Association between Venom Immunotherapy and Changes in Serum Protein—Peptide Patterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of the Study Group
2.2. Measurement of sIgE and sIgG4 Serum Levels
2.3. Pre-Treatment of the Serum Samples
2.4. MALDI-TOF MS Proteomic Profiling
2.5. NanoLC-MALDI-TOF/TOF MS Identification of Discriminative Proteomic Features
2.6. Data Analysis
3. Results
3.1. Serum sIgG4 Levels before and on the 90th and 180th Days of VIT
3.2. MALDI-TOF MS Protein–Peptide Profiling and Identification of the Discriminatory Features
3.3. Comparison of Protein–Peptide Profiles Derived from Allergic Patients Undergoing VIT Classified as Low and High IgG4 Responder Groups
- The ratio of serum sIgG4 levels at two time-points < 15 (sIgG4 value of the 90th/180th day divided by the sIgG4 value of baseline)
- Differences between sIgG4 on the 90th/180th day and baseline < 6
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bilò, M.B.; Bonifazi, F. The natural history and epidemiology of insect venom allergy: Clinical implications. Clin. Exp. Allergy 2009, 39, 1467–1476. [Google Scholar] [CrossRef]
- Bilò, B.M.; Bonifazi, F. Epidemiology of insect-venom anaphylaxis. Curr. Opin. Allergy Clin. Immunol. 2008, 8, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Incorvaia, C.; Mauro, M.; Gritti, B.L.; Makri, E.; Ridolo, E. Venom immunotherapy in patients with allergic reactions to insect stings. Expert Rev. Clin. Immunol. 2018, 14, 53–59. [Google Scholar] [CrossRef]
- Dhami, S.; Zaman, H.; Varga, E.-M.; Sturm, G.J.; Muraro, A.; Akdis, C.A.; Antolín-Amérigo, D.; Bilò, M.B.; Bokanovic, D.; Calderon, M.A.; et al. Allergen immunotherapy for insect venom allergy: A systematic review and meta-analysis. Allergy Eur. J. Allergy Clin. Immunol. 2017, 72, 342–365. [Google Scholar] [CrossRef]
- Kosnik, M.; Korosec, P. Venom immunotherapy: Clinical efficacy, safety and contraindications. Expert Rev. Clin. Immunol. 2015, 11, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Sturm, G.J.; Varga, E.M.; Roberts, G.; Mosbech, H.; Bilò, M.B.; Akdis, C.A.; Antolin-Amerigo, D.; Cichocka-Jarosz, E.; Gawlik, R.; Jakob, T.; et al. EAACI guidelines on allergen immunotherapy: Hymenoptera venom allergy. Allergy 2018, 73, 744–764. [Google Scholar] [CrossRef]
- Goldberg, A.; Confino-Cohen, R. Bee venom immunotherapy—How early is it effective? Allergy Eur. J. Allergy Clin. Immunol. 2010, 65, 391–395. [Google Scholar] [CrossRef]
- Heneberg, P.; Riegerová, K.; Kučera, P. Pimecrolimus Is a Potent Inhibitor of Allergic Reactions to Hymenopteran Venom Ex-tracts and Birch Pollen Allergen In Vitro. PLoS ONE 2015, 10, e0142953. [Google Scholar] [CrossRef] [PubMed]
- Heneberg, P.; Riegerová, K.; Říhová, A.; Šimčíková, D.; Kučera, P. Updates on the surface antigens of basophils: CD16 on baso-phils of patients with respiratory or insect venom allergy and the rejection of CD203c and CD63 externalization decoupling by bisindolylmaleimides. Clin. Exp. Allergy 2019, 49, 54–67. [Google Scholar] [CrossRef]
