VLP-Based COVID-19 Vaccines: An Adaptable Technology against the Threat of New Variants
Abstract
:1. Introduction
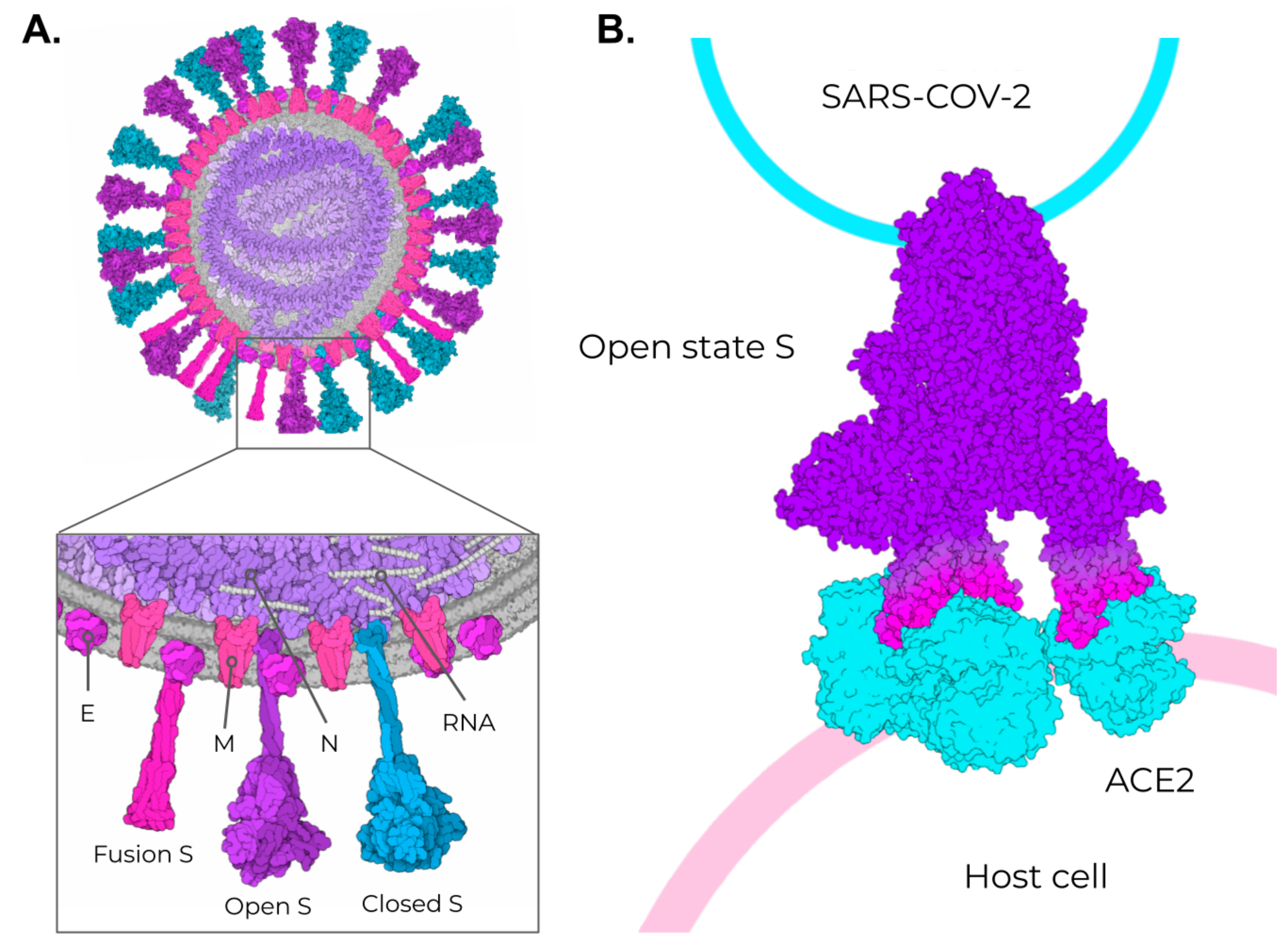
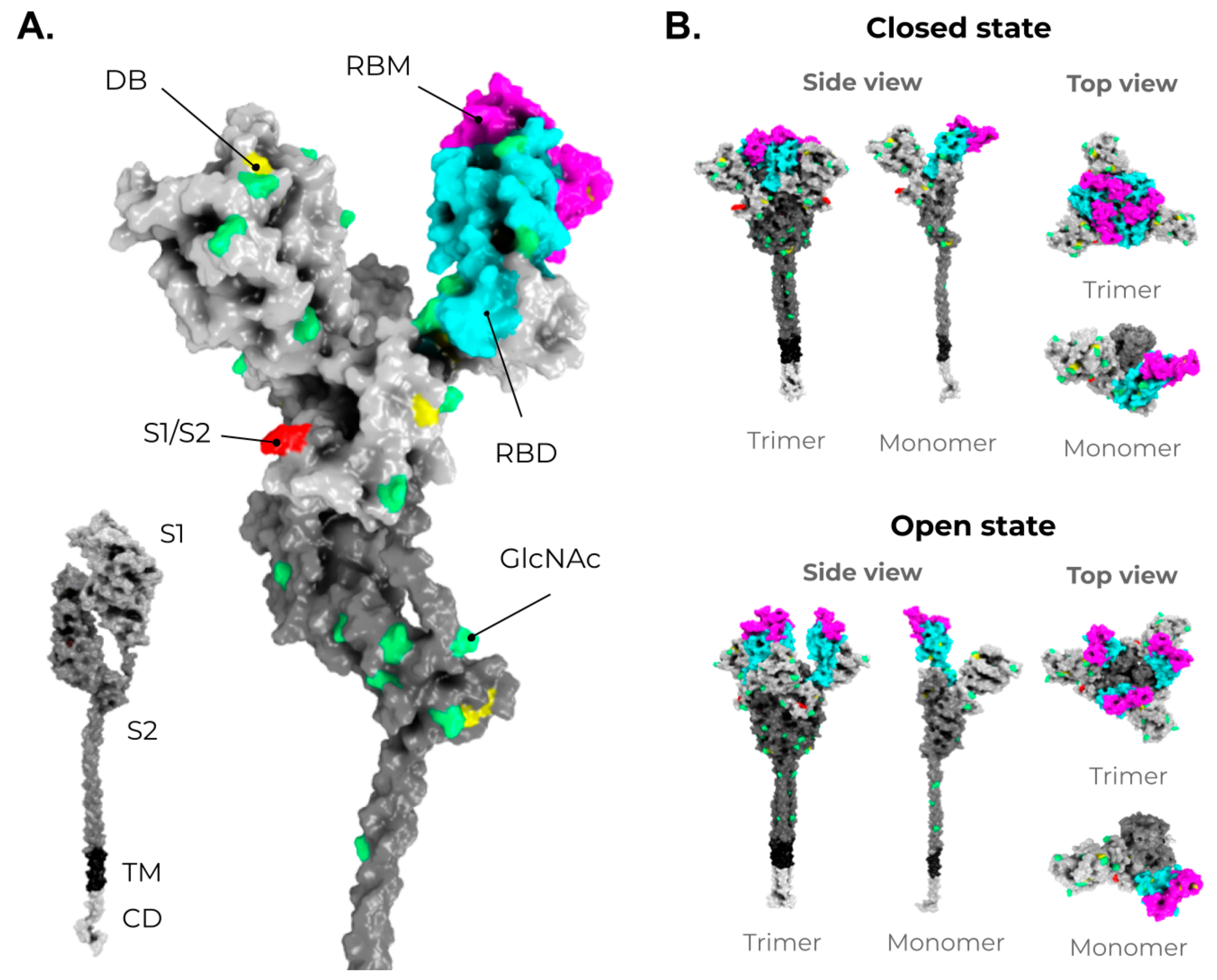
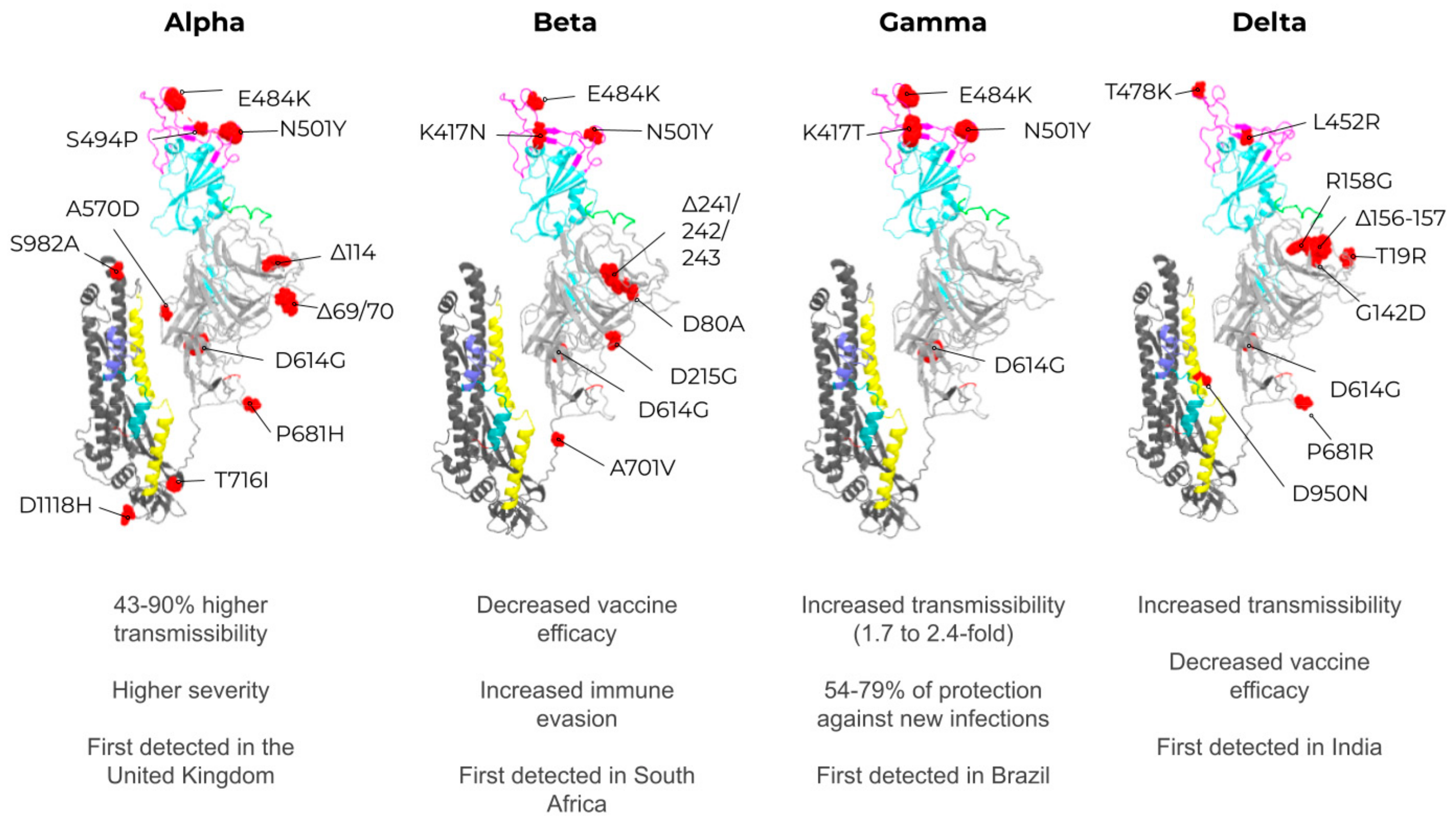
2. SARS-CoV-2, VOCs, and Structural Vaccinology
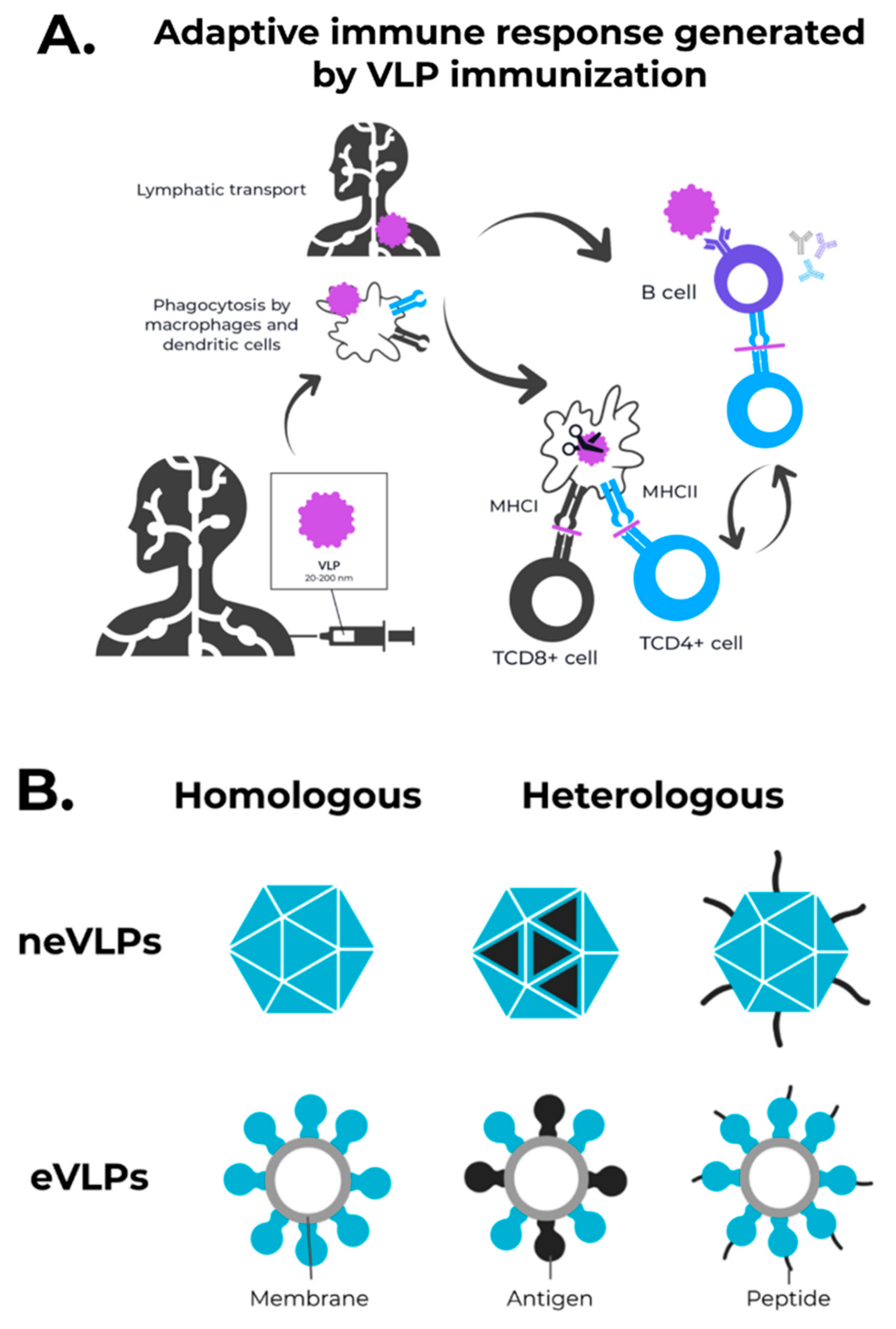
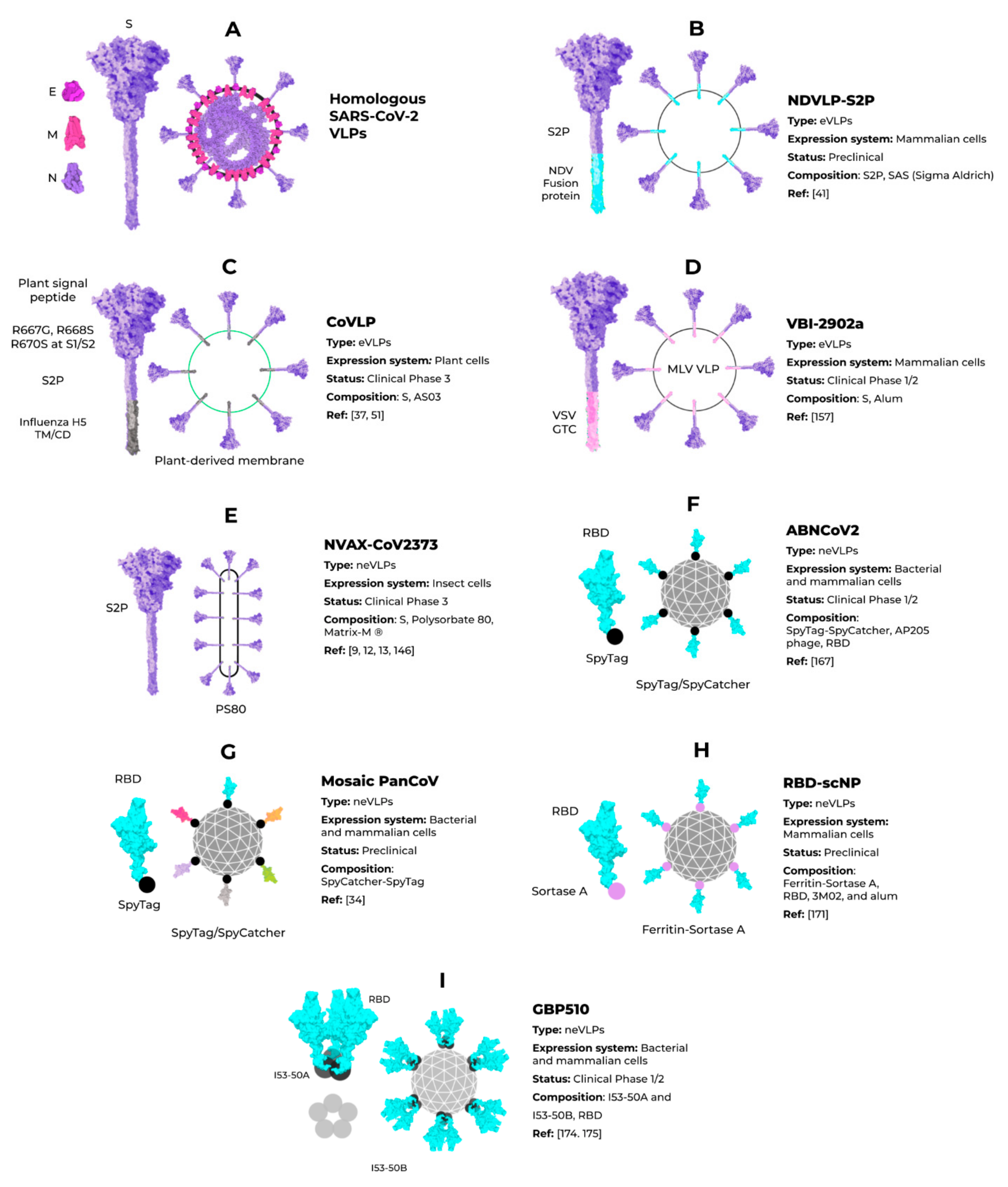
3. Enveloped VLPs against SARS-CoV-2
4. Non-Enveloped VLPs against SARS-CoV-2
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Weekly Epidemiological Update on COVID-19—22 June 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---22-june-2021 (accessed on 23 June 2021).
- Li, Y.; Tenchov, R.; Smoot, J.; Liu, C.; Watkins, S.; Zhou, Q. A Comprehensive Review of the Global Efforts on COVID-19 Vaccine Development. ACS Cent. Sci. 2021, 7, 512–533. [Google Scholar] [CrossRef] [PubMed]
- WHO Issues Emergency Use Listing for Eighth COVID-19 Vaccine. Available online: https://www.who.int/news/item/03-11-2021-who-issues-emergency-use-listing-for-eighth-covid-19-vaccine (accessed on 7 November 2021).
- COVID-19 Vaccines. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines (accessed on 7 November 2021).
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an Inactivated Vaccine Candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, G.; Pan, H.; Li, C.; Hu, Y.; Chu, K.; Han, W.; Chen, Z.; Tang, R.; Yin, W.; et al. Safety, Tolerability, and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine in Healthy Adults Aged 18–59 Years: A Randomised, Double-Blind, Placebo-Controlled, Phase 1/2 Clinical Trial. Lancet Infect. Dis. 2021, 21, 181–192. [Google Scholar] [CrossRef]
- Bueno, S.M.; Abarca, K.; González, P.A.; Gálvez, N.M.; Soto, J.A.; Duarte, L.F.; Schultz, B.M.; Pacheco, G.A.; González, L.A.; Vázquez, Y.; et al. Interim Report: Safety and Immunogenicity of an Inactivated Vaccine against SARS-CoV-2 in Healthy Chilean Adults in a Phase 3 Clinical Trial. medRxiv 2021. [Google Scholar] [CrossRef]
- Tanriover, M.D.; Doğanay, H.L.; Akova, M.; Güner, H.R.; Azap, A.; Akhan, S.; Köse, Ş.; Erdinç, F.Ş.; Akalın, E.H.; Tabak, Ö.F.; et al. Efficacy and Safety of an Inactivated Whole-Virion SARS-CoV-2 Vaccine (CoronaVac): Interim Results of a Double-Blind, Randomised, Placebo-Controlled, Phase 3 Trial in Turkey. Lancet 2021, 398, 213–222. [Google Scholar] [CrossRef]
- Xia, S.; Duan, K.; Zhang, Y.; Zhao, D.; Zhang, H.; Xie, Z.; Li, X.; Peng, C.; Zhang, Y.; Zhang, W.; et al. Effect of an Inactivated Vaccine Against SARS-CoV-2 on Safety and Immunogenicity Outcomes: Interim Analysis of 2 Randomized Clinical Trials. JAMA 2020, 324, 951–960. [Google Scholar] [CrossRef]
- Al Kaabi, N.; Zhang, Y.; Xia, S.; Yang, Y.; Al Qahtani, M.M.; Abdulrazzaq, N.; Al Nusair, M.; Hassany, M.; Jawad, J.S.; Abdalla, J.; et al. Effect of 2 Inactivated SARS-CoV-2 Vaccines on Symptomatic COVID-19 Infection in Adults: A Randomized Clinical Trial. JAMA 2021, 326, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Ella, R.; Reddy, S.; Blackwelder, W.; Potdar, V.; Yadav, P.; Sarangi, V.; Aileni, V.K.; Kanungo, S.; Rai, S.; Reddy, P.; et al. Efficacy, Safety, and Lot to Lot Immunogenicity of an Inactivated SARS-CoV-2 Vaccine (BBV152): A, Double-Blind, Randomised, Controlled Phase 3 Trial. medRxiv 2021. [Google Scholar] [CrossRef]
- Ella, R.; Vadrevu, K.M.; Jogdand, H.; Prasad, S.; Reddy, S.; Sarangi, V.; Ganneru, B.; Sapkal, G.; Yadav, P.; Abraham, P.; et al. Safety and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine, BBV152: A Double-Blind, Randomised, Phase 1 Trial. Lancet. Infect. Dis. 2021, 21, 637–646. [Google Scholar] [CrossRef]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and Immunogenicity of the ChAdOx1 NCoV-19 Vaccine against SARS-CoV-2: A Preliminary Report of a Phase 1/2, Single-Blind, Randomised Controlled Trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Falsey, A.R.; Sobieszczyk, M.E.; Hirsch, I.; Sproule, S.; Robb, M.L.; Corey, L.; Neuzil, K.M.; Hahn, W.; Hunt, J.; Mulligan, M.J.; et al. Phase 3 Safety and Efficacy of AZD1222 (ChAdOx1 NCoV-19) COVID-19 Vaccine. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and Efficacy of the ChAdOx1 NCoV-19 Vaccine (AZD1222) against SARS-CoV-2: An Interim Analysis of Four Randomised Controlled Trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Sadoff, J.; Le Gars, M.; Shukarev, G.; Heerwegh, D.; Truyers, C.; de Groot, A.M.; Stoop, J.; Tete, S.; Van Damme, W.; Leroux-Roels, I.; et al. Interim Results of a Phase 1–2a Trial of Ad26.COV2.S COVID-19 Vaccine. N. Engl. J. Med. 2021, 384, 1824–1835. [Google Scholar] [CrossRef]
