramaniam, V.; Miorin, L.; Sourisseau, M.; Schwarz, M.C.; Sánchez-Seco, M.P.; Evans, M.J.; Best, S.M.; et al. Zika virus targets human stat2 to inhibit type i interferon signaling. Cell Host Microbe 2016, 19, 882–890. [Google Scholar] [CrossRef]
- Hertzog, J.; Dias Junior, A.G.; Rigby, R.E.; Donald, C.L.; Mayer, A.; Sezgin, E.; Song, C.; Jin, B.; Hublitz, P.; Eggeling, C.; et al. Infection with a brazilian isolate of zika virus generates rig-i stimulatory rna and the viral ns5 protein blocks type i ifn induction and signaling. Eur. J. Immunol. 2018, 48, 1120–1136. [Google Scholar] [CrossRef]
- Zhao, Z.; Tao, M.; Han, W.; Fan, Z.; Imran, M.; Cao, S.; Ye, J. Nuclear localization of zika virus ns5 contributes to suppression of type i interferon production and response. J. Gen. Virol. 2019. [Google Scholar] [CrossRef]
- Conde, J.N.; Schutt, W.R.; Mladinich, M.; Sohn, S.Y.; Hearing, P.; Mackow, E.R. Ns5 sumoylation directs nuclear responses that permit zika virus to persistently infect human brain microvascular endothelial cells. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Göertz, G.P.; Abbo, S.R.; Fros, J.J.; Pijlman, G.P. Functional rna during zika virus infection. Virus Res. 2018, 254, 41–53. [Google Scholar] [CrossRef]
- Kümmerer, B.M. Establishment and application of flavivirus replicons. Adv. Exp. Med. Biol. 2018, 1062, 165–173. [Google Scholar] [PubMed]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; Torres, S.; van der Aa, L.; Liu, W.J.; Palmenberg, A.C.; Shi, P.Y.; Hall, R.A.; et al. A highly structured, nuclease-resistant, noncoding rna produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef]
- Silva, P.A.; Pereira, C.F.; Dalebout, T.J.; Spaan, W.J.; Bredenbeek, P.J. An rna pseudoknot is required for production of yellow fever virus subgenomic rna by the host nuclease xrn1. J. Virol. 2010, 84, 11395–11406. [Google Scholar] [CrossRef]
- Moon, S.L.; Anderson, J.R.; Kumagai, Y.; Wilusz, C.J.; Akira, S.; Khromykh, A.A.; Wilusz, J. A noncoding rna produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease xrn1 and alters host mrna stability. RNA 2012, 18, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- Funk, A.; Truong, K.; Nagasaki, T.; Torres, S.; Floden, N.; Balmori Melian, E.; Edmonds, J.; Dong, H.; Shi, P.Y.; Khromykh, A.A. Rna structures required for production of subgenomic flavivirus rna. J. Virol. 2010, 84, 11407–11417. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, B.M.; Laurence, H.M.; Massey, A.R.; Costantino, D.A.; Xie, X.; Yang, Y.; Shi, P.Y.; Nix, J.C.; Beckham, J.D.; Kieft, J.S. Zika virus produces noncoding rnas using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 2016, 354, 1148–1152. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Carballeda, J.M.; Villordo, S.M.; Aguirre, S.; Pallarés, H.M.; Maestre, A.M.; Sánchez-Vargas, I.; Blair, C.D.; Fabri, C.; Morales, M.A.; et al. Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding rnas and fitness in human cells. PLoS Pathog. 2017, 13, e1006265. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Carballeda, J.M.; Filomatori, C.V.; Gamarnik, A.V. Rna structure duplications and flavivirus host adaptation. Trends Microbiol. 2016, 24, 270–283. [Google Scholar] [CrossRef]
- Donald, C.L.; Brennan, B.; Cumberworth, S.L.; Rezelj, V.V.; Clark, J.J.; Cordeiro, M.T.; Freitas de Oliveira França, R.; Pena, L.J.; Wilkie, G.S.; Da Silva Filipe, A.; et al. Full genome sequence and sfrna interferon antagonist activity of zika virus from recife, brazil. PLoS Negl. Trop. Dis. 2016, 10, e0005048. [Google Scholar] [CrossRef]
- Shan, C.; Muruato, A.E.; Nunes, B.T.D.; Luo, H.; Xie, X.; Medeiros, D.B.A.; Wakamiya, M.; Tesh, R.B.; Barrett, A.D.; Wang, T.; et al. A live-attenuated zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat. Med. 2017, 23, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Aliota, M.T.; Caine, E.A.; Walker, E.C.; Larkin, K.E.; Camacho, E.; Osorio, J.E. Characterization of lethal zika virus infection in ag129 mice. PLoS Negl. Trop. Dis. 2016, 10, e0004682. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Shin, O.S. Advances in zika virus–host cell interaction: Current knowledge and future perspectives. Int. J. Mol. Sci. 2019, 20, 1101. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Balasubramaniam, V.R.; Brown, J.A.; Mena, I.; Grant, A.; Bardina, S.V.; Maringer, K.; Schwarz, M.C.; Maestre, A.M.; Sourisseau, M.; et al. A novel zika virus mouse model reveals strain specific differences in virus pathogenesis and host inflammatory immune responses. PLoS Pathog. 2017, 13, e1006258. [Google Scholar] [CrossRef]