- Rodríguez Trabado, A.; Cámara Hijón, C.; Ramos Cantariño, A.; Romero-Chala, S.; García-Trujillo, J.A.; Fernández Pereira, L.M. Short-, Intermediate-, and Long-Term Changes in Basophil Reactivity Induced by Venom Immunotherapy. Allergy Asthma Immunol. Res. 2016, 8, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Žitnik, S.E.K.; Vesel, T.; Avčin, T.; Šilar, M.; Košnik, M.; Korošec, P. Monitoring honeybee venom immunotherapy in children with the basophil activation test. Pediatr. Allergy Immunol. 2012, 23, 167–172. [Google Scholar] [CrossRef]
- Ebo, D.G.; Hagendorens, M.M.; Schuerwegh, A.J.; Beirens, L.M.-N.; Bridts, C.H.; De Clerck, L.S.; Stevens, W.J. Flow-assisted quantification of in vitro activated basophils in the diagnosis of wasp venom allergy and follow-up of wasp venom immunotherapy. Cytom. Part B Clin. Cytom. 2007, 72, 196–203. [Google Scholar] [CrossRef]
- Bidad, K.; Nawijn, M.C.; van Oosterhout, A.J.M.; van der Heide, S.; Oude Elberink, J.N.G. Basophil activation test in the diagnosis and monitoring of mastocytosis patients with wasp venom allergy on immunotherapy. Cytom. Part B Clin. Cytom. 2014, 86, 183–190. [Google Scholar] [CrossRef]
- Eržen, R.; Košnik, M.; Šilar, M.; Korošec, P. Basophil response and the induction of a tolerance in venom immunotherapy: A long-term sting challenge study. Allergy 2012, 67, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Lerch, E.; Müller, U.R. Long-term protection after stopping venom immunotherapy: Results of re-stings in 200 patients. J. Allergy Clin. Immunol. 1998, 101, 606–612. [Google Scholar] [CrossRef]
- Akdis, M.; Akdis, C.A. Mechanisms of allergen-specific immunotherapy: Multiple suppressor factors at work in immune toler-ance to allergens. J. Allergy Clin. Immunol. 2014, 133, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, C.; Kucuksezer, U.C.; Akdis, M.; Akdis, C.A. Mechanisms of immunotherapy to wasp and bee venom. Clin. Exp. Allergy 2011, 41, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Graham, F.; Bégin, P.; Paradis, L.; Lacombe-Barrios, J.; Paradis, J.; Des Roches, A. Comparison of ImmunoCAP and Immulite serum specific IgE assays for the assessment of egg allergy. Allergy Asthma Clin. Immunol. 2016, 12, 129. [Google Scholar] [CrossRef]
- Bonifazi, F.; Jutel, M.; Bilo, B.M.; Birnbaum, J.; Muller, U. Prevention and treatment of hymenoptera venom allergy: Guidelines for clinical practice. Allergy 2005, 60, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Swiatly, A.; Horala, A.; Hajduk, J.; Matysiak, J.; Nowak-Markwitz, E.; Kokot, Z.J. MALDI-TOF-MS analysis in discovery and identification of serum proteomic patterns of ovarian cancer. BMC Cancer 2017, 17, 472. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Wen, Q.; Li, Z.-J.; Xu, R.-C.; Peng, F.-F.; Yu, X.-Q. Optimization and evaluation of magnetic bead separation combined with matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF MS) for proteins profiling of peri-toneal dialysis effluent. Int. J. Mol. Sci. 2014, 15, 1162–1175. [Google Scholar] [CrossRef]