- Bos, R.; Rutten, L.; van der Lubbe, J.E.M.; Bakkers, M.J.G.; Hardenberg, G.; Wegmann, F.; Zuijdgeest, D.; de Wilde, A.H.; Koornneef, A.; Verwilligen, A.; et al. Ad26 Vector-Based COVID-19 Vaccine Encoding a Prefusion-Stabilized SARS-CoV-2 Spike Immunogen Induces Potent Humoral and Cellular Immune Responses. NPJ Vaccines 2020, 5, 1–11. [Google Scholar] [CrossRef]
- Mercado, N.B.; Zahn, R.; Wegmann, F.; Loos, C.; Chandrashekar, A.; Yu, J.; Liu, J.; Peter, L.; McMahan, K.; Tostanoski, L.H.; et al. Single-Shot Ad26 Vaccine Protects against SARS-CoV-2 in Rhesus Macaques. Nature 2020, 586, 583–588. [Google Scholar] [CrossRef]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 MRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Augusto, G.; Bachmann, M.F. The 3Ds in Virus-like Particle Based-vaccines: “Design, Delivery and Dynamics”. Immunol. Rev. 2020, 296, 155–168. [Google Scholar] [CrossRef]
- Bachmann, M.; Rohrer, U.; Kundig, T.; Burki, K.; Hengartner, H.; Zinkernagel, R. The Influence of Antigen Organization on B Cell Responsiveness. Science 1993, 262, 1448–1451. [Google Scholar] [CrossRef]
- Cubas, R.; Zhang, S.; Kwon, S.; Sevick-Muraca, E.M.; Li, M.; Chen, C.; Yao, Q. Virus-like Particle (VLP) Lymphatic Trafficking and Immune Response Generation After Immunization by Different Routes. J. Immunother. 2009, 32, 118–128. [Google Scholar] [CrossRef] [Green Version]
- Mohsen, M.; Gomes, A.; Vogel, M.; Bachmann, M. Interaction of Viral Capsid-Derived Virus-Like Particles (VLPs) with the Innate Immune System. Vaccines 2018, 6, 37. [Google Scholar] [CrossRef] [Green Version]
- Win, S.J.; Ward, V.K.; Dunbar, P.R.; Young, S.L.; Baird, M.A. Cross-presentation of Epitopes on Virus-like Particles via the MHC I Receptor Recycling Pathway. Immunol. Cell Biol. 2011, 89, 681–688. [Google Scholar] [CrossRef]
- Bangaru, S.; Ozorowski, G.; Turner, H.L.; Antanasijevic, A.; Huang, D.; Wang, X.; Torres, J.L.; Diedrich, J.K.; Tian, J.-H.; Portnoff, A.D.; et al. Structural Analysis of Full-Length SARS-CoV-2 Spike Protein from an Advanced Vaccine Candidate. Science 2020, 370, 1089–1094. [Google Scholar] [CrossRef]
- Block, S.L.; Nolan, T.; Sattler, C.; Barr, E.; Giacoletti, K.E.D.; Marchant, C.D.; Castellsagué, X.; Rusche, S.A.; Lukac, S.; Bryan, J.T.; et al. Comparison of the Immunogenicity and Reactogenicity of a Prophylactic Quadrivalent Human Papillomavirus (Types 6, 11, 16, and 18) L1 Virus-Like Particle Vaccine in Male and Female Adolescents and Young Adult Women. Pediatrics 2006, 118, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.A.; Gnanapragasam, P.N.P.; Lee, Y.E.; Hoffman, P.R.; Ou, S.; Kakutani, L.M.; Keeffe, J.R.; Wu, H.-J.; Howarth, M.; West, A.P.; et al. Mosaic Nanoparticles Elicit Cross-Reactive Immune Responses to Zoonotic Coronaviruses in Mice. Science 2021, 371, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1–2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef]
- Tian, J.H.; Patel, N.; Haupt, R.; Zhou, H.; Weston, S.; Hammond, H.; Logue, J.; Portnoff, A.D.; Norton, J.; Guebre-Xabier, M.; et al. SARS-CoV-2 Spike Glycoprotein Vaccine Candidate NVX-CoV2373 Immunogenicity in Baboons and Protection in Mice. Nat. Commun. 2021, 12, 372. [Google Scholar] [CrossRef]
- Ward, B.J.; Makarkov, A.; Séguin, A.; Pillet, S.; Trépanier, S.; Dhaliwall, J.; Libman, M.D.; Vesikari, T.; Landry, N. Efficacy, Immunogenicity, and Safety of a Plant-Derived, Quadrivalent, Virus-like Particle Influenza Vaccine in Adults (18–64 Years) and Older Adults (≥65 Years): Two Multicentre, Randomised Phase 3 Trials. Lancet 2020, 396, 1491–1503. [Google Scholar] [CrossRef]