- Smith, D.R.; Hollidge, B.; Daye, S.; Zeng, X.; Blancett, C.; Kuszpit, K.; Bocan, T.; Koehler, J.W.; Coyne, S.; Minogue, T.; et al. Neuropathogenesis of zika virus in a highly susceptible immunocompetent mouse model after antibody blockade of type i interferon. PLoS Negl. Trop. Dis. 2017, 11, e0005296. [Google Scholar] [CrossRef]
- Jagger, B.W.; Miner, J.J.; Cao, B.; Arora, N.; Smith, A.M.; Kovacs, A.; Mysorekar, I.U.; Coyne, C.B.; Diamond, M.S. Gestational stage and ifn-λ signaling regulate zikv infection in utero. Cell Host Microbe 2017, 22, 366–376.e363. [Google Scholar] [CrossRef]
- Casazza, R.L.; Lazear, H.M. Antiviral immunity backfires: Pathogenic effects of type i interferon signaling in fetal development. Sci. Immunol. 2018, 3, 19. [Google Scholar] [CrossRef]
- Pallarés, H.M.; Costa Navarro, G.S.; Villordo, S.M.; Merwaiss, F.; de Borba, L.; Gonzalez Lopez Ledesma, M.M.; Ojeda, D.S.; Henrion-Lacritick, A.; Morales, M.A.; Fabri, C.; et al. Zika virus subgenomic flavivirus rna generation requires cooperativity between duplicated rna structures that are essential for productive infection in human cells. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Slonchak, A.; Hugo, L.E.; Freney, M.E.; Hall-Mendelin, S.; Amarilla, A.A.; Torres, F.J.; Setoh, Y.X.; Peng, N.Y.G.; Sng, J.D.J.; Hall, R.A.; et al. Zika virus noncoding rna suppresses apoptosis and is required for virus transmission by mosquitoes. Nat. Commun. 2020, 11, 2205. [Google Scholar] [CrossRef]
- Fensterl, V.; Sen, G.C. Interferon-induced ifit proteins: Their role in viral pathogenesis. J. Virol. 2015, 89, 2462–2468. [Google Scholar] [CrossRef]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.Y.; Schneller, S.; Zust, R.; Dong, H.; et al. 2’-o methylation of the viral mrna cap evades host restriction by ifit family members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef]
- Szretter, K.J.; Daniels, B.P.; Cho, H.; Gainey, M.D.; Yokoyama, W.M.; Gale, M., Jr.; Virgin, H.W.; Klein, R.S.; Sen, G.C.; Diamond, M.S. 2′-o methylation of the viral mrna cap by west nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012, 8, e1002698. [Google Scholar] [CrossRef] [PubMed]
- Züst, R.; Dong, H.; Li, X.F.; Chang, D.C.; Zhang, B.; Balakrishnan, T.; Toh, Y.X.; Jiang, T.; Li, S.H.; Deng, Y.Q.; et al. Rational design of a live attenuated dengue vaccine: 2’-o-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog. 2013, 9, e1003521. [Google Scholar] [CrossRef] [PubMed]
- Coutard, B.; Barral, K.; Lichière, J.; Selisko, B.; Martin, B.; Aouadi, W.; Lombardia, M.O.; Debart, F.; Vasseur, J.J.; Guillemot, J.C.; et al. Zika virus methyltransferase: Structure and functions for drug design perspectives. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Fink, K.; Züst, R.; Lim, S.P.; Qin, C.F.; Shi, P.Y. Flavivirus rna methylation. J. Gen. Virol. 2014, 95, 763–778. [Google Scholar] [CrossRef]
- Egloff, M.P.; Benarroch, D.; Selisko, B.; Romette, J.L.; Canard, B. An rna cap (nucleoside-2′-o-)-methyltransferase in the flavivirus rna polymerase ns5: Crystal structure and functional characterization. Embo J. 2002, 21, 2757–2768. [Google Scholar] [CrossRef] [PubMed]
- Coloma, J.; Jain, R.; Rajashankar, K.R.; García-Sastre, A.; Aggarwal, A.K. Structures of ns5 methyltransferase from zika virus. Cell Rep. 2016, 16, 3097–3102. [Google Scholar] [CrossRef]
- Duan, W.; Song, H.; Wang, H.; Chai, Y.; Su, C.; Qi, J.; Shi, Y.; Gao, G.F. The crystal structure of zika virus ns5 reveals conserved drug targets. Embo J. 2017, 36, 919–933. [Google Scholar] [CrossRef]
- Stephen, P.; Baz, M.; Boivin, G.; Lin, S.X. Structural insight into ns5 of zika virus leading to the discovery of mtase inhibitors. J. Am. Chem. Soc. 2016, 138, 16212–16215. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Chang, D.C.; Xie, X.; Toh, Y.X.; Chung, K.Y.; Zou, G.; Lescar, J.; Lim, S.P.; Shi, P.Y. Biochemical and genetic characterization of dengue virus methyltransferase. Virology 2010, 405, 568–578. [Google Scholar] [CrossRef]
- Li, S.H.; Dong, H.; Li, X.F.; Xie, X.; Zhao, H.; Deng, Y.Q.; Wang, X.Y.; Ye, Q.; Zhu, S.Y.; Wang, H.J.; et al. Rational design of a flavivirus vaccine by abolishing viral rna 2’-o methylation. J. Virol. 2013, 87, 5812–5819. [Google Scholar] [CrossRef]