- Klupczynska, A.; Swiatly, A.; Hajduk, J.; Matysiak, J.; Dyszkiewicz, W.; Pawlak, K.; Kokot, Z.J. Identification of Serum Peptidome Signa-tures of Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2016, 17, 410. [Google Scholar] [CrossRef]
- Matuszewska, E.; Matysiak, J.; Kycler, Z.; Kokot, Z.J.; Matysiak, J. Proteomic features characterization of Hymenoptera venom allergy. Allergy Asthma Clin. Immunol. 2019, 15, 77. [Google Scholar] [CrossRef]
- Naik, R.S.; Wala, M.S. Inflammation, Allergy and Asthma, Complex Immune Origin Diseases: Mechanisms and Therapeutic Agents. Recent Pat. Inflamm. Allergy Drug. Discov. 2013, 7, 62–95. [Google Scholar] [CrossRef]
- Gabay, C.; Kushner, I. Acute-Phase Proteins and Other Systemic Responses to Inflammation. N. Engl. J. Med. 1999, 340, 448–454. [Google Scholar] [CrossRef]
- Smiley, S.T.; King, J.A.; Hancock, W.W. Fibrinogen Stimulates Macrophage Chemokine Secretion through Toll-Like Receptor 4. J. Immunol. 2001, 167, 2887–2894. [Google Scholar] [CrossRef] [PubMed]
- Davalos, D.; Akassoglou, K. Fibrinogen as a key regulator of inflammation in disease. Semin. Immunopathol. 2012, 34, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.Y.; Smith, T.D.; Meli, V.S.; Tran, T.N.; Botvinick, E.L.; Liu, W.F. Differential regulation of macrophage inflammatory activation by fibrin and fibrinogen. Acta Biomater. 2017, 47, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Rhim, T.; Choi, Y.-S.; Nam, B.-Y.; Uh, S.T.; Park, J.S.; Kim, Y.-H.; Paik, Y.-K.; Park, C.-S. Plasma protein profiles in early asthmatic responses to inhalation allergen challenge. Allergy Eur. J. Allergy Clin. Immunol. 2009, 64, 47–54. [Google Scholar] [CrossRef]
- Wang, H.; Gottfries, J.; Barrenäs, F.; Benson, M. Identification of novel biomarkers in seasonal allergic rhinitis by combining pro-teomic, multivariate and pathway analysis. PLoS ONE 2011, 6, e23563. [Google Scholar] [CrossRef]
- Całkosiński, I.; Dobrzyński, M.; Całkosińska, M.; Seweryn, E.; Bronowicka-Szydełko, A.; Dzierzba, K.; Ceremuga, I.; Gamian, A. Characterization of an in ammatory response. Postepy Hig. Med. Dosw. 2009, 63, 395–408. [Google Scholar]
- Pulani, D.; Rudan, I. The Past Decade: Fibrinogen. Coll. Antropol. 2005, 29, 341–349. [Google Scholar]
- Choo, Y.M.; Lee, K.S.; Yoon, H.J.; Kim, B.Y.; Sohn, M.R.; Roh, J.Y.; Je, Y.H.; Kim, N.J.; Kim, I.; Woo, S.D.; et al. Dual function of a bee venom serine protease: Prophenoloxi-dase-activating factor in arthropods and fibrin(ogen)olytic enzyme in mammals. PLoS ONE 2010, 5, e10393. [Google Scholar] [CrossRef]
- Matysiak, J.; Hajduk, J.; Mayer, F.; Hebeler, R.; Kokot, Z.J. Hyphenated LC–MALDI–ToF/ToF and LC–ESI–QToF approach in pro-teomic characterization of honeybee venom. J. Pharm. Biomed. Anal. 2016, 12, 69–76. [Google Scholar] [CrossRef]