- Braun, M.; Jandus, C.; Maurer, P.; Hammann-Haenni, A.; Schwarz, K.; Bachmann, M.F.; Speiser, D.E.; Romero, P. Virus-like Particles Induce Robust Human T-Helper Cell Responses: Cellular Immune Response. Eur. J. Immunol. 2012, 42, 330–340. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like Particles: Preparation, Immunogenicity and Their Roles as Nanovaccines and Drug Nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Li, M.; Cripe, T.P.; Estes, P.A.; Lyon, M.K.; Rose, R.C.; Garcea, R.L. Expression of the Human Papillomavirus Type 11 L1 Capsid Protein in Escherichia Coli: Characterization of Protein Domains Involved in DNA Binding and Capsid Assembly. J. Virol. 1997, 71, 2988–2995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Shi, W.; Abiona, O.M.; Nazzari, A.; Olia, A.S.; Ou, L.; Phung, E.; Stephens, T.; Tsybovsky, Y.; Verardi, R.; et al. Newcastle Disease Virus-Like Particles Displaying Prefusion-Stabilized SARS-CoV-2 Spikes Elicit Potent Neutralizing Responses. Vaccines 2021, 9, 73. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent Neutralizing Antibodies from COVID-19 Patients Define Multiple Targets of Vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef]
- Dormitzer, P.R.; Ulmer, J.B.; Rappuoli, R. Structure-Based Antigen Design: A Strategy for next Generation Vaccines. Trends Biotechnol. 2008, 26, 659–667. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Xia, H.; Zhang, X.; Fontes-Garfias, C.R.; Swanson, K.A.; Cai, H.; Sarkar, R.; Chen, W.; Cutler, M.; et al. Neutralizing Activity of BNT162b2-Elicited Serum. N. Engl. J. Med. 2021, 384, 1466–1468. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and Structures of a Rationally Designed Prefusion MERS-CoV Spike Antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wec, A.Z.; Wrapp, D.; Herbert, A.S.; Maurer, D.P.; Haslwanter, D.; Sakharkar, M.; Jangra, R.K.; Dieterle, M.E.; Lilov, A.; Huang, D.; et al. Broad Neutralization of SARS-Related Viruses by Human Monoclonal Antibodies. Science 2020, 369, 731–736. [Google Scholar] [CrossRef] [PubMed]
- LCS, C.; BM, R.; GW, B. Production of GP64-Free Virus-like Particles from Baculovirus-Infected Insect Cells. J. Gen. Virol. 2018, 99, 265–274. [Google Scholar] [CrossRef]
- Plescia, C.B.; David, E.A.; Patra, D.; Sengupta, R.; Amiar, S.; Su, Y.; Stahelin, R. V SARS-CoV-2 Viral Budding and Entry Can Be Modeled Using BSL-2 Level Virus-like Particles. J. Biol. Chem. 2021, 296, 100103. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Shi, M.; Li, J.; Song, P.; Li, N. Corrigendum: Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol. 2020, 8, 1026. [Google Scholar] [CrossRef] [PubMed]
- Atcheson, E.; Cabral-Miranda, G.; Salman, A.M.; Reyes-Sandoval, A. Discovery of Four New B-Cell Protective Epitopes for Malaria Using Q Beta Virus-like Particle as Platform. NPJ Vaccines 2020, 5, 92. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.-Y.; Couture, M.; D’Aoust, M.-A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 Randomized Trial of a Plant-Derived Virus-like Particle Vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Liu, X.; Xu, Q.; Wang, Z.; Chen, J.; Li, T.; Zheng, Q.; Yu, H.; Gu, Y.; Li, S.; et al. Recent Progress on the Versatility of Virus-Like Particles. Vaccines 2020, 8, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.B.; Kim, J.-H.; Hur, W.; Choi, J.E.; Kim, S.M.; Park, D.J.; Kang, B.-Y.; Lee, G.W.; Yoon, S.K. Liver-Specific Gene Delivery Using Engineered Virus-Like Particles of Hepatitis E Virus. Sci. Rep. 2019, 9, 1616. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.-H.; Chen, P.-Y.; Wei, K.-C.; Huang, C.-W.; Shiue, Y.-L.; Huang, C.-Y.; Yang, H.-W. Convection-Enhanced Delivery of a Virus-Like Nanotherapeutic Agent with Dual-Modal Imaging for Besiegement and Eradication of Brain Tumors. Theranostics 2019, 9, 1752–1763. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.; Steinmetz, N.F. Delivery of SiRNA Therapeutics Using Cowpea Chlorotic Mottle Virus-like Particles. Biomater. Sci. 2019, 7, 3138–3142. [Google Scholar] [CrossRef] [PubMed]
- Azizgolshani, O.; Garmann, R.F.; Cadena-Nava, R.; Knobler, C.M.; Gelbart, W.M. Reconstituted Plant Viral Capsids Can Release Genes to Mammalian Cells. Virology 2013, 441, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Crooke, S.N.; Schimer, J.; Raji, I.; Wu, B.; Oyelere, A.K.; Finn, M.G. Lung Tissue Delivery of Virus-Like Particles Mediated by Macrolide Antibiotics. Mol. Pharm. 2019, 16, 2947–2955. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Sue, S.-C.; Yu, T.; Hsieh, C.-M.; Tsai, C.-K.; Chiang, Y.-C.; Lee, S.; Hsiao, H.; Wu, W.-J.; Chang, W.-L.; et al. Modular Organization of SARS Coronavirus Nucleocapsid Protein. J. Biomed. Sci. 2006, 13, 59–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yacoby, I.; Shamis, M.; Bar, H.; Shabat, D.; Benhar, I. Targeting Antibacterial Agents by Using Drug-Carrying Filamentous Bacteriophages. Antimicrob. Agents Chemother. 2006, 50, 2087–2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dashti, N.H.; Abidin, R.S.; Sainsbury, F. Programmable In Vitro Coencapsidation of Guest Proteins for Intracellular Delivery by Virus-like Particles. ACS Nano 2018, 12, 4615–4623. [Google Scholar] [CrossRef] [PubMed]