- Ray, D.; Shah, A.; Tilgner, M.; Guo, Y.; Zhao, Y.; Dong, H.; Deas, T.S.; Zhou, Y.; Li, H.; Shi, P.Y. West nile virus 5’-cap structure is formed by sequential guanine n-7 and ribose 2‘-o methylations by nonstructural protein 5. J. Virol. 2006, 80, 8362–8370. [Google Scholar] [CrossRef]
- Zhou, Y.; Ray, D.; Zhao, Y.; Dong, H.; Ren, S.; Li, Z.; Guo, Y.; Bernard, K.A.; Shi, P.Y.; Li, H. Structure and function of flavivirus ns5 methyltransferase. J. Virol. 2007, 81, 3891–3903. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. Mirna nomenclature: A view incorporating genetic origins, biosynthetic pathways, and sequence variants. Trends Genet. 2015, 31, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, M.A. Micrornas and the immune response. Trends Immunol. 2008, 29, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. Micrornas: Small rnas with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Dang, J.W.; Tiwari, S.K.; Qin, Y.; Rana, T.M. Genome-wide integrative analysis of zika-virus-infected neuronal stem cells reveals roles for micrornas in cell cycle and stemness. Cell Rep. 2019, 27, 3618–3628.e3615. [Google Scholar] [CrossRef]
- Baril, M.; Racine, M.E.; Penin, F.; Lamarre, D. Mavs dimer is a crucial signaling component of innate immunity and the target of hepatitis c virus ns3/4a protease. J. Virol. 2009, 83, 1299–1311. [Google Scholar] [CrossRef]
- Icardi, L.; Mori, R.; Gesellchen, V.; Eyckerman, S.; De Cauwer, L.; Verhelst, J.; Vercauteren, K.; Saelens, X.; Meuleman, P.; Leroux-Roels, G.; et al. The sin3a repressor complex is a master regulator of stat transcriptional activity. Proc. Natl. Acad. Sci. USA 2012, 109, 12058–12063. [Google Scholar] [CrossRef]
- Kozak, R.A.; Majer, A.; Biondi, M.J.; Medina, S.J.; Goneau, L.W.; Sajesh, B.V.; Slota, J.A.; Zubach, V.; Severini, A.; Safronetz, D.; et al. Microrna and mrna dysregulation in astrocytes infected with zika virus. Viruses 2017, 9, 297. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Sen, U.; Vrati, S. Regulated ire1-dependent decay pathway is activated during japanese encephalitis virus-induced unfolded protein response and benefits viral replication. J. Gen. Virol. 2014, 95, 71–79. [Google Scholar] [CrossRef]
- Liang, Q.; Luo, Z.; Zeng, J.; Chen, W.; Foo, S.S.; Lee, S.A.; Ge, J.; Wang, S.; Goldman, S.A.; Zlokovic, B.V.; et al. Zika virus ns4a and ns4b proteins deregulate akt-mtor signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 2016, 19, 663–671. [Google Scholar] [CrossRef]
- Hetz, C.; Papa, F.R. The unfolded protein response and cell fate control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Zhang, W.; Sun, J.; Fu, Z.; Ke, X.; Zheng, C.; Zhang, Y.; Li, P.; Liu, Y.; Hu, Q.; et al. Zikv infection activates the ire1-xbp1 and atf6 pathways of unfolded protein response in neural cells. J. Neuroinflammation 2018, 15, 275. [Google Scholar] [CrossRef]
- Azouz, F.; Arora, K.; Krause, K.; Nerurkar, V.R.; Kumar, M. Integrated microrna and mrna profiling in zika virus-infected neurons. Viruses 2019, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Pareek, S.; Roy, S.; Kumari, B.; Jain, P.; Banerjee, A.; Vrati, S. Mir-155 induction in microglial cells suppresses japanese encephalitis virus replication and negatively modulates innate immune responses. J. Neuroinflammation 2014, 11, 97. [Google Scholar] [CrossRef]
- Jiang, M.; Broering, R.; Trippler, M.; Wu, J.; Zhang, E.; Zhang, X.; Gerken, G.; Lu, M.; Schlaak, J.F. Microrna-155 controls toll-like receptor 3- and hepatitis c virus-induced immune responses in the liver. J. Viral Hepat. 2014, 21, 99–110. [Google Scholar] [CrossRef]
- Dickey, L.L.; Hanley, T.M.; Huffaker, T.B.; Ramstead, A.G.; O’Connell, R.M.; Lane, T.E. Microrna 155 and viral-induced neuroinflammation. J. Neuroimmunol. 2017, 308, 17–24. [Google Scholar] [CrossRef]
- Thounaojam, M.C.; Kaushik, D.K.; Kundu, K.; Basu, A. Microrna-29b modulates japanese encephalitis virus-induced microglia activation by targeting tumor necrosis factor alpha-induced protein 3. J. Neurochem. 2014, 129, 143–154. [Google Scholar] [CrossRef]
- Buggele, W.A.; Horvath, C.M. Microrna profiling of sendai virus-infected a549 cells identifies mir-203 as an interferon-inducible regulator of ifit1/isg56. J. Virol. 2013, 87, 9260–9270. [Google Scholar] [CrossRef]
