Complete Genome Sequence, Genome Stability and Phylogeny of the Vaccine Strain Mycobacterium bovis BCG SL222 Sofia
Abstract
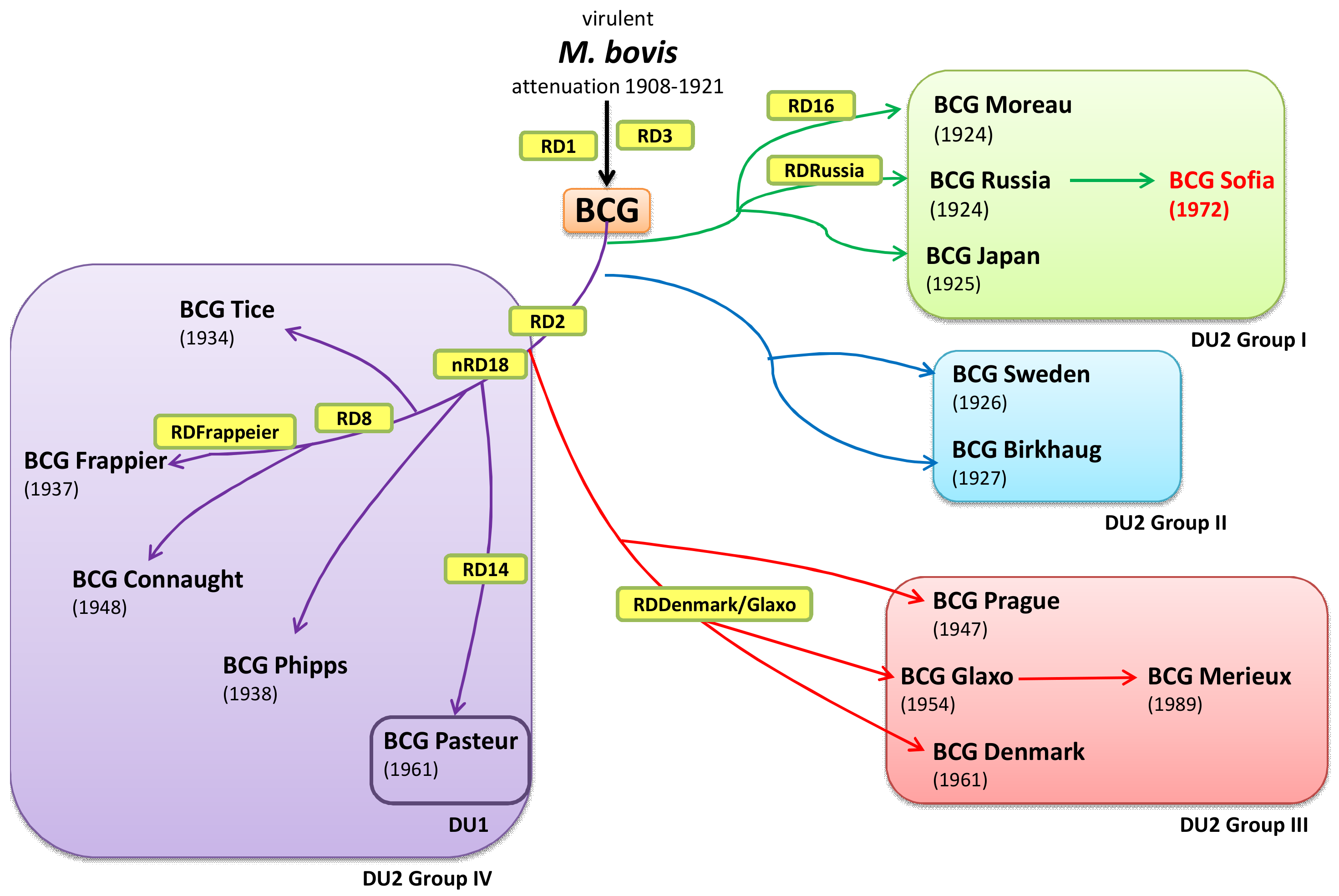
1. Introduction
2. Materials and Methods
2.1. Origin of the BCG SL222 Sofia Sub-Strain
2.2. DNA Isolation and Complete Genome Sequencing Approach
2.3. Genome Sequencing and Assembly
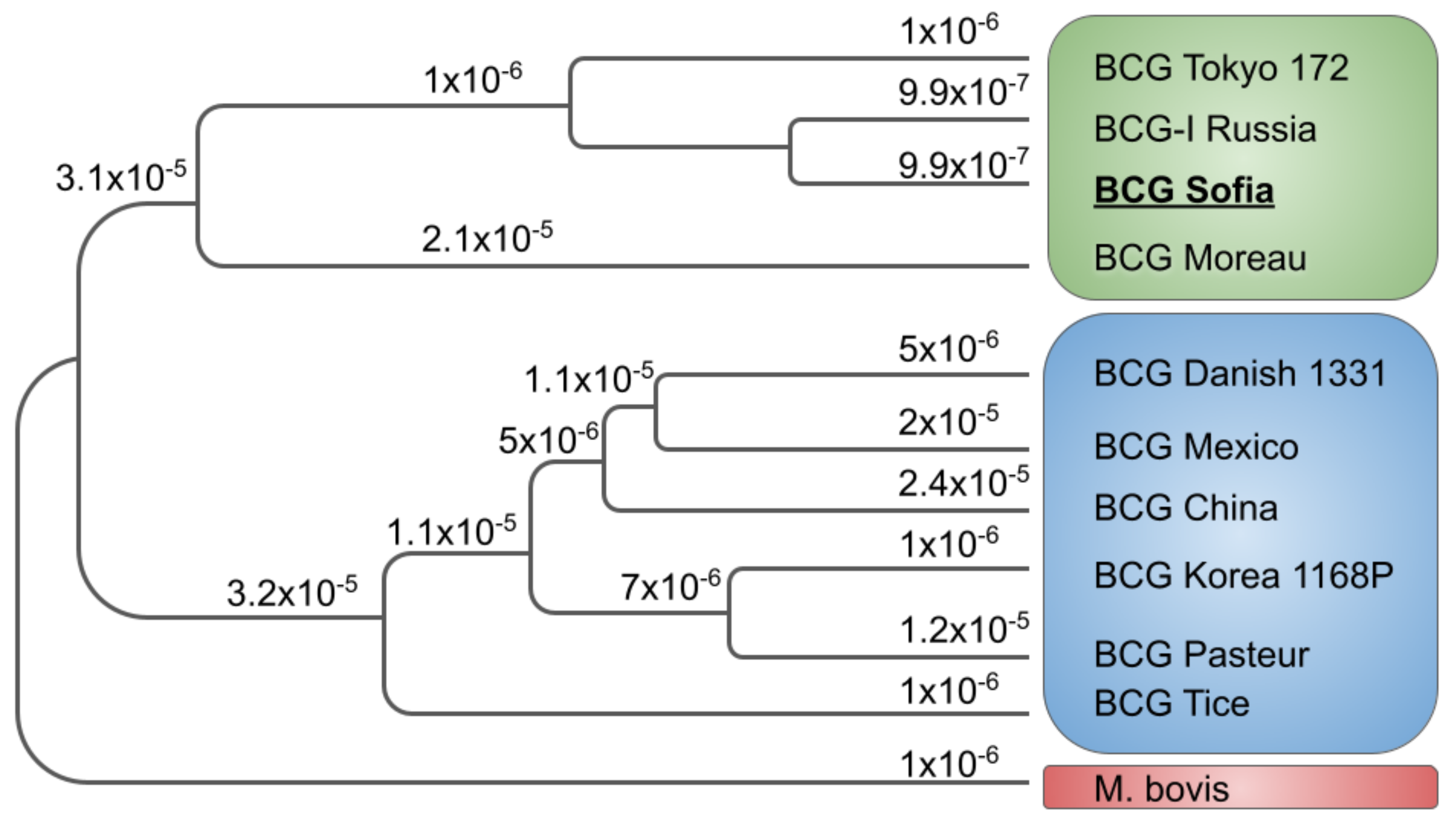
2.4. Phylogeny Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luca, S.; Mihaescu, T. History of BCG vaccine. Maedica 2013, 8, 53–58. [Google Scholar] [PubMed]
- Abdallah, A.M.; Behr, M.A. Evolution and Strain Variation in BCG. In Strain Variation in the Mycobacterium Tuberculosis Complex: Its Role in Biology, Epidemiology and Control; Advances in Experimental Medicine and Biology; Gagneux, S., Ed.; Springer: Cham, Switzerland, 2017; Volume 1019. [Google Scholar] [CrossRef]
- Mostowy, S.; Tsolaki, A.G.; Small, P.M.; Behr, M.A. The in vitro evolution of BCG vaccines. Vaccine 2003, 21, 4270–4274. [Google Scholar] [CrossRef]
- Mangtani, P.; Nguipdop-Djomo, P.; Keogh, R.H.; Trinder, L.; Smith, P.G.; Fine, P.E.; Sterne, J.; Abubakar, I.; Vynnycky, E.; Watson, J.; et al. Observational study to estimate the changes in the effectiveness of bacillus Calmette-Guérin (BCG) vaccination with time since vaccination for preventing tuberculosis in the UK. Health Technol. Assess. 2017, 39, 1–54. [Google Scholar] [CrossRef] [PubMed]