- Koyani, R.; Pérez-Robles, J.; Cadena-Nava, R.D.; Vazquez-Duhalt, R. Biomaterial-Based Nanoreactors, an Alternative for Enzyme Delivery. Nanotechnol. Rev. 2017, 6, 405–419. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, L.; Cadena-Nava, R.D.; Palomares, L.A.; Ruiz-Garcia, J.; Koay, M.S.T.; Cornelissen, J.J.M.T.; Vazquez-Duhalt, R. Chemotherapy Pro-Drug Activation by Biocatalytic Virus-like Nanoparticles Containing Cytochrome P450. Enzyme Microb. Technol. 2014, 60, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. In Coronaviruses; Maier, H.J., Bickerton, E., Britton, P., Eds.; Springer: New York, NY, USA, 2015; Volume 1282, pp. 1–23. ISBN 978-1-4939-2437-0. [Google Scholar]
- Neuman, B.W.; Joseph, J.S.; Saikatendu, K.S.; Serrano, P.; Chatterjee, A.; Johnson, M.A.; Liao, L.; Klaus, J.P.; Yates, J.R.; Wuthrich, K.; et al. Proteomics Analysis Unravels the Functional Repertoire of Coronavirus Nonstructural Protein 3. J. Virol. 2008, 82, 5279–5294. [Google Scholar] [CrossRef] [Green Version]
- de Haan, C.A.; Vennema, H.; Rottier, P.J. Assembly of the Coronavirus Envelope: Homotypic Interactions between the M Proteins. J. Virol. 2000, 74, 4967–4978. [Google Scholar] [CrossRef] [PubMed]
- Godeke, G.-J.; de Haan, C.A.M.; Rossen, J.W.A.; Vennema, H.; Rottier, P.J.M. Assembly of Spikes into Coronavirus Particles Is Mediated by the Carboxy-Terminal Domain of the Spike Protein. J. Virol. 2000, 74, 1566–1571. [Google Scholar] [CrossRef] [Green Version]
- Pervushin, K.; Tan, E.; Parthasarathy, K.; Lin, X.; Jiang, F.L.; Yu, D.; Vararattanavech, A.; Soong, T.W.; Liu, D.X.; Torres, J. Structure and Inhibition of the SARS Coronavirus Envelope Protein Ion Channel. PLoS Pathog. 2009, 5, e1000511. [Google Scholar] [CrossRef] [PubMed]
- Godet, M.; L’Haridon, R.; Vautherot, J.-F.; Laude, H. TGEV Corona Virus ORF4 Encodes a Membrane Protein That Is Incorporated into Virions. Virology 1992, 188, 666–675. [Google Scholar] [CrossRef]
- Liu, D.X.; Inglis, S.C. Association of the Infectious Bronchitis Virus 3c Protein with the Virion Envelope. Virology 1991, 185, 911–917. [Google Scholar] [CrossRef]
- Boscarino, J.A.; Logan, H.L.; Lacny, J.J.; Gallagher, T.M. Envelope Protein Palmitoylations Are Crucial for Murine Coronavirus Assembly. J. Virol. 2008, 82, 2989–2999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machamer, C.E.; Youn, S. The transmembrane domain of the infectious bronchitis virus E protein is required for efficient virus release. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2006; Volume 581, pp. 193–198. ISBN 9780387262024. [Google Scholar]
- Ruch, T.R.; Machamer, C.E. The Hydrophobic Domain of Infectious Bronchitis Virus E Protein Alters the Host Secretory Pathway and Is Important for Release of Infectious Virus. J. Virol. 2011, 85, 675–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruch, T.R.; Machamer, C.E. A Single Polar Residue and Distinct Membrane Topologies Impact the Function of the Infectious Bronchitis Coronavirus E Protein. PLoS Pathog. 2012, 8, e1002674. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Lescar, J.; Tam, J.P.; Liu, D.X. Expression of SARS-Coronavirus Envelope Protein in Escherichia Coli Cells Alters Membrane Permeability. Biochem. Biophys. Res. Commun. 2004, 325, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Maurya, V.K.; Prasad, A.K.; Bhatt, M.L.B.; Saxena, S.K. Structural, Glycosylation and Antigenic Variation between 2019 Novel Coronavirus (2019-NCoV) and SARS Coronavirus (SARS-CoV). Virusdisease 2020, 31, 13–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, L.; Mckinlay, C.; Gage, P.; Ewart, G. SARS Coronavirus E Protein Forms Cation-Selective Ion Channels. Virology 2004, 330, 322–331. [Google Scholar] [CrossRef] [Green Version]