- Zhang, S.; Li, J.; Li, J.; Yang, Y.; Kang, X.; Li, Y.; Wu, X.; Zhu, Q.; Zhou, Y.; Hu, Y. Up-regulation of microrna-203 in influenza a virus infection inhibits viral replication by targeting dr1. Sci. Rep. 2018, 8, 6797. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.N.; Holanda, G.M.; Pinto Silva, E.V.; Casseb, S.M.M.; Melo, K.F.L.; Carvalho, C.A.M.; Lima, J.A.; Vasconcelos, P.F.C.; Cruz, A.C.R. Zika virus alters the expression profile of microrna-related genes in liver, lung, and kidney cell lineages. Viral Immunol. 2018, 31, 583–588. [Google Scholar] [CrossRef]
- Xu, Y.P.; Qiu, Y.; Zhang, B.; Chen, G.; Chen, Q.; Wang, M.; Mo, F.; Xu, J.; Wu, J.; Zhang, R.R.; et al. Zika virus infection induces rnai-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 2019, 29, 265–273. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, C.; Xi, Z. Enhancement of rnai by a small molecule antibiotic enoxacin. Cell Res. 2008, 18, 1077–1079. [Google Scholar] [CrossRef]
- Nowakowski, T.J.; Pollen, A.A.; Di Lullo, E.; Sandoval-Espinosa, C.; Bershteyn, M.; Kriegstein, A.R. Expression analysis highlights axl as a candidate zika virus entry receptor in neural stem cells. Cell Stem Cell 2016, 18, 591–596. [Google Scholar] [CrossRef]
- Tang, H.; Hammack, C.; Ogden, S.C.; Wen, Z.; Qian, X.; Li, Y.; Yao, B.; Shin, J.; Zhang, F.; Lee, E.M.; et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 2016, 18, 587–590. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-region-specific organoids using mini-bioreactors for modeling zikv exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef]
- Netea, M.G.; Schlitzer, A.; Placek, K.; Joosten, L.A.B.; Schultze, J.L. Innate and adaptive immune memory: An evolutionary continuum in the host’s response to pathogens. Cell Host Microbe 2019, 25, 13–26. [Google Scholar] [CrossRef]
- Gourbal, B.; Pinaud, S.; Beckers, G.J.M.; Van Der Meer, J.W.M.; Conrath, U.; Netea, M.G. Innate immune memory: An evolutionary perspective. Immunol. Rev. 2018, 283, 21–40. [Google Scholar] [CrossRef]
- Netea, M.G.; Domínguez-Andrés, J.; Barreiro, L.B.; Chavakis, T.; Divangahi, M.; Fuchs, E.; Joosten, L.A.B.; van der Meer, J.W.M.; Mhlanga, M.M.; Mulder, W.J.M.; et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020, 20, 375–388. [Google Scholar] [CrossRef]
- Netea, M.G.; Quintin, J.; van der Meer, J.W. Trained immunity: A memory for innate host defense. Cell Host Microbe 2011, 9, 355–361. [Google Scholar] [CrossRef]
- Maloney, B.E.; Perera, K.D.; Saunders, D.R.D.; Shadipeni, N.; Fleming, S.D. Interactions of viruses and the humoral innate immune response. Clin. Immunol. 2020, 212, 108351. [Google Scholar] [CrossRef]
- Mesin, L.; Ersching, J.; Victora, G.D. Germinal center b cell dynamics. Immunity 2016, 45, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Elong Ngono, A.; Vizcarra, E.A.; Tang, W.W.; Sheets, N.; Joo, Y.; Kim, K.; Gorman, M.J.; Diamond, M.S.; Shresta, S. Mapping and role of the cd8(+) t cell response during primary zika virus infection in mice. Cell Host Microbe 2017, 21, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Pardy, R.D.; Rajah, M.M.; Condotta, S.A.; Taylor, N.G.; Sagan, S.M.; Richer, M.J. Analysis of the t cell response to zika virus and identification of a novel cd8+ t cell epitope in immunocompetent mice. PLoS Pathog. 2017, 13, e1006184. [Google Scholar] [CrossRef]
- Hassert, M.; Harris, M.G.; Brien, J.D.; Pinto, A.K. Identification of protective cd8 t cell responses in a mouse model of zika virus infection. Front. Immunol. 2019, 10, 1678. [Google Scholar] [CrossRef]
- Grifoni, A.; Pham, J.; Sidney, J.; O’Rourke, P.H.; Paul, S.; Peters, B.; Martini, S.R.; de Silva, A.D.; Ricciardi, M.J.; Magnani, D.M.; et al. Prior dengue virus exposure shapes t cell immunity to zika virus in humans. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Weiskopf, D.; Angelo, M.A.; de Azeredo, E.L.; Sidney, J.; Greenbaum, J.A.; Fernando, A.N.; Broadwater, A.; Kolla, R.V.; De Silva, A.D.; de Silva, A.M.; et al. Comprehensive analysis of dengue virus-specific responses supports an hla-linked protective role for cd8+ t cells. Proc. Natl. Acad. Sci. USA 2013, 110, E2046–E2053. [Google Scholar] [CrossRef]
- Weiskopf, D.; Angelo, M.A.; Sidney, J.; Peters, B.; Shresta, S.; Sette, A. Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection. J. Virol. 2014, 88, 11383–11394. [Google Scholar] [CrossRef] [PubMed]
