Vaccine Hyporesponse Induced by Individual Antibiotic Treatment in Mice and Non-Human Primates Is Diminished upon Recovery of the Gut Microbiome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Rhesus Macaques
2.3. Antibiotic Treatments
2.4. Immunizations, Vaccine Antigens and Adjuvants
2.5. Collection of Stool and Tissues
2.6. Antigen-Specific Antibody Titers
2.7. Pharmacokinetics
2.8. T-cell Activity Quantification
2.9. DNA Extraction and 16S rRNA Gene Sequencing
2.10. Shotgun Metagenomic Sequencing and KEGG Pathway Enrichment Analysis
2.11. RNA Extraction, Sequencing, and Differential Gene Expression Analysis
2.12. GO Term Analysis and Clustering
2.13. Statistical Analyses
3. Results
3.1. Microbiome Perturbation by Different Antibiotics Differentially Modulates Vaccine Immune Response
3.2. Antibiotic-Mediated Vaccine Hyporesponse Is Associated with Altered Microbiome Functionality
3.3. Vancomycin-Induced Vaccine Hyporesponse Is Minimized by Allowing Sufficient Time for the Microbiome to Recover
3.4. Vancomycin Disrupts Vaccine Outcome in Rhesus Macaques
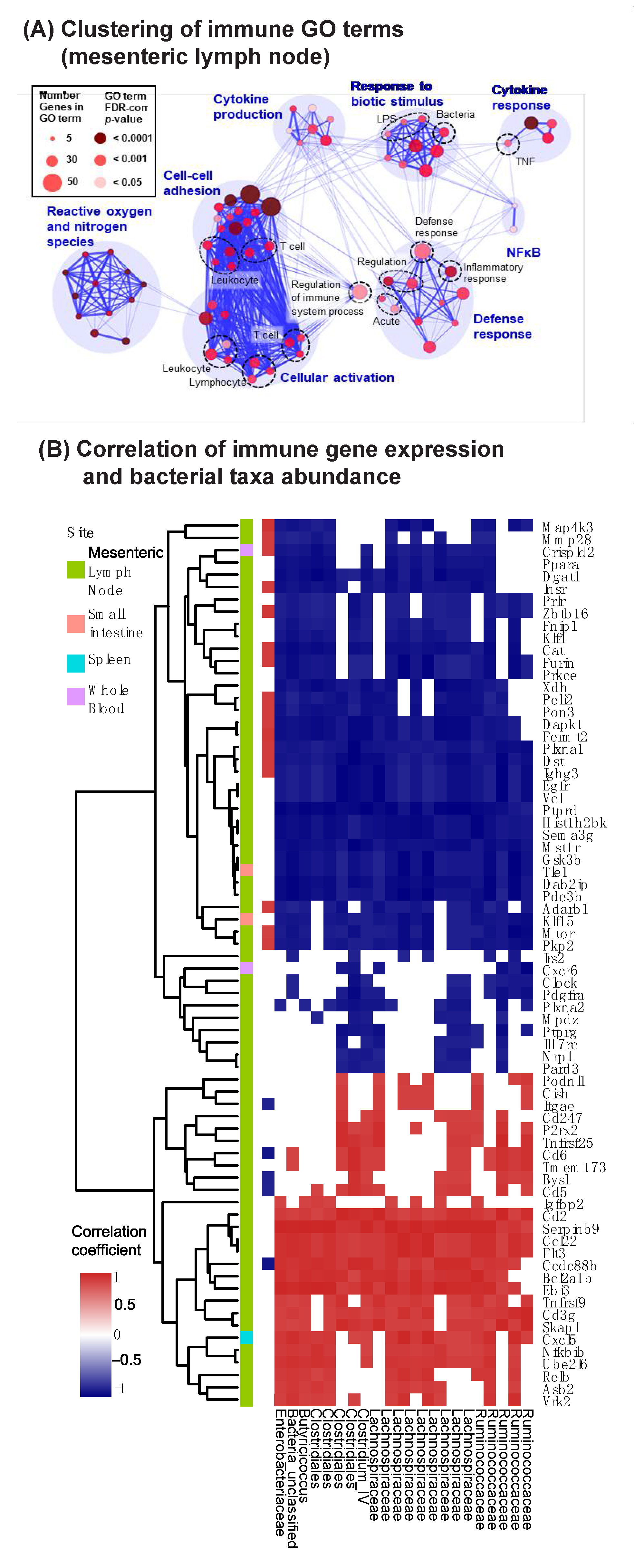
3.5. Vancomycin Impacts Key Innate Immune Pathways in the Mesenteric Lymph Nodes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nakaya, H.I.; Hagan, T.; Duraisingham, S.S.; Lee, E.K.; Kwissa, M.; Rouphael, N.; Frasca, D.; Gersten, M.; Mehta, A.K.; Gaujoux, R.; et al. Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity 2015, 43, 1186–1198. [Google Scholar] [CrossRef] [Green Version]
- Hagan, T.; Cortese, M.; Rouphael, N.; Boudreau, C.; Linde, C.; Maddur, M.S.; Das, J.; Wang, H.; Guthmiller, J.; Zheng, N.Y.; et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 2019, 178, 1313–1328.e1313. [Google Scholar] [CrossRef]
- Parker, E.P.; Ramani, S.; Lopman, B.A.; Church, J.A.; Iturriza-Gómara, M.; Prendergast, A.J.; Grassly, N.C. Causes of impaired oral vaccine efficacy in developing countries. Future Microbiol. 2018, 13, 97–118. [Google Scholar] [CrossRef]
- Fourati, S.; Cristescu, R.; Loboda, A.; Talla, A.; Filali, A.; Railkar, R.; Schaeffer, A.K.; Favre, D.; Gagnon, D.; Peretz, Y.; et al. Pre-vaccination inflammation and B-cell signalling predict age-related hyporesponse to hepatitis B vaccination. Nat. Commun. 2016, 7, 10369. [Google Scholar] [CrossRef]
- Crooke, S.N.; Ovsyannikova, I.G.; Poland, G.A.; Kennedy, R.B. Immunosenescence and human vaccine immune responses. Immun. Ageing 2019, 16, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulendran, B. Systems vaccinology: Probing humanity’s diverse immune systems with vaccines. Proc. Natl. Acad. Sci. USA 2014, 111, 12300–12306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiedermann, U.; Garner-Spitzer, E.; Wagner, A. Primary vaccine failure to routine vaccines: Why and what to do? Hum. Vaccin Immunother. 2016, 12, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [CrossRef] [Green Version]