- Singh Tomar, P.P.; Arkin, I.T. SARS-CoV-2 E Protein Is a Potential Ion Channel That Can Be Inhibited by Gliclazide and Memantine. Biochem. Biophys. Res. Commun. 2020, 530, 10–14. [Google Scholar] [CrossRef]
- Schoeman, D.; Fielding, B.C. Coronavirus Envelope Protein: Current Knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [Green Version]
- DeDiego, M.L.; Pewe, L.; Alvarez, E.; Rejas, M.T.; Perlman, S.; Enjuanes, L. Pathogenicity of Severe Acute Respiratory Coronavirus Deletion Mutants in HACE-2 Transgenic Mice. Virology 2008, 376, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Nieto-Torres, J.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Castaño-Rodriguez, C.; Fernandez-Delgado, R.; Torres, J.; Aguilella, V.M.; Enjuanes, L. Severe Acute Respiratory Syndrome Coronavirus E Protein Transports Calcium Ions and Activates the NLRP3 Inflammasome. Virology 2015, 485, 330–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teoh, K.-T.; Siu, Y.-L.; Chan, W.-L.; Schlüter, M.A.; Liu, C.-J.; Peiris, J.S.M.; Bruzzone, R.; Margolis, B.; Nal, B. The SARS Coronavirus E Protein Interacts with PALS1 and Alters Tight Junction Formation and Epithelial Morphogenesis. Mol. Biol. Cell 2010, 21, 3838–3852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.C.; Yi, Z.; Hogue, B.G. Identification of Mouse Hepatitis Coronavirus A59 Nucleocapsid Protein Phosphorylation Sites. Virus Res. 2007, 126, 139–148. [Google Scholar] [CrossRef]
- Li, F.Q.; Xiao, H.; Tam, J.P.; Liu, D.X. Sumoylation of the Nucleocapsid Protein of Severe Acute Respiratory Syndrome Coronavirus. FEBS Lett. 2005, 579, 2387–2396. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.; Chen, C.-M.M.; Chiang, M.; Hsu, Y.; Huang, T. Transient Oligomerization of the SARS-CoV N Protein–Implication for Virus Ribonucleoprotein Packaging. PLoS ONE 2013, 8, e65045. [Google Scholar] [CrossRef]
- Kuo, L.; Koetzner, C.A.; Hurst, K.R.; Masters, P.S. Recognition of the Murine Coronavirus Genomic RNA Packaging Signal Depends on the Second RNA-Binding Domain of the Nucleocapsid Protein. J. Virol. 2014, 88, 4451–4465. [Google Scholar] [CrossRef] [Green Version]
- Fan, H.; Ooi, A.; Tan, Y.W.; Wang, S.; Fang, S.; Liu, D.X.; Lescar, J. The Nucleocapsid Protein of Coronavirus Infectious Bronchitis Virus: Crystal Structure of Its N-Terminal Domain and Multimerization Properties. Structure 2005, 13, 1859–1868. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Yang, M.; Hong, Z.; Zhang, L.; Huang, Z.; Chen, X.; He, S.; Zhou, Z.; Zhou, Z.; Chen, Q.; et al. Crystal Structure of SARS-CoV-2 Nucleocapsid Protein RNA Binding Domain Reveals Potential Unique Drug Targeting Sites. Acta Pharm. Sin. B 2020, 10, 1228–1238. [Google Scholar] [CrossRef]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef]
- Caldas, L.A.; Carneiro, F.A.; Higa, L.M.; Monteiro, F.L.; da Silva, G.P.; da Costa, L.J.; Durigon, E.L.; Tanuri, A.; de Souza, W. Ultrastructural Analysis of SARS-CoV-2 Interactions with the Host Cell via High Resolution Scanning Electron Microscopy. Sci. Rep. 2020, 10, 16099. [Google Scholar] [CrossRef]
- Hurst, K.R.; Ye, R.; Goebel, S.J.; Jayaraman, P.; Masters, P.S. An Interaction between the Nucleocapsid Protein and a Component of the Replicase-Transcriptase Complex Is Crucial for the Infectivity of Coronavirus Genomic RNA. J. Virol. 2010, 84, 10276–10288. [Google Scholar] [CrossRef] [Green Version]
- Gardner, A.; Autin, L.; Barbaro, B.; Olson, A.J.; Goodsell, D.S. CellPAINT: Interactive Illustration of Dynamic Mesoscale Cellular Environments. IEEE Comput. Graph. Appl. 2018, 38, 51–64. [Google Scholar] [CrossRef]
- Tomasello, G.; Armenia, I.; Molla, G. The Protein Imager: A Full-Featured Online Molecular Viewer Interface with Server-Side HQ-Rendering Capabilities. Bioinformatics 2020, 36, 2909–2911. [Google Scholar] [CrossRef]
- Zhao, P.; Praissman, J.L.; Grant, O.C.; Cai, Y.; Xiao, T.; Rosenbalm, K.E.; Aoki, K.; Kellman, B.P.; Bridger, R.; Barouch, D.H.; et al. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28, 586–601.e6. [Google Scholar] [CrossRef]