- Matysiak, J.; Hajduk, J.; Pietrzak, Ł.; Schmelzer, C.E.; Kokot, Z.J. Shotgun proteome analysis of honeybee venom using targeted enrichment strategies. Toxicon 2014, 90, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Choo, Y.M.; Lee, K.S.; Yoon, H.J.; Qiu, Y.; Wan, H.; Sohn, M.R.; Sohn, H.D.; Jin, B.R. Antifibrinolytic Role of a Bee Venom Serine Protease Inhibitor That Acts as a Plasmin Inhibitor. PLoS ONE 2012, 7, e32269. [Google Scholar] [CrossRef] [PubMed]
- Oikonomopoulou, K.; Ricklin, D.; Ward, P.A.; Lambris, J.D. Interactions between coagulation and complement—Their role in inflammation. Semin. Immunopathol. 2012, 34, 151–165. [Google Scholar] [CrossRef]
- Dunkelberger, J.R.; Song, W.C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef]
- Markiewski, M.M.; Lambris, J.D. The Role of Complement in Inflammatory Diseases from Behind the Scenes into the Spotlight. Am. J. Pathol. 2007, 171, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Pidde-Queiroz, G.; de Fatima Furtado, M.; Filgueiras, C.F.; Pessoa, L.A.; Spadafora-Ferreira, M.; van den Berg, C.W.; Tambourgi, D.V. Human comple-ment activation and anaphylatoxins generation induced by snake venom toxins from Bothrops genus. Mol. Immunol. 2010, 47, 2537–2544. [Google Scholar] [CrossRef] [PubMed]
- Garantziotis, S.; Hollingsworth, J.W.; Ghanayem, R.B.; Timberlake, S.; Zhuo, L.; Kimata, K.; Schwartz, D.A. Inter-alpha-Trypsin Inhibitor Attenuates Complement Activation and Complement-Induced Lung Injury. J. Immunol. 2007, 179, 4187–4192. [Google Scholar] [CrossRef]
- Subbannayya, Y.; Mir, S.A.; Renuse, S.; Manda, S.S.; Pinto, S.M.; Puttamallesh, V.N.; Solanki, H.S.; Manju, H.; Syed, N.; Sharma, R.; et al. Identification of differentially expressed serum proteins in gastric adenocarcinoma. J. Proteom. 2015, 127, 80–88. [Google Scholar] [CrossRef]
- Cortelazzo, A.; Guerranti, R.; Bini, L.; Hope-Onyekwere, N.; Muzzi, C.; Leoncini, R.; Pagani, R. Effects of snake venom proteases on hu-man fibrinogen chains. Blood Transfus. 2010, 8 (Suppl. S3), S120–S125. [Google Scholar] [PubMed]
- Pandya, B.V.; Rubin, R.N.; Olexa, S.A.; Budzynski, A.Z. Unique degradation of human fibrinogen by proteases from western dia-mondback rattlesnake (Crotalus atrox) venom. Toxicon 1983, 21, 515–526. [Google Scholar] [CrossRef]
- Patra, A.; Kalita, B.; Chanda, A.; Mukherjee, A.K. Proteomics and antivenomics of Echis carinatus carinatus venom: Correlation with pharmacological properties and pathophysiology of envenomation. Sci. Rep. 2017, 7, 17119. [Google Scholar] [CrossRef] [PubMed]
- Galera, C.; Soohun, N.; Zankar, N.; Caimmi, S.; Gallen, C.; Demoly, P. Severe anaphylaxis to bee venom immunotherapy: Efficacy of pretreatment and concurrent treatment with omalizumab. J. Investig. Allergol. Clin. Immunol. 2009, 19, 225–229. [Google Scholar]
- Xu, Q.; Wu, N.; Cui, L.; Wu, Z.; Qiu, G. Filamin B: The next hotspot in skeletal research? J. Genet. Genom. 2017, 44, 335–342. [Google Scholar] [CrossRef]
- Baldassarre, M.; Razinia, Z.; Burande, C.F.; Lamsoul, I.; Lutz, P.G.; Calderwood, D.A. Filamins Regulate Cell Spreading and Initiation of Cell Migration. PLoS ONE 2009, 4, e7830. [Google Scholar] [CrossRef]