- Ursell, L.K.; Haiser, H.J.; Van Treuren, W.; Garg, N.; Reddivari, L.; Vanamala, J.; Dorrestein, P.C.; Turnbaugh, P.J.; Knight, R. The intestinal metabolome: An intersection between microbiota and host. Gastroenterology 2014, 146, 1470–1476. [Google Scholar] [CrossRef] [Green Version]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalová, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef] [Green Version]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [Green Version]
- Ussar, S.; Fujisaka, S.; Kahn, C.R. Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome. Mol. Metab. 2016, 5, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.C.; Olson, C.A.; Hsiao, E.Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 2017, 20, 145–155. [Google Scholar] [CrossRef]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef] [Green Version]
- Hannigan, G.D.; Grice, E.A. Microbial ecology of the skin in the era of metagenomics and molecular microbiology. Cold Spring Harb. Perspect. Med. 2013, 3, a015362. [Google Scholar] [CrossRef] [Green Version]
- Eloe-Fadrosh, E.A.; McArthur, M.A.; Seekatz, A.M.; Drabek, E.F.; Rasko, D.A.; Sztein, M.B.; Fraser, C.M. Impact of oral typhoid vaccination on the human gut microbiota and correlations with s. Typhi-specific immunological responses. PLoS ONE 2013, 8, e62026. [Google Scholar] [CrossRef]
- Huda, M.N.; Lewis, Z.; Kalanetra, K.M.; Rashid, M.; Ahmad, S.M.; Raqib, R.; Qadri, F.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Stool microbiota and vaccine responses of infants. Pediatrics 2014, 134, e362–e372. [Google Scholar] [CrossRef] [Green Version]
- Harris, V.C.; Armah, G.; Fuentes, S.; Korpela, K.E.; Parashar, U.; Victor, J.C.; Tate, J.; de Weerth, C.; Giaquinto, C.; Wiersinga, W.J.; et al. Significant Correlation Between the Infant Gut Microbiome and Rotavirus Vaccine Response in Rural Ghana. J. Infect. Dis. 2017, 215, 34–41. [Google Scholar] [CrossRef]
- Harris, V.; Ali, A.; Fuentes, S.; Korpela, K.; Kazi, M.; Tate, J.; Parashar, U.; Wiersinga, W.J.; Giaquinto, C.; de Weerth, C.; et al. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes 2018, 9, 93–101. [Google Scholar] [CrossRef]
- Parker, E.P.K.; Praharaj, I.; Zekavati, A.; Lazarus, R.P.; Giri, S.; Operario, D.J.; Liu, J.; Houpt, E.; Iturriza-Gómara, M.; Kampmann, B.; et al. Influence of the intestinal microbiota on the immunogenicity of oral rotavirus vaccine given to infants in south India. Vaccine 2018, 36, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Z.; Ravindran, R.; Chassaing, B.; Carvalho, F.A.; Maddur, M.S.; Bower, M.; Hakimpour, P.; Gill, K.P.; Nakaya, H.I.; Yarovinsky, F.; et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014, 41, 478–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynn, M.A.; Tumes, D.J.; Choo, J.M.; Sribnaia, A.; Blake, S.J.; Leong, L.E.X.; Young, G.P.; Marshall, H.S.; Wesselingh, S.L.; Rogers, G.B.; et al. Early-Life Antibiotic-Driven Dysbiosis Leads to Dysregulated Vaccine Immune Responses in Mice. Cell Host Microbe 2018, 23, 653–660.e655. [Google Scholar] [CrossRef] [Green Version]