- Muus, C.; Luecken, M.D.; Eraslan, G.; Sikkema, L.; Waghray, A.; Heimberg, G.; Kobayashi, Y.; Vaishnav, E.D.; Subramanian, A.; Smillie, C.; et al. Single-Cell Meta-Analysis of SARS-CoV-2 Entry Genes across Tissues and Demographics. Nat. Med. 2021, 27, 546–559. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 Entry Factors Are Highly Expressed in Nasal Epithelial Cells Together with Innate Immune Genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Wicht, O.; van Kuppeveld, F.J.M.; He, Q.; Rottier, P.J.M.; Bosch, B.-J. A Single Point Mutation Creating a Furin Cleavage Site in the Spike Protein Renders Porcine Epidemic Diarrhea Coronavirus Trypsin Independent for Cell Entry and Fusion. J. Virol. 2015, 89, 8077–8081. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Mercurio, I.; Tragni, V.; Busto, F.; De Grassi, A.; Pierri, C.L. Protein Structure Analysis of the Interactions between SARS-CoV-2 Spike Protein and the Human ACE2 Receptor: From Conformational Changes to Novel Neutralizing Antibodies. Cell. Mol. Life Sci. 2021, 78, 1501–1522. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Plotkin, S.; Robinson, J.M.; Cunningham, G.; Iqbal, R.; Larsen, S. The Complexity and Cost of Vaccine Manufacturing—An Overview. Vaccine 2017, 35, 4064–4071. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, T.M. Murine Coronavirus Membrane Fusion Is Blocked by Modification of Thiols Buried within the Spike Protein. J. Virol. 1996, 70, 4683–4690. [Google Scholar] [CrossRef] [Green Version]
- Lavillette, D.; Barbouche, R.; Yao, Y.; Boson, B.; Cosset, F.-L.; Jones, I.M.; Fenouillet, E. Significant Redox Insensitivity of the Functions of the SARS-CoV Spike Glycoprotein. J. Biol. Chem. 2006, 281, 9200–9204. [Google Scholar] [CrossRef] [Green Version]
- Costello, D.A.; Millet, J.K.; Hsia, C.-Y.; Whittaker, G.R.; Daniel, S. Single Particle Assay of Coronavirus Membrane Fusion with Proteinaceous Receptor-Embedded Supported Bilayers. Biomaterials 2013, 34, 7895–7904. [Google Scholar] [CrossRef]
- Giron, C.C.; Laaksonen, A.; Barroso da Silva, F.L. Up State of the SARS-CoV-2 Spike Homotrimer Favors an Increased Virulence for New Variants. Front. Med. Technol. 2021, 3. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Zheng, W.; Huang, X.; Bell, E.W.; Zhou, X.; Zhang, Y. Protein Structure and Sequence Reanalysis of 2019-NCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J. Proteome Res. 2020, 19, 1351–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 5 July 2021).
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science 2020, 372. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger The PyMOL Molecular Graphics System. Available online: https://pymol.org/2/ (accessed on 5 July 2021).
- Zhang Modeling of the SARS-CoV-2 Genome Using I-TASSER. Available online: https://zhanggroup.org/COVID-19/ (accessed on 24 August 2021).
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.d.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S.; et al. Convergent Evolution of SARS-CoV-2 Spike Mutations, L452R, E484Q, and P681R, in the Second Wave of COVID-19 in Maharashtra, India. BioRxiv 2021. [Google Scholar] [CrossRef]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G Variant Exhibits Efficient Replication Ex Vivo and Transmission in Vivo. Science 2020, eabe8499. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Emary, K.R.W.; Golubchik, T.; Aley, P.K.; Ariani, C.V.; Angus, B.J.; Bibi, S.; Blane, B.; Bonsall, D.; Cicconi, P.; Charlton, S.; et al. Efficacy of ChAdOx1 NCoV-19 (AZD1222) Vaccine Against SARS-CoV-2 VOC 202012/01 (B.1.1.7). SSRN Electron. J. 2021. [Google Scholar] [CrossRef]
- Pritchard, E.; Matthews, P.C.; Stoesser, N.; Eyre, D.W.; Gethings, O.; Vihta, K.D.; Jones, J.; House, T.; VanSteenHouse, H.; Bell, I.; et al. Impact of Vaccination on New SARS-CoV-2 Infections in the United Kingdom. Nat. Med. 2021, 1–9. [Google Scholar] [CrossRef]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.; et al. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. Cell Host Microbe 2021, 29, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Lam, E.C.; St. Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. Cell 2021, 184, 2372–2383.e9. [Google Scholar] [CrossRef]
- Coutinho, R.M.; Marquitti, F.M.D.; Ferreira, L.S.; Borges, M.E.; Paixão da Silva, R.L.; Canton, O.; Portella, T.P.; Poloni, S.; Franco, C.; Plucinski, M.M.; et al. Model-Based Estimation of Transmissibility and Reinfection of SARS-CoV-2 P.1 Variant of the SARS-CoV-2. MedRxiv 2021. [Google Scholar] [CrossRef]
- Mullen, J.L.; Tsueng, G.; the Center for Viral Systems Biology. Outbreak.Info. Available online: outbreak.info (accessed on 7 November 2021).