- Baltz, A.G.; Munschauer, M.; Schwanhäusser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-Bound Proteome and Its Global Occupancy Profile on Protein-Coding Transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef]
- Mattson, L.; Lentini, A.; Gawel, D.R.; Badam, T.V.S.; Benson, M.; Ledin, T.; Nestor, C.E.; Gustafsson, M.; Serra-Musach, J.; Bjorkander, J.; et al. Potential Involvement of Type I Interferon Signaling in Immunotherapy in Seasonal Allergic Rhinitis. J. Immunol. Res. 2016, 2016, 5153184. [Google Scholar] [CrossRef] [PubMed]
- Van Horn, S.R.G.; Farrar, J.D. Interferon at the crossroads of allergy and viral infections. J. Leukoc. Biol. 2015, 98, 185–194. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Choi, J.S.; Lee, J.Y.; Yu, K.R.; Ka, S.H.; Cho, Y.; Choi, E.-J.; Baek, S.H.; Seol, J.H.; Park, D.; et al. Filamin B Serves as a Molecular Scaffold for Type I Interferon-induced c-Jun NH2-terminal Kinase Signaling Pathway. Mol. Biol. Cell 2008, 19, 5116–5130. [Google Scholar] [CrossRef][Green Version]
- Jeon, Y.J.; Choi, J.S.; Lee, J.Y.; Yu, K.R.; Kim, S.M.; Ka, S.H.; Oh, K.H.; Kim, K.; Zhang, D.; Bang, O.S.; et al. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 2009, 10, 374–380. [Google Scholar] [CrossRef]
- Whitmarsh, A.J. Filamin B: A scaffold for interferon signalling. EMBO Rep. 2009, 10, 349–351. [Google Scholar] [CrossRef] [PubMed]
- Narain, N.R.; Diers, A.R.; Lee, A.; Lao, S.; Chan, J.Y.; Schofield, S.; Andreazi, J.; Ouro-Djobo, R.; Jimenez, J.J.; Friss, T.; et al. Identification of Filamin-A and -B as potential biomarkers for prostate cancer. Futur. Sci. OA 2017, 3, FSO161. [Google Scholar] [CrossRef]
- Pecci, A.; Ma, X.; Savoia, A.; Adelstein, R.S. MYH9: Structure, functions and role of non-muscle myosin IIA in human disease. Gene 2018, 664, 152–167. [Google Scholar] [CrossRef]
- Vicente-Manzanares, M.; Ma, X.; Adelstein, R.S.; Horwitz, A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 778–790. [Google Scholar] [CrossRef]
- Heissler, S.M.; Manstein, D.J. Nonmuscle myosin-2: Mix and match. Cell. Mol. Life Sci. 2012, 70, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, R.; Chen, X.-X.; Zhi, Y.; Deng, R.; Zhou, E.-M.; Qiao, S.; Zhang, G. Nonmuscle Myosin Heavy Chain IIA Recognizes Sialic Acids on Si-alylated RNA Viruses to Suppress Proinflammatory Responses via the DAP12-Syk Pathway. mBio 2019, 10, e00574-19. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.; Xue, B.; Li, L.; Nan, Y.; Zhang, L.; Li, K.; Zhao, Q.; Hiscox, J.A.; Stewart, J.P.; Wu, C.; et al. Direct Interaction Between CD163 N-Terminal Domain and MYH9 C-Terminal Domain Contributes to Porcine Reproductive and Respiratory Syndrome Virus Internalization by Permissive Cells. Front. Microbiol. 2019, 10, 1815. [Google Scholar] [CrossRef]