- Weiskopf, D.; Angelo, M.A.; Grifoni, A.; O’Rourke, P.H.; Sidney, J.; Paul, S.; De Silva, A.D.; Phillips, E.; Mallal, S.; Premawansa, S.; et al. Hla-drb1 alleles are associated with different magnitudes of dengue virus-specific cd4+ t-cell responses. J. Infect. Dis. 2016, 214, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Elong Ngono, A.; Shresta, S. Cross-reactive t cell immunity to dengue and zika viruses: New insights into vaccine development. Front. Immunol. 2019, 10, 1316. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.; Sammon, M.; Garg, M. Dengue, zika and chikungunya: Emerging arboviruses in the new world. West. J. Emerg. Med. 2016, 17, 671–679. [Google Scholar] [CrossRef]
- Zimmerman, M.G.; Wrammert, J.; Suthar, M.S. Cross-reactive antibodies during zika virus infection: Protection, pathogenesis, and placental seeding. Cell Host Microbe 2020, 27, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef]
- Ades, A.E.; Thorne, C.; Soriano-Arandes, A.; Peckham, C.S.; Brown, D.W.; Lang, D.; Morris, J.G.; Christie, C.D.C.; Giaquinto, C. Researching zika in pregnancy: Lessons for global preparedness. Lancet Infect. Dis. 2020, 20, e61–e68. [Google Scholar] [CrossRef]
- Erbelding, E.; Cassetti, C. Zika virus and future research directions. J. Infect. Dis. 2017, 216, S991–S994. [Google Scholar] [CrossRef]
- Slon Campos, J.L.; Mongkolsapaya, J.; Screaton, G.R. The immune response against flaviviruses. Nat. Immunol. 2018, 19, 1189–1198. [Google Scholar] [CrossRef]
- Wen, J.; Tang, W.W.; Sheets, N.; Ellison, J.; Sette, A.; Kim, K.; Shresta, S. Identification of zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive cd8(+) t cells. Nat. Microbiol. 2017, 2, 17036. [Google Scholar] [CrossRef]
- Wen, J.; Elong Ngono, A.; Regla-Nava, J.A.; Kim, K.; Gorman, M.J.; Diamond, M.S.; Shresta, S. Dengue virus-reactive cd8(+) t cells mediate cross-protection against subsequent zika virus challenge. Nat. Commun. 2017, 8, 1459. [Google Scholar] [CrossRef]
- Regla-Nava, J.A.; Elong Ngono, A.; Viramontes, K.M.; Huynh, A.T.; Wang, Y.T.; Nguyen, A.T.; Salgado, R.; Mamidi, A.; Kim, K.; Diamond, M.S.; et al. Cross-reactive dengue virus-specific cd8(+) t cells protect against zika virus during pregnancy. Nat. Commun. 2018, 9, 3042. [Google Scholar] [CrossRef]
- Herrera, B.B.; Tsai, W.Y.; Chang, C.A.; Hamel, D.J.; Wang, W.K.; Lu, Y.; Mboup, S.; Kanki, P.J. Sustained specific and cross-reactive t cell responses to zika and dengue virus ns3 in west africa. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Lim, M.Q.; Kumaran, E.A.P.; Tan, H.C.; Lye, D.C.; Leo, Y.S.; Ooi, E.E.; MacAry, P.A.; Bertoletti, A.; Rivino, L. Cross-reactivity and anti-viral function of dengue capsid and ns3-specific memory t cells toward zika virus. Front. Immunol. 2018, 9, 2225. [Google Scholar] [CrossRef]
- Huber, R.G.; Lim, X.N.; Ng, W.C.; Sim, A.Y.L.; Poh, H.X.; Shen, Y.; Lim, S.Y.; Sundstrom, K.B.; Sun, X.; Aw, J.G.; et al. Structure mapping of dengue and zika viruses reveals functional long-range interactions. Nat. Commun. 2019, 10, 1408. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.; Gresh, L.; Ojeda, S.; Katzelnick, L.C.; Sanchez, N.; Mercado, J.C.; Chowell, G.; Lopez, B.; Elizondo, D.; Coloma, J.; et al. Prior dengue virus infection and risk of zika: A pediatric cohort in nicaragua. PLoS Med. 2019, 16, e1002726. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Barraquer, I.; Costa, F.; Nascimento, E.J.M.; Nery, N.J.; Castanha, P.M.S.; Sacramento, G.A.; Cruz, J.; Carvalho, M.; De Olivera, D.; Hagan, J.E.; et al. Impact of preexisting dengue immunity on zika virus emergence in a dengue endemic region. Science 2019, 363, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, C.; Fischer, C.; Feldmann, M.; Sarno, M.; Luz, E.; Moreira-Soto, A.; Cabral, R.; Netto, E.M.; Brites, C.; Kümmerer, B.M.; et al. Cross-protection of dengue virus infection against congenital zika syndrome, northeastern brazil. Emerg. Infect. Dis. 2019, 25, 1485–1493. [Google Scholar] [CrossRef] [PubMed]
- Hassert, M.; Wolf, K.J.; Schwetye, K.E.; DiPaolo, R.J.; Brien, J.D.; Pinto, A.K. Cd4+t cells mediate protection against zika associated severe disease in a mouse model of infection. PLoS Pathog. 2018, 14, e1007237. [Google Scholar] [CrossRef] [PubMed]