- Elbe, S.; Buckland-Merrett, G. Data, Disease and Diplomacy: GISAID’s Innovative Contribution to Global Health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Lustig, Y.; Zuckerman, N.; Nemet, I.; Atari, N.; Kliker, L.; Regev-Yochay, G.; Sapir, E.; Mor, O.; Alroy-Preis, S.; Mendelson, E.; et al. Neutralising Capacity against Delta (B.1.617.2) and Other Variants of Concern Following Comirnaty (BNT162b2, BioNTech/Pfizer) Vaccination in Health Care Workers, Israel. Euro Surveill. 2021, 26. [Google Scholar] [CrossRef] [PubMed]
- Bernal, J.L.; Andrews, N.; Gower, C.; Stowe, J.; Robertson, C.; Tessier, E.; Simmons, R.; Cottrell, S.; Roberts, R.; O’Doherty, M.; et al. Early Effectiveness of COVID-19 Vaccination with BNT162b2 MRNA Vaccine and ChAdOx1 Adenovirus Vector Vaccine on Symptomatic Disease, Hospitalisations and Mortality in Older Adults in England. medRxiv 2021. [Google Scholar] [CrossRef]
- Campbell, F.; Archer, B.; Laurenson-Schafer, H.; Jinnai, Y.; Konings, F.; Batra, N.; Pavlin, B.; Vandemaele, K.; Van Kerkhove, M.D.; Jombart, T.; et al. Increased Transmissibility and Global Spread of SARS-CoV-2 Variants of Concern as at June 2021. Eurosurveillance 2021, 26. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.; Datir, R.; Kemp, S.; Papa, G.; Rakshit, P.; Singh, S.; Meng, B.; Pandey, R.; Ponnusamy, K.; Radhakrishnan, V.S.; et al. SARS-CoV-2 B.1.617 Emergence and Sensitivity to Vaccine-Elicited Antibodies. bioRxiv 2021. [Google Scholar] [CrossRef]
- Brown, C.M. Outbreak of SARS-CoV-2 Infections, Including COVID-19 Vaccine Breakthrough Infections, Associated with Large Public Gatherings—Barnstable County, Massachusetts, July 2021. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 1059–1062. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention CDC COVID Data Tracker. Available online: https://covid.cdc.gov/covid-data-tracker/#variant-proportions (accessed on 12 August 2021).
- Frost, S.D.W.; Magalis, B.R.; Kosakovsky Pond, S.L. Neutral Theory and Rapidly Evolving Viral Pathogens. Mol. Biol. Evol. 2018, 35, 1348–1354. [Google Scholar] [CrossRef]
- Bernal, J.L.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 Variant. medRxiv 2021. [Google Scholar] [CrossRef]
- Guirakhoo, F.; Kuo, L.; Peng, J.; Huang, J.H.; Kuo, B.; Lin, F.; Liu, K.; Liu, Z.; Wu, G.; Ding, S.; et al. A Novel SARS-CoV-2 Multitope Protein/Peptide Vaccine Candidate Is Highly Immunogenic and Prevents Lung Infection in an Adeno Associated Virus Human Angiotensin-Converting Enzyme 2 (AAV HACE2) Mouse Model. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fischman, S.; Ofran, Y. Computational Design of Antibodies. Curr. Opin. Struct. Biol. 2018, 51, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Sormanni, P.; Aprile, F.A.; Vendruscolo, M. Third Generation Antibody Discovery Methods: In Silico Rational Design. Chem. Soc. Rev. 2018, 47, 9137–9157. [Google Scholar] [CrossRef]
- Zhao, J.; Nussinov, R.; Wu, W.-J.; Ma, B. In Silico Methods in Antibody Design. Antibodies 2018, 7, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poveda-Cuevas, S.A.; Etchebest, C.; Barroso Da Silva, F.L. Identification of Electrostatic Epitopes in Flavivirus by Computer Simulations: The PROCEEDpKa Method. J. Chem. Inf. Model. 2020, 60, 944–963. [Google Scholar] [CrossRef] [PubMed]
- Robson, B. Techniques Assisting Peptide Vaccine and Peptidomimetic Design. Sidechain Exposure in the SARS-CoV-2 Spike Glycoprotein. Comput. Biol. Med. 2021, 128, 104124. [Google Scholar] [CrossRef] [PubMed]
- Oli, A.N.; Obialor, W.O.; Ifeanyichukwu, M.O.; Odimegwu, D.C.; Okoyeh, J.N.; Emechebe, G.O.; Adejumo, S.A.; Ibeanu, G.C. Immunoinformatics and Vaccine Development: An Overview. ImmunoTargets Ther. 2020, 9, 13–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nezafat, N.; Sadraeian, M.; Rahbar, M.R.; Khoshnoud, M.J.; Mohkam, M.; Gholami, A.; Banihashemi, M.; Ghasemi, Y. Production of a Novel Multi-Epitope Peptide Vaccine for Cancer Immunotherapy in TC-1 Tumor-Bearing Mice. Biologicals 2015, 43, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Vicente, T.; Roldão, A.; Peixoto, C.; Carrondo, M.J.T.; Alves, P.M. Large-Scale Production and Purification of VLP-Based Vaccines. J. Invertebr. Pathol. 2011, 107 Suppl, S42–S48. [Google Scholar] [CrossRef]
- Garçon, N.; Vaughn, D.W.; Didierlaurent, A.M. Development and Evaluation of AS03, an Adjuvant System Containing α-Tocopherol and Squalene in an Oil-in-Water Emulsion. Expert Rev. Vaccines 2012, 11, 349–366. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A. Effectiveness of the BNT162b2 COVID-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Dagan, N.; Barda, N.; Kepten, E.; Miron, O.; Perchik, S.; Katz, M.A.; Hernán, M.A.; Lipsitch, M.; Reis, B.; Balicer, R.D. BNT162b2 MRNA COVID-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021, 384, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Haas, E.J.; Angulo, F.J.; McLaughlin, J.M.; Anis, E.; Singer, S.R.; Khan, F.; Brooks, N.; Smaja, M.; Mircus, G.; Pan, K.; et al. Impact and Effectiveness of MRNA BNT162b2 Vaccine against SARS-CoV-2 Infections and COVID-19 Cases, Hospitalisations, and Deaths Following a Nationwide Vaccination Campaign in Israel: An Observational Study Using National Surveillance Data. Lancet 2021, 397, 1819–1829. [Google Scholar] [CrossRef]
- Flood, A.; Estrada, M.; McAdams, D.; Ji, Y.; Chen, D. Development of a Freeze-Dried, Heat-Stable Influenza Subunit Vaccine Formulation. PLoS ONE 2016, 11, e0164692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, L.; Zheng, T.; Xu, K.; Han, Y.; Xu, L.; Huang, E.; An, Y.; Cheng, Y.; Li, S.; Liu, M.; et al. A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS. Cell 2020, 182, 722–733.e11. [Google Scholar] [CrossRef] [PubMed]
- de Villiers, E.M. Heterogeneity of the Human Papillomavirus Group. J. Virol. 1989, 63, 4898–4903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fluckiger, A.-C.; Ontsouka, B.; Bozic, J.; Diress, A.; Ahmed, T.; Berthoud, T.; Tran, A.; Duque, D.; Liao, M.; Mccluskie, L.; et al. An Enveloped Virus-like Particle Vaccine Expressing a Stabilized Prefusion Form of the SARS-CoV-2 Spike Protein Elicits Potent Immunity after a Single Dose. bioRxiv 2021. [Google Scholar] [CrossRef]