- Cernuschi, T.; Malvolti, S.; Nickels, E.; Friede, M. Bacillus Calmette-Guérin (BCG) vaccine: A global assessment of demand and supply balance. Vaccine 2018, 36, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Stefanova, T. Quality control and safety assessment of BCG vaccines in the post-genomic era. Biotechnol. Biotechnol. Equip. 2014, 28, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Chouchkova, M. Analysis of the Biological Activity of the AntiTB BCG Vaccine. Ph.D. Thesis, National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria, 1981. [Google Scholar]
- Engibarov, A.; Chouchkova, M.; Koychev, C. Studies on the quality control of BCG vaccine: Laboratory examinations and post vaccination control in the field. Dev. Biol. Stand. 1986, 58, 163–171. [Google Scholar]
- Narvskaya, O.; Starkova, D.; Levi, D.; Alexandrova, N.; Molchanov, V.; Chernyaeva, E.; Vyazovaya, A.; Mushkin, A.; Zhuravlev, V.; Solovieva, N.; et al. First insight into the whole-genome sequence variations in Mycobacterium bovis BCG-I (Russia) vaccine seed lots and their progeny clinical isolates from children with BCG-induced adverse events. BMC Genom. 2020, 21, 567. [Google Scholar] [CrossRef] [PubMed]