- Elong Ngono, A.; Young, M.P.; Bunz, M.; Xu, Z.; Hattakam, S.; Vizcarra, E.; Regla-Nava, J.A.; Tang, W.W.; Yamabhai, M.; Wen, J.; et al. Cd4+ t cells promote humoral immunity and viral control during zika virus infection. PLoS Pathog. 2019, 15, e1007474. [Google Scholar]
- Nancy, P.; Tagliani, E.; Tay, C.S.; Asp, P.; Levy, D.E.; Erlebacher, A. Chemokine gene silencing in decidual stromal cells limits t cell access to the maternal-fetal interface. Science 2012, 336, 1317–1321. [Google Scholar] [CrossRef]
- Shan, C.; Xie, X.; Luo, H.; Muruato, A.E.; Liu, Y.; Wakamiya, M.; La, J.H.; Chung, J.M.; Weaver, S.C.; Wang, T.; et al. Maternal vaccination and protective immunity against zika virus vertical transmission. Nat. Commun. 2019, 10, 5677. [Google Scholar] [CrossRef] [PubMed]
- Elong Ngono, A.; Syed, T.; Nguyen, A.V.; Regla-Nava, J.A.; Susantono, M.; Spasova, D.; Aguilar, A.; West, M.; Sparks, J.; Gonzalez, A.; et al. Cd8(+) t cells mediate protection against zika virus induced by an ns3-based vaccine. Sci. Adv. 2020, 6, eabb2154. [Google Scholar] [CrossRef] [PubMed]
- Sariol, C.A.; Nogueira, M.L.; Vasilakis, N. A tale of two viruses: Does heterologous flavivirus immunity enhance zika disease? Trends Microbiol. 2018, 26, 186–190. [Google Scholar] [CrossRef]
- Rathore, A.P.S.; Saron, W.A.A.; Lim, T.; Jahan, N.; St John, A.L. Maternal immunity and antibodies to dengue virus promote infection and zika virus-induced microcephaly in fetuses. Sci. Adv. 2019, 5, eaav3208. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Singh, G.; Acklin, J.A.; Lee, S.; Duehr, J.E.; Chokola, A.N.; Frere, J.J.; Hoffman, K.W.; Foster, G.A.; Krysztof, D.; et al. Dengue virus immunity increases zika virus-induced damage during pregnancy. Immunity 2019, 50, 751–762.e755. [Google Scholar] [CrossRef]
- Zimmerman, M.G.; Quicke, K.M.; O’Neal, J.T.; Arora, N.; Machiah, D.; Priyamvada, L.; Kauffman, R.C.; Register, E.; Adekunle, O.; Swieboda, D.; et al. Cross-reactive dengue virus antibodies augment zika virus infection of human placental macrophages. Cell Host Microbe 2018, 24, 731–742.e736. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Olsen, P.C.; Costa, F.; Wang, Q.; Oliveira, T.Y.; Nery, N., Jr.; Aromolaran, A.; do Rosário, M.S.; Sacramento, G.A.; Cruz, J.S.; et al. Risk of zika microcephaly correlates with features of maternal antibodies. J. Exp. Med. 2019, 216, 2302–2315. [Google Scholar] [CrossRef]
- Diamond, M.S.; Ledgerwood, J.E.; Pierson, T.C. Zika virus vaccine development: Progress in the face of new challenges. Annu. Rev. Med. 2019, 70, 121–135. [Google Scholar] [CrossRef]
- Grubor-Bauk, B.; Wijesundara, D.K.; Masavuli, M.; Abbink, P.; Peterson, R.L.; Prow, N.A.; Larocca, R.A.; Mekonnen, Z.A.; Shrestha, A.; Eyre, N.S.; et al. Ns1 DNA vaccination protects against zika infection through t cell-mediated immunity in immunocompetent mice. Sci. Adv. 2019, 5, eaax2388. [Google Scholar] [CrossRef] [PubMed]
- Bullard, B.L.; Corder, B.N.; Gorman, M.J.; Diamond, M.S.; Weaver, E.A. Efficacy of a t cell-biased adenovirus vector as a zika virus vaccine. Sci. Rep. 2018, 8, 18017. [Google Scholar] [CrossRef] [PubMed]
- Counotte, M.J.; Kim, C.R.; Wang, J.; Bernstein, K.; Deal, C.D.; Broutet, N.J.N.; Low, N. Sexual transmission of zika virus and other flaviviruses: A living systematic review. PLoS Med. 2018, 15, e1002611. [Google Scholar] [CrossRef] [PubMed]
- Spadoni, I.; Fornasa, G.; Rescigno, M. Organ-specific protection mediated by cooperation between vascular and epithelial barriers. Nat. Rev. Immunol. 2017, 17, 761–773. [Google Scholar] [CrossRef] [PubMed]
- El Costa, H.; Gouilly, J.; Mansuy, J.M.; Chen, Q.; Levy, C.; Cartron, G.; Veas, F.; Al-Daccak, R.; Izopet, J.; Jabrane-Ferrat, N. Zika virus reveals broad tissue and cell tropism during the first trimester of pregnancy. Sci. Rep. 2016, 6, 35296. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.R.; Roby, J.A.; Dobyns, W.B.; Rajagopal, L.; Gale, M., Jr.; Adams Waldorf, K.M. Immune evasion strategies used by zika virus to infect the fetal eye and brain. Viral Immunol. 2020, 33, 22–37. [Google Scholar] [CrossRef]
- Adams Waldorf, K.M.; Stencel-Baerenwald, J.E.; Kapur, R.P.; Studholme, C.; Boldenow, E.; Vornhagen, J.; Baldessari, A.; Dighe, M.K.; Thiel, J.; Merillat, S.; et al. Fetal brain lesions after subcutaneous inoculation of zika virus in a pregnant nonhuman primate. Nat. Med. 2016, 22, 1256–1259. [Google Scholar] [CrossRef]