- Högger, P.; Sorg, C. Soluble CD163 Inhibits Phorbol Ester-Induced Lymphocyte Proliferation. Biochem. Biophys. Res. Commun. 2001, 288, 841–843. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Lin, S.; Chen, C.; Wang, C.; Ma, Q.; Jiang, B. Identification of Kininogen-1 as a Serum Biomarker for the Early De-tection of Advanced Colorectal Adenoma and Colorectal Cancer. PLoS ONE 2013, 8, e70519. [Google Scholar] [CrossRef]
- Yousef, G.M.; Diamandis, E.P. The New Human Tissue Kallikrein Gene Family: Structure, Function, and Association to Disease. Endocr. Rev. 2001, 22, 184–204. [Google Scholar] [CrossRef]
- Golias, C.; Charalabopoulos, A.; Stagikas, D.; Charalabopoulos, K.; Batistatou, A. The kinin system-bradykinin: Biological effects and clinical implications. Multiple role of the kinin system-bradykinin. Hippokratia 2007, 11, 124–128. [Google Scholar]
- Couture, R.; Harrisson, M.; Vianna, R.M.; Cloutier, F. Kinin receptors in pain and inflammation. Eur. J. Pharmacol. 2001, 429, 161–176. [Google Scholar] [CrossRef]
- Lopes-Ferreira, M.; da Silva Emim, J.A.; Oliveira, V.; Puzer, L.; Cezari, M.H.; da Silva Araujo, M.; Juliano, L.; Jose Lapa, A.; Souccar, C.; Moura-da-Silva, A. Kininogenase activity of Thalassophryne nattereri fish venom. Biochem. Pharmacol. 2004, 68, 2151–2157. [Google Scholar] [CrossRef]
- Vaiyapuri, S.; Harrison, R.A.; Bicknell, A.B.; Gibbins, J.M.; Hutchinson, G. Purification and Functional Characterisation of Rhinocerase, a Novel Serine Protease from the Venom of Bitis gabonica rhinoceros. PLoS ONE 2010, 5, e9687. [Google Scholar] [CrossRef]
- Bilò, M.B.; Martini, M.; Corsi, A.; Tontini, C.; Antonicelli, L. Venom immunotherapy in Europe and the United States. Allergo J. Int. 2020, 29, 29–37. [Google Scholar] [CrossRef]
- Tracy, J.M.; Golden, D.B. Hymenoptera Venom Extracts in Clinical Practice. J. Allergy Clin. Immunol. Pr. 2018, 6, 1856–1862. [Google Scholar] [CrossRef]
- Galli, S.J.; Tsai, M.; Piliponsky, A.M. The development of allergic inflammation. Nature 2008, 454, 445–454. [Google Scholar] [CrossRef]
- Boyle, R.J.; Elremeli, M.; Hockenhull, J.; Cherry, M.G.; Bulsara, M.K.; Daniels, M.; Elberink, J.O. Venom immunotherapy for preventing allergic reactions to insect stings. Cochrane Database Syst. Rev. 2012, 10, CD008838. [Google Scholar] [CrossRef]
- Ruëff, F.; Vos, B.; Oude Elberink, J.; Bender, A.; Chatelain, R.; Dugas-Breit, S.; Horny, H.-P.; Kuchenhoff, H.; Linhardt, A.; Mastnik, S.; et al. Predictors of clinical effectiveness of Hymenoptera venom immunotherapy. Clin. Exp. Allergy 2014, 44, 736–746. [Google Scholar] [CrossRef]
- Jarkvist, J.; Salehi, C.; Akin, C.; Gülen, T. Venom immunotherapy in patients with clonal mast cell disorders: IgG4 correlates with protection. Allergy Eur. J. Allergy Clin. Immunol. 2020, 75, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Albanesi, M.; Nico, A.; Sinisi, A.; Giliberti, L.; Rossi, M.P.; Rossini, M.; Kourtis, G.; Rucco, A.S.; LoConte, F.; Muolo, L.; et al. A 13-year real-life study on efficacy, safety and biological effects of Vespula venom immunotherapy. Clin. Mol. Allergy 2018, 16, 2. [Google Scholar] [CrossRef] [PubMed]