- Sotnikova, E.A.; Shitikov, E.A.; Malakhova, M.V.; Kostryukova, E.S.; Ilina, E.N.; Atrasheuskaya, A.V.; Ignatyev, G.M.; Vinokurova, N.V.; Gorbachyov, V.Y. Complete genome sequence of Mycobacterium bovis strain BCG-I (Russia). Genome Announc. 2016, 4, e00182-16. [Google Scholar] [CrossRef]
- Levi, D.T.; Obukhov, Y.I.; Aleksandrova, N.V.; Volkova, R.A.; Elbert, E.V.; Alvarez Figeroa, M.V.; Prokopenko, A.V.; Ludanniy, R.I. Identity and stability assessment of BCG vaccine by multiplex PCR. BIOpreparations Prev. Diagn. Treat. 2016, 16, 49–54. (In Russian) [Google Scholar]
- Mustafa, A.S. BCG as a Vector for Novel Recombinant Vaccines against Infectious Diseases and Cancers. Vaccines 2020, 8, 736. [Google Scholar] [CrossRef]
- Stefanova, T. Genetic stability of BCG production in Bulgaria. Probl. Infect. Parasit. Dis. 2015, 43, 30–35. [Google Scholar]
- Ho, M.M.; Markey, K.; Rigsby, P.; Hockley, J.; Corbel, M.J. Report of an International collaborative study to establish the first WHO reference reagents for BCG vaccines of three different sub-strains. Vaccine 2010, 29, 512–518. [Google Scholar] [CrossRef]
- Markey, K.; Rigsby, P.; Hockley, J.; Ho, M.M. WHO Expert Committee on Biological Standardization: International Collaborative Study to Evaluate and Establish the 1st WHO Reference Reagents for BCG Vaccines of Russian BCG-I Sub-Strain; World Health Organization WHO/BS/10.2148; WHO: Geneva, Switzerland, 2010; Available online: https://apps.who.int/iris/handle/10665/70582 (accessed on 9 March 2021).