- Keating, G.M.; Noble, S. Recombinant Hepatitis B Vaccine (Engerix-B??): A Review of Its Immunogenicity and Protective Efficacy Against Hepatitis B. Drugs 2003, 63, 1021–1051. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.M.; Wormald, M.R.; Stanfield, R.L.; Huang, M.; Mattsson, N.; Speir, J.A.; DiGennaro, J.A.; Fetrow, J.S.; Dwek, R.A.; Wilson, I.A. Roles for Glycosylation of Cell Surface Receptors Involved in Cellular Immune Recognition. J. Mol. Biol. 1999, 293, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Monie, A.; Hung, C.-F.; Roden, R.; Wu, T.-C. Cervarix: A Vaccine for the Prevention of HPV 16, 18-Associated Cervical Cancer. Biologics 2008, 2, 97–105. [Google Scholar] [PubMed]
- Giannini, S.L.; Hanon, E.; Moris, P.; Van Mechelen, M.; Morel, S.; Dessy, F.; Fourneau, M.A.; Colau, B.; Suzich, J.; Losonksy, G.; et al. Enhanced Humoral and Memory B Cellular Immunity Using HPV16/18 L1 VLP Vaccine Formulated with the MPL/Aluminium Salt Combination (AS04) Compared to Aluminium Salt Only. Vaccine 2006, 24, 5937–5949. [Google Scholar] [CrossRef] [PubMed]
- Paavonen, J.; Jenkins, D.; Bosch, F.X.; Naud, P.; Salmerón, J.; Wheeler, C.M.; Chow, S.-N.; Apter, D.L.; Kitchener, H.C.; Castellsague, X.; et al. Efficacy of a Prophylactic Adjuvanted Bivalent L1 Virus-like-Particle Vaccine against Infection with Human Papillomavirus Types 16 and 18 in Young Women: An Interim Analysis of a Phase III Double-Blind, Randomised Controlled Trial. Lancet 2007, 369, 2161–2170. [Google Scholar] [CrossRef]
- Harper, D.M.; Franco, E.L.; Wheeler, C.M.; Moscicki, A.-B.; Romanowski, B.; Roteli-Martins, C.M.; Jenkins, D.; Schuind, A.; Costa Clemens, S.A.; Dubin, G. Sustained Efficacy up to 4·5 Years of a Bivalent L1 Virus-like Particle Vaccine against Human Papillomavirus Types 16 and 18: Follow-up from a Randomised Control Trial. Lancet 2006, 367, 1247–1255. [Google Scholar] [CrossRef] [Green Version]
- Walls, A.C.; Fiala, B.; Schäfer, A.; Wrenn, S.; Pham, M.N.; Murphy, M.; Tse, L.V.; Shehata, L.; O’Connor, M.A.; Chen, C.; et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell 2020, 183, 1367–1382.e17. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Walls, A.C.; Golden, N.; Atyeo, C.; Fischinger, S.; Li, C.; Aye, P.; Navarro, M.J.; Lai, L.; Edara, V.V.; et al. Adjuvanting a Subunit COVID-19 Vaccine to Induce Protective Immunity. Nature 2021, 594, 253–258. [Google Scholar] [CrossRef]
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 Covid-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Fougeroux, C.; Goksøyr, L.; Idorn, M.; Soroka, V.; Myeni, S.K.; Dagil, R.; Janitzek, C.M.; Søgaard, M.; Aves, K.-L.; Horsted, E.W.; et al. Capsid-like Particles Decorated with the SARS-CoV-2 Receptor-Binding Domain Elicit Strong Virus Neutralization Activity. Nat. Commun. 2021, 12, 324. [Google Scholar] [CrossRef]
- Zhou, H.; Ji, J.; Chen, X.; Bi, Y.; Li, J.; Wang, Q.; Hu, T.; Song, H.; Zhao, R.; Chen, Y.; et al. Identification of Novel Bat Coronaviruses Sheds Light on the Evolutionary Origins of SARS-CoV-2 and Related Viruses. Cell 2021, 184, 4380–4391.e14. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and Evolution of Pathogenic Coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappazzo, C.G.; Tse, L.V.; Kaku, C.I.; Wrapp, D.; Sakharkar, M.; Huang, D.; Deveau, L.M.; Yockachonis, T.J.; Herbert, A.S.; Battles, M.B.; et al. Broad and Potent Activity against SARS-like Viruses by an Engineered Human Monoclonal Antibody. Science 2021, 371, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.E.; Zhang, K.; Sanyal, M.; Tang, S.; Weidenbacher, P.A.; Li, S.; Pham, T.D.; Pak, J.E.; Chiu, W.; Kim, P.S. A Single Immunization with Spike-Functionalized Ferritin Vaccines Elicits Neutralizing Antibody Responses against SARS-CoV-2 in Mice. ACS Cent. Sci. 2021, 7, 183–199. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhou, H.; Wang, W.; Tan, W.; Fu, Y.-X.; Zhu, M. A Novel Method for Synthetic Vaccine Construction Based on Protein Assembly. Sci. Rep. 2015, 4, 7266. [Google Scholar] [CrossRef] [PubMed]
- Bale, J.B.; Gonen, S.; Liu, Y.; Sheffler, W.; Ellis, D.; Thomas, C.; Cascio, D.; Yeates, T.O.; Gonen, T.; King, N.P.; et al. Accurate Design of Megadalton-Scale Two-Component Icosahedral Protein Complexes. Science 2016, 353, 389–394. [Google Scholar] [CrossRef] [Green Version]
| Name | Platform | Adjuvant | Dosage | Efficacy * | References |
|---|---|---|---|---|---|
| Coronavac (Sinovac) | Inactivated | Alum | 2 doses | 83.5% (95% CI, 65.4–92.1) | [8,9,10,11] |
| BBIBP-CorV (Sinopharm) | Inactivated | Alum | 2 doses | 72.8% (95% CI, 58.1–82.4) | [12,13] |
| BBV152-Covaxin (Bharat Biotech) | Inactivated | Alum | 2 doses | 77.8% (95% CI, 65.2–86.4) | [14,15] |
| AZD1222–Vaxzevria (Oxford/AstraZeneca) | Viral vector | No | 2 doses | 74.0% (95% CI, 65.3–80.5) | [16,17,18] |
| Covishield (Oxford/AstraZeneca formulation) | Viral vector | No | 2 doses | 74.0% (95% CI, 65.3–80.5) | [16,17,18] |
| Ad26.COV2.S (Johnson &Johnson-Janssen) | Viral vector | No | 1 dose | 66.9% (95% CI, 59.0–73.4) | [19,20,21,22] |
| mRNA-1273 (Moderna) | mRNA | No | 2 doses | 94.1% (95% CI, 89.3–96.8) | [23,24] |
| BNT162b-Comirnaty (Pfizer/BioNTech) | mRNA | No | 2 doses | 95% (95% CI, 90.3–97.6) | [25,26] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prates-Syed, W.A.; Chaves, L.C.S.; Crema, K.P.; Vuitika, L.; Lira, A.; Côrtes, N.; Kersten, V.; Guimarães, F.E.G.; Sadraeian, M.; Barroso da Silva, F.L.; et al. VLP-Based COVID-19 Vaccines: An Adaptable Technology against the Threat of New Variants. Vaccines 2021, 9, 1409. https://doi.org/10.3390/vaccines9121409
Prates-Syed WA, Chaves LCS, Crema KP, Vuitika L, Lira A, Côrtes N, Kersten V, Guimarães FEG, Sadraeian M, Barroso da Silva FL, et al. VLP-Based COVID-19 Vaccines: An Adaptable Technology against the Threat of New Variants. Vaccines. 2021; 9(12):1409. https://doi.org/10.3390/vaccines9121409
Chicago/Turabian StylePrates-Syed, Wasim A., Lorena C. S. Chaves, Karin P. Crema, Larissa Vuitika, Aline Lira, Nelson Côrtes, Victor Kersten, Francisco E. G. Guimarães, Mohammad Sadraeian, Fernando L. Barroso da Silva, and et al. 2021. "VLP-Based COVID-19 Vaccines: An Adaptable Technology against the Threat of New Variants" Vaccines 9, no. 12: 1409. https://doi.org/10.3390/vaccines9121409
APA StylePrates-Syed, W. A., Chaves, L. C. S., Crema, K. P., Vuitika, L., Lira, A., Côrtes, N., Kersten, V., Guimarães, F. E. G., Sadraeian, M., Barroso da Silva, F. L., Cabral-Marques, O., Barbuto, J. A. M., Russo, M., Câmara, N. O. S., & Cabral-Miranda, G. (2021). VLP-Based COVID-19 Vaccines: An Adaptable Technology against the Threat of New Variants. Vaccines, 9(12), 1409. https://doi.org/10.3390/vaccines9121409





_Sadraeian.png)