- Arzt, L.; Bokanovic, D.; Schrautzer, C.; Laipold, K.; Möbs, C.; Pfützner, W.; Herzog, S.A.; Vollmann, J.; Reider, N.; Bohle, B.; et al. Immunological differences between insect venom-allergic patients with and without immunotherapy and asymptomatically sensitized subjects. Allergy Eur. J. Allergy Clin. Immunol. 2018, 73, 1223–1231. [Google Scholar] [CrossRef] [PubMed]
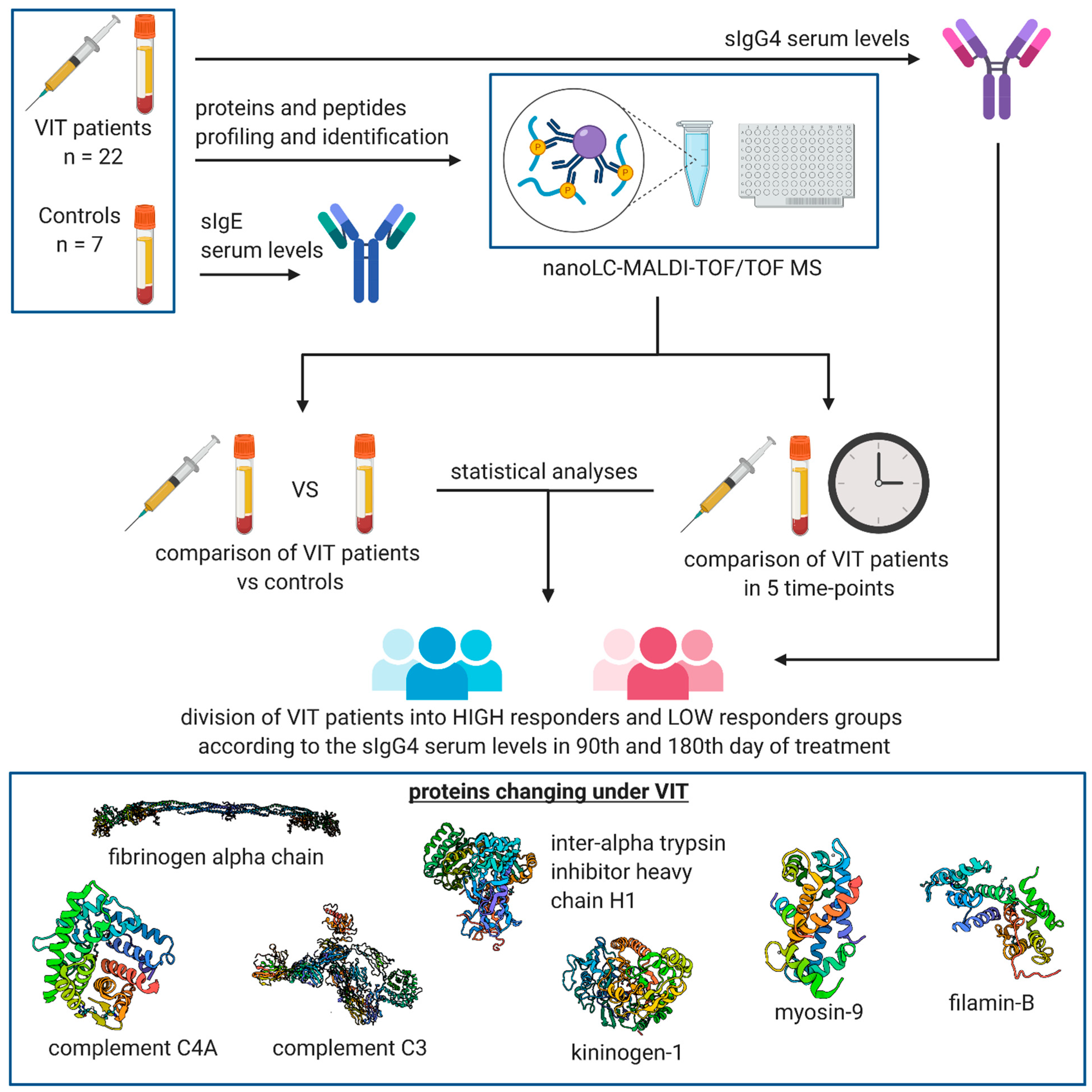

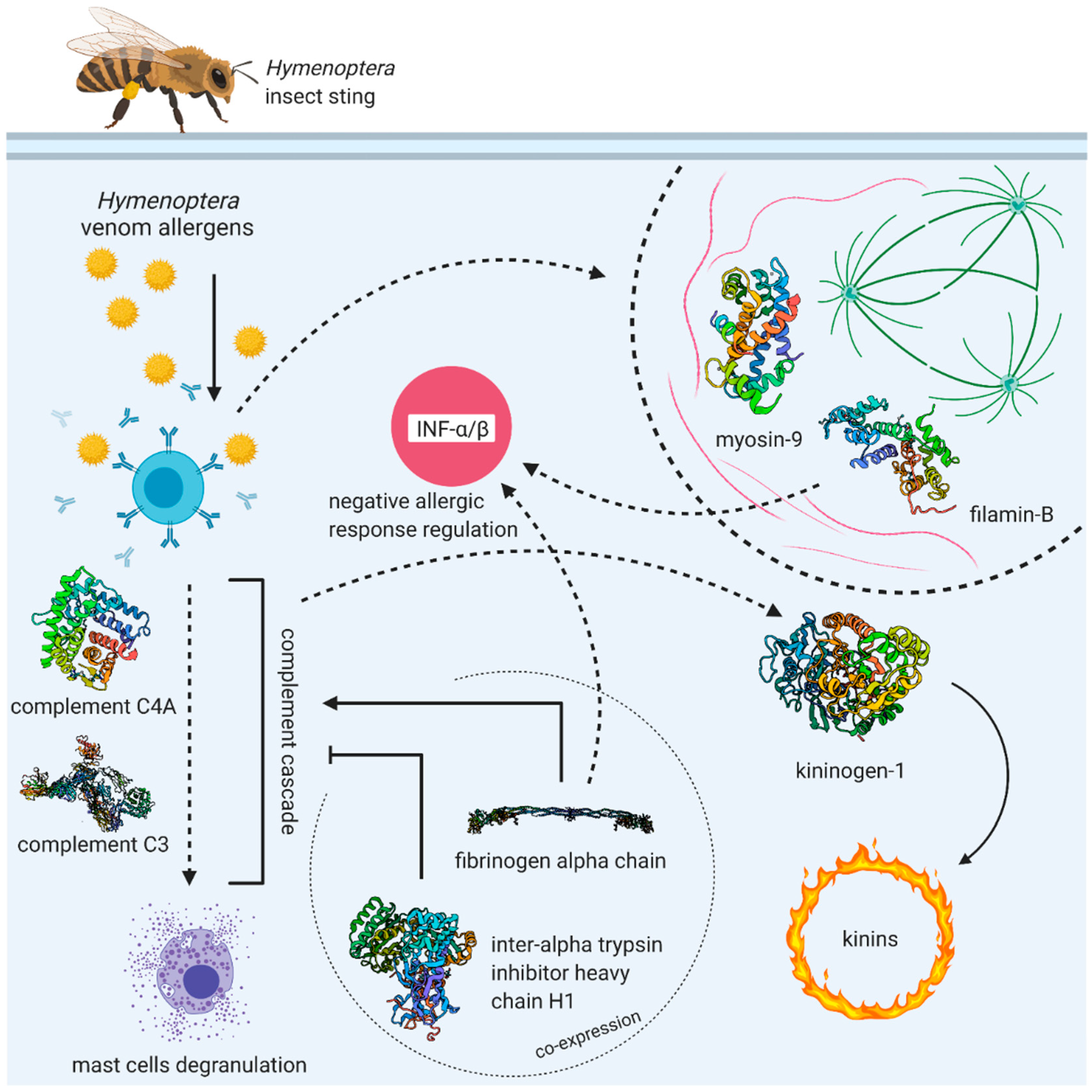
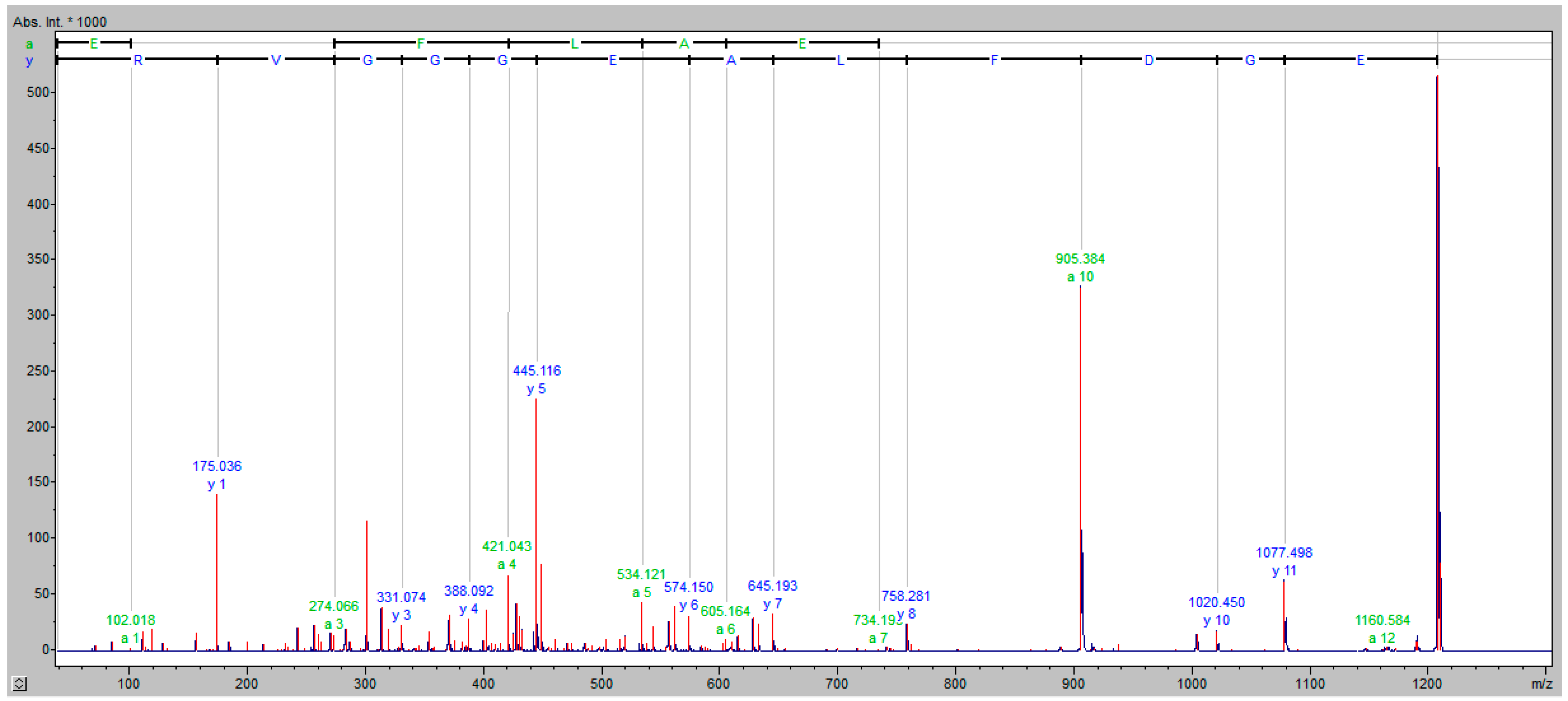
| Patient Number | Age (Years) | Insect That Causes an Allergic Reaction | Grade of the Allergic Reaction according to the Mueller Scale | Additional Diseases |
|---|---|---|---|---|
| Study Group | ||||
| UR-1 | 64 | Wasp | IV | Hypertension |
| UR-2 | 58 | Wasp | III | Allergic rhinitis, asthma |
| UR-3 | 38 | Wasp | III | Asthma |
| UR-4 | 28 | Wasp | III | Asthma |
| UR-5 | 31 | Wasp | III | - |
| UR-7 | 64 | Wasp | III | Asthma, hypertension |
| UR-8 | 54 | Wasp | III | Allergic rhinitis, asthma |
| UR-9 | 67 | Wasp | IV | Hypertension |
| UR-10 | 52 | Wasp | III | Tachycardia |
| UR-11 | 57 | Wasp | III | - |
| UR-19 | 48 | Wasp | III | - |
| UR-28 | 51 | Bee | III | - |
| UR-29 | 62 | Hornet | IV | - |
| UR-31 | 50 | Hornet | IV | Hypertension |
| UR-32 | 47 | Bee | III | - |
| UR-33 | 42 | Wasp | IV | - |
| UR-34 | 37 | Wasp | III | Asthma |
| UR-35 | 45 | Bee | III | - |
| UR-36 | 53 | Wasp | III | - |
| UR-37 | 40 | Bee | III | - |
| UR-38 | 46 | Wasp | III | - |
| UR-40 | 40 | Wasp | III | Allergic rhinitis, asthma |
| Control Group | ||||
| UR-13 | 29 | Wasp | III | Allergic rhinitis |
| UR-14 | 30 | Wasp | I | - |
| UR-20 | 48 | Wasp | III | - |
| UR-21 | 61 | Wasp | IV | Hypertension |
| UR-23 | 40 | Wasp | III | - |
| UR-24 | 51 | Wasp | III | Allergic rhinitis, hypertension, ischemic heart disease |
| UR-25 | 36 | Wasp | I | Allergic rhinitis, hypothyroidism |
| Precursor Ion m/z | Protein Name | 1st Day | 11th Day | 90th Day | |||
|---|---|---|---|---|---|---|---|
| p-Value | FDR | p-Value | FDR | p-Value | FDR | ||
| 1617.57 | Fibrinogen alpha chain | 0.00080919 | 0.029131 | >0.05 | >0.05 | 0.0010915 | 0.0056133 |
| 1449.73 | Complement C4-A | 0.0019183 | 0.03357 | >0.05 | >0.05 | >0.05 | >0.05 |
| 2554.56 | Fibrinogen alpha chain | 0.0027975 | 0.03357 | >0.05 | >0.05 | >0.05 | >0.05 |
| 3315.61 | x | 0.0042908 | 0.038618 | >0.05 | >0.05 | 0.018102 | 0.047737 |
| 1077.93 | Fibrinogen alpha chain | >0.05 | >0.05 | 0.0018253 | 0.044054 | 0.00084769 | 0.0050861 |
| 1020.92 | Fibrinogen alpha chain | >0.05 | >0.05 | 0.0031328 | 0.044054 | 0.00072854 | 0.0050861 |
| 1519.87 | Complement C3 | 0.030118 | >0.05 | 0.0036712 | 0.044054 | 0.00012064 | 0.0014477 |
| 1466.66 | Fibrinogen alpha chain | 0.04616 | >0.05 | >0.05 | >0.05 | 1.1921 × 10−5 | 0.00028324 |
| 1207.25 | Fibrinogen alpha chain | >0.05 | >0.05 | >0.05 | >0.05 | 1.5736 × 10−5 | 0.00028324 |
| 1351.37 | Fibrinogen alpha chain | 0.031164 | >0.05 | >0.05 | >0.05 | 0.0005094 | 0.0045846 |
| 2093.20 | Fibrinogen alpha chain | >0.05 | >0.05 | >0.05 | >0.05 | 0.0035243 | 0.015859 |