- WHO. 07/274: BCG Vaccine of Russian BCG-I Sub-Strain (1st WHO Reference Reagent); NIBSC Code: 07/274; Version 3.0, 17/04/2013; WHO: Geneva, Switzerland, 2013; Available online: https://www.nibsc.org/documents/ifu/07-274.pdf (accessed on 9 March 2021).
- Guérin, C. Le BCG et la prevention de la tuberculose. Rev. At. 1948, 27, 183–188. [Google Scholar]
- Van Deinse, F.; Senechal, F. BCG on Sauton’s medium; effect of a long series of subcultures on the morphological and biological properties of BCG cultures. Bull. World Health Organ. 1950, 2, 347–354. [Google Scholar] [PubMed]
- Ausubel, M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Smith, J.A.; Seidman, J.G.; Struhl, K. Current Protocols in Molecular Biology; Greene Publishing Associates: Brooklyn, NY, USA, 1988. [Google Scholar]
- Van Soolingen, D.; de Haas, P.E.; Hermans, P.W.; van Embden, J.D. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 1994, 235, 196–205. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Otrashevskaya, E.V.; Vinokurova, V.N.; Shitikov, E.A.; Sotnikova, E.A.; Perevyshina, T.A.; Kolchenko, S.A.; Butusova, T.B.; Kostryukova, E.S.; Ilina, E.N.; Ignalev, G.M. M. bovis BCG-1 (Russia) sub-strain genome stability investigation within the entire production process. J. Microbiol. Epidemiol. Immunobiol. 2018, 2, 58–67. (In Russian) [Google Scholar] [CrossRef]
- Stefanova, T.; Chouchkova, M.; Hinds, J.; Butcher, P.; Inwald, J.; Dale, J.; Hewinson, R.G.; Gordon, S.V. Genetic composition of Mycobacterium bovis BCG Sub-strain Sofia. J. Clin. Microbiol. 2003, 41, 5349. [Google Scholar] [CrossRef][Green Version]
- Voronina, O.L.; Kunda, M.S.; Aksenova, E.I.; Semenov, A.N.; Ryzova, N.N.; Lunin, V.G.; Gintsburg, A.L. Mosaic structure of Mycobacterium bovis BCG genomes as a representation of phage sequences’ mobility. BMC Genom. 2016, 17, 1009. [Google Scholar] [CrossRef][Green Version]
- Abdallah, A.M.; Hill-Cawthorne, G.A.; Otto, T.D.; Coll, F.; Guerra-Assuncao, J.A.; Gao, G.; Naem, R.; Ansari, H.; Malas, T.B.; Adrob, S.A.; et al. Genomic expression catalogue of a global collection of BCG vaccine strains show evidence for highly diverged metabolic and cell-wall adaptations. Sci. Rep. 2015, 5, 15443. [Google Scholar] [CrossRef]
- Keller, P.M.; Böttger, E.C.; Sander, P. Tuberculosis vaccine strain Mycobacterium bovis BCG Russia is a natural recA mutant. BMC Microbiol. 2008, 8, 120. [Google Scholar] [CrossRef]
- Borgers, K.; Ou, J.Y.; Zheng, P.X.; Tiles, P.; Van Hecke, A.; Plets, E.; Michielsen, G.; Festijens, N.; Callewaert, N.; Lin, Y.C. Reference genome and comparative genome analysis for the WHO reference strain for Mycobacterium bovis BCG Danish, the present tuberculosis vaccine. BMC Genom. 2019, 20, 561. [Google Scholar] [CrossRef]
- Moorlag, S.J.; Arts, R.J.; van Crevel, R.; Netea, M.G. Nonspecific effects of BCG vaccine on viral infections. Clin. Microbiol. Infect. 2019, 25, 1473. [Google Scholar] [CrossRef]
- Van Puffelen, J.H.; Keating, S.T.; Oosterwijk, E.; van der Heijden, A.G.; Netea, M.G.; Joosten, L.; Vermenulen, S.H. Trained immunity as a molecular mechanism for BCG immunotherapy in bladder cancer. Nat. Rev. Urol. 2020, 17, 513–525. [Google Scholar] [CrossRef]