- Chiu, C.F.; Chu, L.W.; Liao, I.C.; Simanjuntak, Y.; Lin, Y.L.; Juan, C.C.; Ping, Y.H. The mechanism of the zika virus crossing the placental barrier and the blood-brain barrier. Front. Microbiol. 2020, 11, 214. [Google Scholar] [CrossRef]
- Wu, K.Y.; Zuo, G.L.; Li, X.F.; Ye, Q.; Deng, Y.Q.; Huang, X.Y.; Cao, W.C.; Qin, C.F.; Luo, Z.G. Vertical transmission of zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res. 2016, 26, 645–654. [Google Scholar] [CrossRef]
- Onorati, M.; Li, Z.; Liu, F.; Sousa, A.M.M.; Nakagawa, N.; Li, M.; Dell’Anno, M.T.; Gulden, F.O.; Pochareddy, S.; Tebbenkamp, A.T.N.; et al. Zika virus disrupts phospho-tbk1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 2016, 16, 2576–2592. [Google Scholar] [CrossRef]
- Adams Waldorf, K.M.; Nelson, B.R.; Stencel-Baerenwald, J.E.; Studholme, C.; Kapur, R.P.; Armistead, B.; Walker, C.L.; Merillat, S.; Vornhagen, J.; Tisoncik-Go, J.; et al. Congenital zika virus infection as a silent pathology with loss of neurogenic output in the fetal brain. Nat. Med. 2018, 24, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Coffey, L.L.; Keesler, R.I.; Pesavento, P.A.; Woolard, K.; Singapuri, A.; Watanabe, J.; Cruzen, C.; Christe, K.L.; Usachenko, J.; Yee, J.; et al. Intraamniotic zika virus inoculation of pregnant rhesus macaques produces fetal neurologic disease. Nat. Commun. 2018, 9, 2414. [Google Scholar] [CrossRef] [PubMed]
- Coletti, A.M.; Singh, D.; Kumar, S.; Shafin, T.N.; Briody, P.J.; Babbitt, B.F.; Pan, D.; Norton, E.S.; Brown, E.C.; Kahle, K.T.; et al. Characterization of the ventricular-subventricular stem cell niche during human brain development. Development 2018, 145, dev170100. [Google Scholar] [CrossRef]
- Satterfield-Nash, A.; Kotzky, K.; Allen, J.; Bertolli, J.; Moore, C.A.; Pereira, I.O.; Pessoa, A.; Melo, F.; Santelli, A.; Boyle, C.A.; et al. Health and development at age 19–24 months of 19 children who were born with microcephaly and laboratory evidence of congenital zika virus infection during the 2015 zika virus outbreak—Brazil, 2017. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 1347–1351. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, B.G.; Werner, H.; Lopes, F.; Hygino da Cruz, L.C., Jr.; Fazecas, T.M.; Daltro, P.A.N.; Nogueira, R.A. Central nervous system effects of intrauterine zika virus infection: A pictorial review. Radiographics 2017, 37, 1840–1850. [Google Scholar] [CrossRef] [PubMed]
- Martins-Filho, P.R.; Souza Tavares, C.S.; Araújo Carvalho, A.C.; Reis, M.; Santos, H.P., Jr.; Santos, V.S. Association between arthrogryposis and mortality in infants with congenital zika syndrome: A systematic review and meta-analysis. Pediatr. Neurol. 2020, 110, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Garcez, P.P.; Loiola, E.C.; Madeiro da Costa, R.; Higa, L.M.; Trindade, P.; Delvecchio, R.; Nascimento, J.M.; Brindeiro, R.; Tanuri, A.; Rehen, S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016, 352, 816–818. [Google Scholar] [CrossRef]
- Simonin, Y.; Loustalot, F.; Desmetz, C.; Foulongne, V.; Constant, O.; Fournier-Wirth, C.; Leon, F.; Molès, J.P.; Goubaud, A.; Lemaitre, J.M.; et al. Zika virus strains potentially display different infectious profiles in human neural cells. EBioMedicine 2016, 12, 161–169. [Google Scholar] [CrossRef]
- Li, H.; Saucedo-Cuevas, L.; Regla-Nava, J.A.; Chai, G.; Sheets, N.; Tang, W.; Terskikh, A.V.; Shresta, S.; Gleeson, J.G. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell 2016, 19, 593–598. [Google Scholar] [CrossRef]
- Figueiredo, C.P.; Barros-Aragão, F.G.Q.; Neris, R.L.S.; Frost, P.S.; Soares, C.; Souza, I.N.O.; Zeidler, J.D.; Zamberlan, D.C.; de Sousa, V.L.; Souza, A.S.; et al. Zika virus replicates in adult human brain tissue and impairs synapses and memory in mice. Nat. Commun. 2019, 10, 3890. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, A. Ocular manifestations of emerging flaviviruses and the blood-retinal barrier. Viruses 2018, 10, 530. [Google Scholar] [CrossRef]
- De Paula Freitas, B.; Zin, A.; Ko, A.; Maia, M.; Ventura, C.V.; Belfort, R., Jr. Anterior-segment ocular findings and microphthalmia in congenital zika syndrome. Ophthalmology 2017, 124, 1876–1878. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, F.S.Y.; Pietrobon, A.J.; Branco, A.; Pereira, N.Z.; Oliveira, L.; Machado, C.M.; Duarte, A.; Sato, M.N. Zika virus infects newborn monocytes without triggering a substantial cytokine response. J. Infect. Dis. 2019, 220, 32–40. [Google Scholar] [CrossRef]