| 4053.21 | m/z > 3500 | >0.05 | >0.05 | >0.05 | >0.05 | 0.0042908 | 0.017163 |
| 1537.73 | Fibrinogen alpha chain | 0.011473 | >0.05 | >0.05 | >0.05 | 0.008316 | 0.029938 |
| 1262.25 | Fibrinogen alpha chain | 0.018957 | >0.05 | >0.05 | >0.05 | 0.014095 | 0.042284 |
| 2660.77 | Fibrinogen alpha chain | >0.05 | >0.05 | >0.05 | >0.05 | 0.014095 | 0.042284 |
| 1420.10 | Filamin-B | >0.05 | >0.05 | >0.05 | >0.05 | 0.018564 | 0.047737 |
| Precursor Ion m/z | Protein Name | 90th Day | 180th Day | ||
|---|---|---|---|---|---|
| p-Value | FDR | p-Value | FDR | ||
| 1945.24 | Kininogen-1 | 0.000254 | 0.009156 | 0.000618 | 0.01857 |
| 2093.20 | Fibrinogen alpha chain | 0.000584 | 0.009573 | 0.001032 | 0.01857 |
| 2660.77 | Fibrinogen alpha chain | 0.000798 | 0.009573 | 0.008021 | 0.035102 |
| 1546.56 | Fibrinogen alpha chain | 0.001546 | 0.012693 | 0.00605 | 0.035102 |
| 1331.52 | Myosin-9 | 0.001806 | 0.012693 | >0.05 | >0.05 |
| 3315.61 | x | 0.002116 | 0.012693 | 0.005189 | 0.035102 |
| 4053.21 | m/z > 3500 | 0.004214 | 0.018932 | 0.008776 | 0.035102 |
| 1519.87 | Complement C3 | 0.004545 | 0.018932 | 0.008021 | 0.035102 |
| 1262.25 | Fibrinogen alpha chain | 0.005259 | 0.018932 | 0.006973 | 0.035102 |
| 1537.73 | Fibrinogen alpha chain | 0.005259 | 0.018932 | 0.006973 | 0.035102 |
| 1207.25 | Fibrinogen alpha chain | 0.006998 | 0.022903 | 0.013758 | 0.045027 |
| 1466.66 | Fibrinogen alpha chain | 0.009238 | 0.027159 | 0.012058 | 0.04341 |
| 1866.3 | Complement C3 | 0.009807 | 0.027159 | 0.018957 | >0.05 |
| 1221.15 | Inter-alpha-trypsin inhibitor heavy chain H1 | 0.016742 | 0.043051 | 0.046026 | >0.05 |
| 1020.92 | Fibrinogen alpha chain | 0.019475 | 0.046741 | 0.022328 | >0.05 |
| 3240.91 | Fibrinogen alpha chain | 0.022615 | 0.048532 | 0.04588 | >0.05 |
| 1617.57 | Fibrinogen alpha chain | 0.022918 | 0.048532 | 0.046123 | >0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matysiak, J.; Matuszewska, E.; Kowalski, M.L.; Kosiński, S.W.; Smorawska-Sabanty, E.; Matysiak, J. Association between Venom Immunotherapy and Changes in Serum Protein—Peptide Patterns. Vaccines 2021, 9, 249. https://doi.org/10.3390/vaccines9030249
Matysiak J, Matuszewska E, Kowalski ML, Kosiński SW, Smorawska-Sabanty E, Matysiak J. Association between Venom Immunotherapy and Changes in Serum Protein—Peptide Patterns. Vaccines. 2021; 9(3):249. https://doi.org/10.3390/vaccines9030249
Chicago/Turabian StyleMatysiak, Joanna, Eliza Matuszewska, Marek L. Kowalski, Sławomir W. Kosiński, Ewa Smorawska-Sabanty, and Jan Matysiak. 2021. "Association between Venom Immunotherapy and Changes in Serum Protein—Peptide Patterns" Vaccines 9, no. 3: 249. https://doi.org/10.3390/vaccines9030249
APA StyleMatysiak, J., Matuszewska, E., Kowalski, M. L., Kosiński, S. W., Smorawska-Sabanty, E., & Matysiak, J. (2021). Association between Venom Immunotherapy and Changes in Serum Protein—Peptide Patterns. Vaccines, 9(3), 249. https://doi.org/10.3390/vaccines9030249