| BCG Sub-Strain | GenBank Accession Number | Length (bp) |
|---|---|---|
| BCG-I Russia-368 | NZ_CP013741 | 4,370,705 |
| BCG SL222 Sofia | CP064405 | 4,370,706 |
| BCG Danish 1331 | NZ_CP039850 | 4,411,814 |
| BCG Tice | NZ_CUWQ01000001 | 4,317,625 |
| BCG China | NZ_CUWG01000001 | 4,318,346 |
| BCG Tokyo 172 | CP014566 | 4,371,707 |
| BCG Moreau RDJ | NZ_AM412059 | 4,339,996 |
| BCG Mexico | CP002095 | 4,350,386 |
| BCG Pasteur 1173P2 | NC008769 | 4,374,550 |
| BCG Korea 1168P | NC020245 | 4,376,727 |
| Mycobacterium bovis AF212297 | LT708304 | 4,349,904 |
| Position (bp) | BCG-I SL368 | BCG SL222 Sofia | Type of Mutation | Ammino Acid Change | Gene |
|---|---|---|---|---|---|
| 841,951 | (G)6 | (G)5 | Non-synonymous deletion | Frameshift Gly -> Arg | PE family protein |
| 2,397,101 | (C)4 | (C)5 | Synonymous insertion | Frameshift Gly -> Gly | PE family protein |
| 3,175,530 | A | G | Synonymous transition | Ala -> Ala | Protein-PII uridylyltransferase |
| 3,912,049 | (G)4 | (G)5 | Synonymous insertion | Frameshift Gly -> Gly | PE family protein |
| DELETIONS > 1000 bp | |||
|---|---|---|---|
| Nucleotide Position | Size (bp) | Affected CDS | Genes/Proteins |
| 1,415,767–1,420,151 | 4384 | 3 ORFs | tbd2, pknH1, pknH2 |
| 1,772,330–1,781,585 | 9255 | 14 ORFs | RD3, probable phage proteins |
| 2,612,546–2,615,131 | 2585 | 2 ORFs | ppe40, ppe71 |
| 4,130,922–4,132,530 | 1608 | 3 ORFs | RDRussia—glutamate-cysteine ligase |
| 4,340,448–4,349,958 | 9510 | 9 ORFs | RD1—esx-1 (secretion associated protein), esat-6, espk, cfp10 |
| INSERTIONS > 800 bp | |||
| 1,761,134–1,761,946 | 812 | 2 ORFs | Putative acyltransferase |
| 3,669,049–3,710,643 | 41,594 | 42 ORFs | DU2 region |
| 4,361,253–4,363,666 | 2413 | 3 ORFs | Type VII secretion protein EccB, insertion not found in BCG-Moreau |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panaiotov, S.; Hodzhev, Y.; Tolchkov, V.; Tsafarova, B.; Mihailov, A.; Stefanova, T. Complete Genome Sequence, Genome Stability and Phylogeny of the Vaccine Strain Mycobacterium bovis BCG SL222 Sofia. Vaccines 2021, 9, 237. https://doi.org/10.3390/vaccines9030237
Panaiotov S, Hodzhev Y, Tolchkov V, Tsafarova B, Mihailov A, Stefanova T. Complete Genome Sequence, Genome Stability and Phylogeny of the Vaccine Strain Mycobacterium bovis BCG SL222 Sofia. Vaccines. 2021; 9(3):237. https://doi.org/10.3390/vaccines9030237
Chicago/Turabian StylePanaiotov, Stefan, Yordan Hodzhev, Vladimir Tolchkov, Borislava Tsafarova, Alexander Mihailov, and Tzvetelina Stefanova. 2021. "Complete Genome Sequence, Genome Stability and Phylogeny of the Vaccine Strain Mycobacterium bovis BCG SL222 Sofia" Vaccines 9, no. 3: 237. https://doi.org/10.3390/vaccines9030237
APA StylePanaiotov, S., Hodzhev, Y., Tolchkov, V., Tsafarova, B., Mihailov, A., & Stefanova, T. (2021). Complete Genome Sequence, Genome Stability and Phylogeny of the Vaccine Strain Mycobacterium bovis BCG SL222 Sofia. Vaccines, 9(3), 237. https://doi.org/10.3390/vaccines9030237