- Sinigaglia, A.; Riccetti, S.; Trevisan, M.; Barzon, L. In silico approaches to zika virus drug discovery. Expert Opin. Drug Discov. 2018, 13, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Devillers, J. Repurposing drugs for use against zika virus infection. Sar Qsar Environ. Res. 2018, 29, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Bernatchez, J.A.; Tran, L.T.; Li, J.; Luan, Y.; Siqueira-Neto, J.L.; Li, R. Drugs for the treatment of zika virus infection. J. Med. Chem. 2020, 63, 470–489. [Google Scholar] [CrossRef] [PubMed]
- Hazlewood, J.E.; Rawle, D.J.; Tang, B.; Yan, K.; Vet, L.J.; Nakayama, E.; Hobson-Peters, J.; Hall, R.A.; Suhrbier, A. A zika vaccine generated using the chimeric insect-specific binjari virus platform protects against fetal brain infection in pregnant mice. Vaccines 2020, 8, 496. [Google Scholar] [CrossRef]
- Hurtado-Monzón, A.M.; Cordero-Rivera, C.D.; Farfan-Morales, C.N.; Osuna-Ramos, J.F.; De Jesús-González, L.A.; Reyes-Ruiz, J.M.; Del Ángel, R.M. The role of anti-flavivirus humoral immune response in protection and pathogenesis. Rev. Med. Virol. 2020, 30, e2100. [Google Scholar] [CrossRef]
- Pattnaik, A.; Sahoo, B.R.; Pattnaik, A.K. Current status of zika virus vaccines: Successes and challenges. Vaccines 2020, 8, 266. [Google Scholar] [CrossRef] [PubMed]
- Martín-Acebes, M.A.; Saiz, J.C.; Jiménez de Oya, N. Antibody-dependent enhancement and zika: Real threat or phantom menace? Front. Cell. Infect. Microbiol. 2018, 8, 44. [Google Scholar] [CrossRef]
- Turco, M.Y.; Moffett, A. Development of the human placenta. Development 2019, 146, dev163428. [Google Scholar] [CrossRef] [PubMed]
- Rossant, J.; Cross, J.C. Placental development: Lessons from mouse mutants. Nat. Rev. Genet. 2001, 2, 538–548. [Google Scholar] [CrossRef] [PubMed]
- França, G.V.; Schuler-Faccini, L.; Oliveira, W.K.; Henriques, C.M.; Carmo, E.H.; Pedi, V.D.; Nunes, M.L.; Castro, M.C.; Serruya, S.; Silveira, M.F.; et al. Congenital zika virus syndrome in brazil: A case series of the first 1501 livebirths with complete investigation. Lancet 2016, 388, 891–897. [Google Scholar] [CrossRef]
- Coyne, C.B.; Lazear, H.M. Zika virus—Reigniting the torch. Nat. Rev. Microbiol. 2016, 14, 707–715. [Google Scholar] [CrossRef]
- Moore, C.A.; Staples, J.E.; Dobyns, W.B.; Pessoa, A.; Ventura, C.V.; Fonseca, E.B.; Ribeiro, E.M.; Ventura, L.O.; Neto, N.N.; Arena, J.F.; et al. Characterizing the pattern of anomalies in congenital zika syndrome for pediatric clinicians. JAMA Pediatr. 2017, 171, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Vhp, L.; Aragão, M.M.; Pinho, R.S.; Hazin, A.N.; Paciorkowski, A.R.; Penalva de Oliveira, A.C.; Masruha, M.R. Congenital zika virus infection: A review with emphasis on the spectrum of brain abnormalities. Curr. Neurol. Neurosci. Rep. 2020, 20, 49. [Google Scholar] [CrossRef] [PubMed]
- Abbink, P.; Stephenson, K.E.; Barouch, D.H. Zika virus vaccines. Nat. Rev. Microbiol. 2018, 16, 594–600. [Google Scholar] [CrossRef] [PubMed]
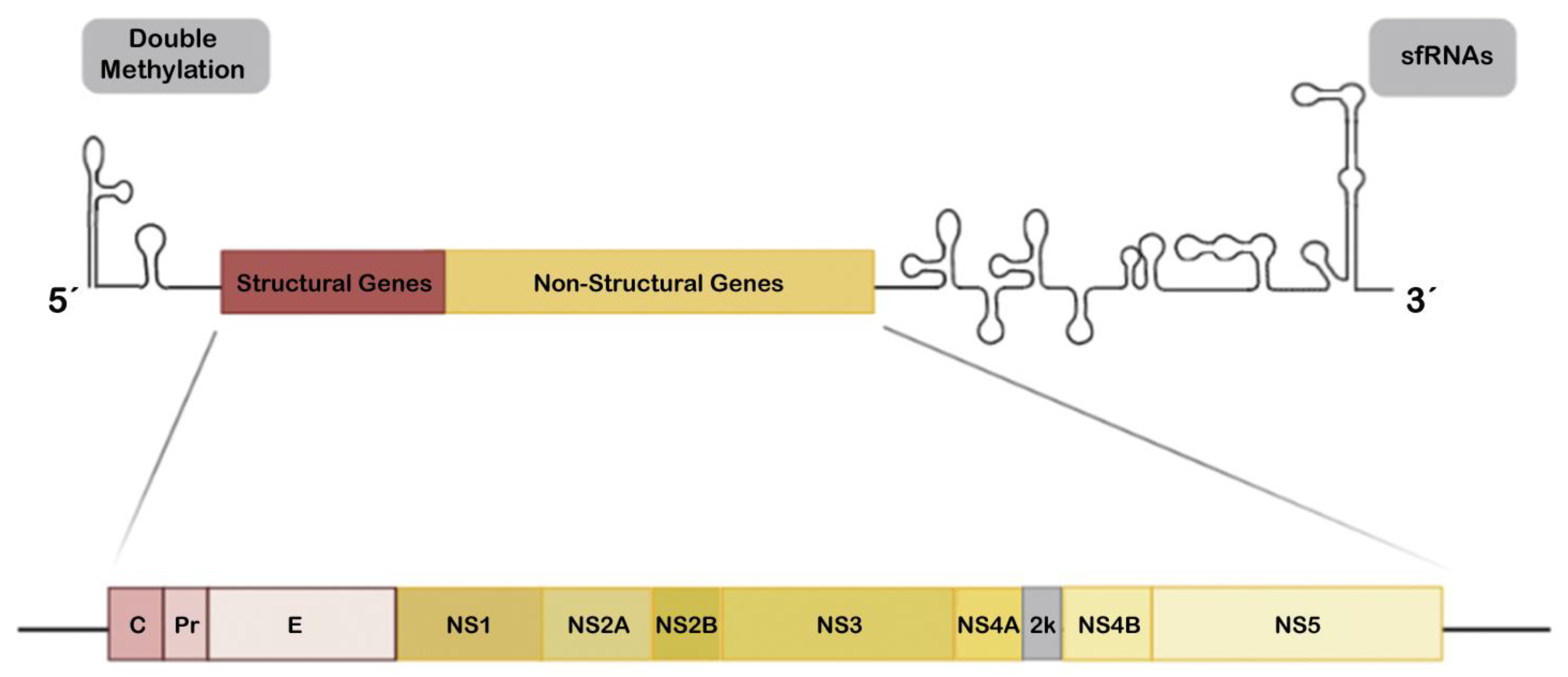
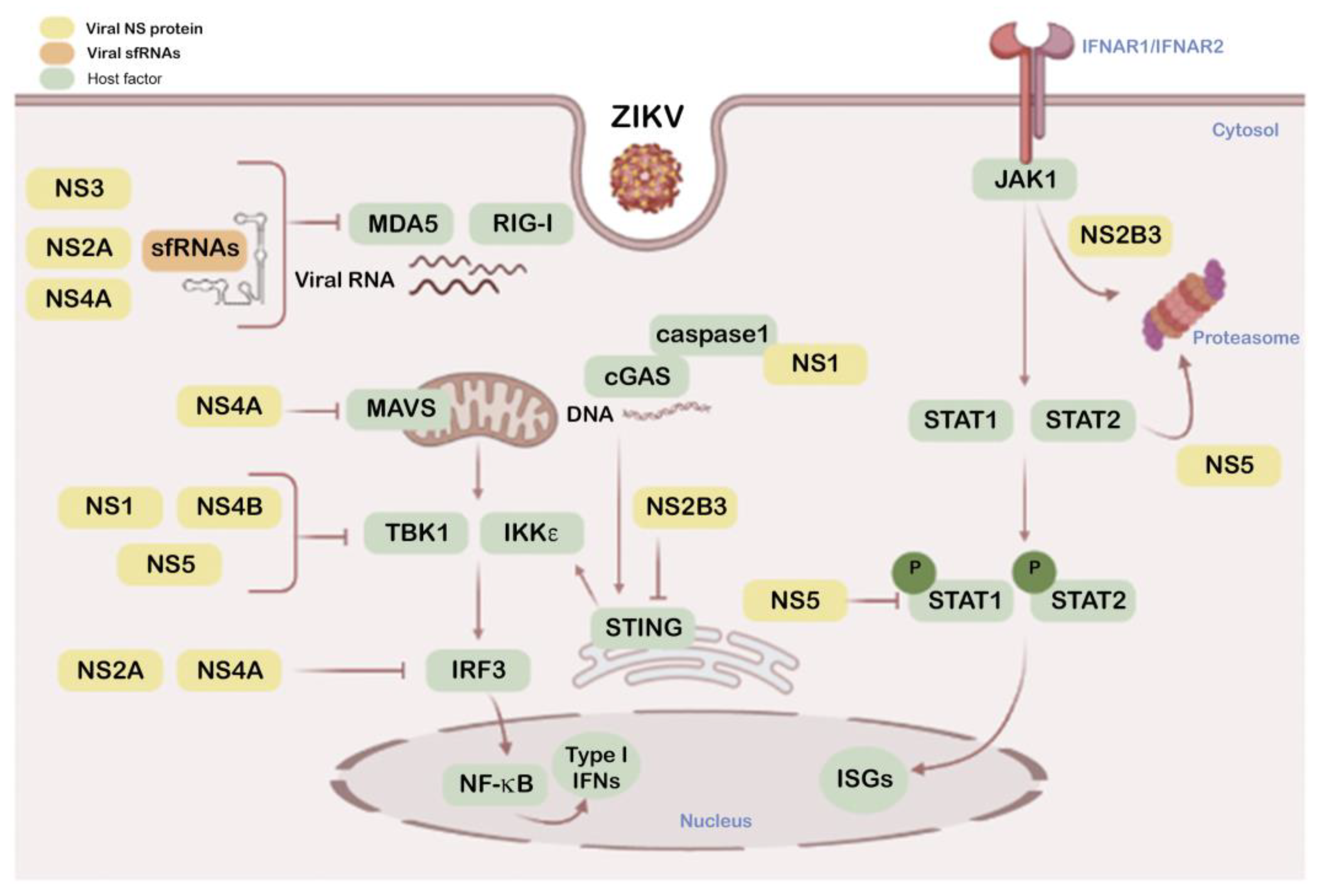
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Estévez-Herrera, J.; Pérez-Yanes, S.; Cabrera-Rodríguez, R.; Márquez-Arce, D.; Trujillo-González, R.; Machado, J.-D.; Madrid, R.; Valenzuela-Fernández, A. Zika Virus Pathogenesis: A Battle for Immune Evasion. Vaccines 2021, 9, 294. https://doi.org/10.3390/vaccines9030294
Estévez-Herrera J, Pérez-Yanes S, Cabrera-Rodríguez R, Márquez-Arce D, Trujillo-González R, Machado J-D, Madrid R, Valenzuela-Fernández A. Zika Virus Pathogenesis: A Battle for Immune Evasion. Vaccines. 2021; 9(3):294. https://doi.org/10.3390/vaccines9030294
Chicago/Turabian StyleEstévez-Herrera, Judith, Silvia Pérez-Yanes, Romina Cabrera-Rodríguez, Daniel Márquez-Arce, Rodrigo Trujillo-González, José-David Machado, Ricardo Madrid, and Agustín Valenzuela-Fernández. 2021. "Zika Virus Pathogenesis: A Battle for Immune Evasion" Vaccines 9, no. 3: 294. https://doi.org/10.3390/vaccines9030294
APA StyleEstévez-Herrera, J., Pérez-Yanes, S., Cabrera-Rodríguez, R., Márquez-Arce, D., Trujillo-González, R., Machado, J.-D., Madrid, R., & Valenzuela-Fernández, A. (2021). Zika Virus Pathogenesis: A Battle for Immune Evasion. Vaccines, 9(3), 294. https://doi.org/10.3390/vaccines9030294