- Lamousé-Smith, E.S.; Tzeng, A.; Starnbach, M.N. The intestinal flora is required to support antibody responses to systemic immunization in infant and germ free mice. PLoS ONE 2011, 6, e27662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, I.C.; Hutchings, C.; Kondova, I.; Hey, A.; Powrie, F.; Beverley, P.; Tchilian, E. Helicobacter hepaticus infection in BALB/c mice abolishes subunit-vaccine-induced protection against M. tuberculosis. Vaccine 2015, 33, 1808–1814. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Kim, Y.G.; Seo, S.U.; Kim, D.J.; Kamada, N.; Prescott, D.; Chamaillard, M.; Philpott, D.J.; Rosenstiel, P.; Inohara, N.; et al. Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat. Med. 2016, 22, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, Y.M.; Kim, W.U.; Park, J.H.; Núñez, G.; Seo, S.U. Recognition of the microbiota by Nod2 contributes to the oral adjuvant activity of cholera toxin through the induction of interleukin-1β. Immunology 2019, 158, 219–229. [Google Scholar] [CrossRef]
- Yang, W.; Xiao, Y.; Huang, X.; Chen, F.; Sun, M.; Bilotta, A.J.; Xu, L.; Lu, Y.; Yao, S.; Zhao, Q.; et al. Microbiota Metabolite Short-Chain Fatty Acids Facilitate Mucosal Adjuvant Activity of Cholera Toxin through GPR43. J. Immunol. 2019, 203, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, G.; Thoryk, E.A.; Cox, K.S.; Meschino, S.; Dubey, S.A.; Vora, K.A.; Celano, R.; Gindy, M.; Casimiro, D.R.; Bett, A.J. A novel lipid nanoparticle adjuvant significantly enhances B cell and T cell responses to sub-unit vaccine antigens. Vaccine 2016, 34, 110–119. [Google Scholar] [CrossRef]
- Thoryk, E.A.; Swaminathan, G.; Meschino, S.; Cox, K.S.; Gindy, M.; Casimiro, D.R.; Bett, A.J. Co-Administration of Lipid Nanoparticles and Sub-Unit Vaccine Antigens Is Required for Increase in Antigen-Specific Immune Responses in Mice. Vaccines 2016, 4, 47. [Google Scholar] [CrossRef] [Green Version]
- Gindy, M.E.; Feuston, B.; Glass, A.; Arrington, L.; Haas, R.M.; Schariter, J.; Stirdivant, S.M. Stabilization of Ostwald ripening in low molecular weight amino lipid nanoparticles for systemic delivery of siRNA therapeutics. Mol. Pharm. 2014, 11, 4143–4153. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D. Secondary structure improves OTU assignments of 16S rRNA gene sequences. ISME J. 2013, 7, 457–460. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Manor, O.; Borenstein, E. MUSiCC: A marker genes based framework for metagenomic normalization and accurate profiling of gene abundances in the microbiome. Genome Biol. 2015, 16, 53. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. clusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinform. 2011, 12, 436. [Google Scholar] [CrossRef] [Green Version]
- Lynn, D.J.; Pulendran, B. The potential of the microbiota to influence vaccine responses. J. Leukoc. Biol. 2018, 103, 225–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez-Castellanos, J.F.; Serrano-Villar, S.; Latorre, A.; Artacho, A.; Ferrús, M.L.; Madrid, N.; Vallejo, A.; Sainz, T.; Martínez-Botas, J.; Ferrando-Martínez, S.; et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal. Immunol. 2015, 8, 760–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, C.T.; Davis-Richardson, A.G.; Giongo, A.; Gano, K.A.; Crabb, D.B.; Mukherjee, N.; Casella, G.; Drew, J.C.; Ilonen, J.; Knip, M.; et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE 2011, 6, e25792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preidis, G.A.; Ajami, N.J.; Wong, M.C.; Bessard, B.C.; Conner, M.E.; Petrosino, J.F. Composition and function of the undernourished neonatal mouse intestinal microbiome. J. Nutr. Biochem. 2015, 26, 1050–1057. [Google Scholar] [CrossRef]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Häsler, R.; et al. Increased Tryptophan Metabolism Is Associated With Activity of Inflammatory Bowel Diseases. Gastroenterology 2017, 153, 1504–1516.e1502. [Google Scholar] [CrossRef] [Green Version]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Li, B.; Zhang, K.; Li, C.; Wang, X.; Chen, Y.; Yang, Y. Characterization and Comparison of Microbiota in the Gastrointestinal Tracts of the Goat (Capra hircus) During Preweaning Development. Front. Microbiol. 2019, 10, 2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilocca, B.; Burbach, K.; Heyer, C.M.E.; Hoelzle, L.E.; Mosenthin, R.; Stefanski, V.; Camarinha-Silva, A.; Seifert, J. Dietary changes in nutritional studies shape the structural and functional composition of the pigs’ fecal microbiome-from days to weeks. Microbiome 2017, 5, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wu, Q.; Zhou, M.; Luo, Z.; Lv, L.; Pei, J.; Wang, C.; Chai, B.; Sui, B.; Huang, F.; et al. Composition of the murine gut microbiome impacts humoral immunity induced by rabies vaccines. Clin. Transl. Med. 2020, 10, e161. [Google Scholar] [CrossRef]
- Candon, S.; Perez-Arroyo, A.; Marquet, C.; Valette, F.; Foray, A.P.; Pelletier, B.; Milani, C.; Ventura, M.; Bach, J.F.; Chatenoud, L. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS ONE 2015, 10, e0125448. [Google Scholar] [CrossRef]
- Tulstrup, M.V.; Christensen, E.G.; Carvalho, V.; Linninge, C.; Ahrné, S.; Højberg, O.; Licht, T.R.; Bahl, M.I. Antibiotic Treatment Affects Intestinal Permeability and Gut Microbial Composition in Wistar Rats Dependent on Antibiotic Class. PLoS ONE 2015, 10, e0144854. [Google Scholar] [CrossRef]
- Jena, P.K.; Sheng, L.; Liu, H.X.; Kalanetra, K.M.; Mirsoian, A.; Murphy, W.J.; French, S.W.; Krishnan, V.V.; Mills, D.A.; Wan, Y.Y. Western Diet-Induced Dysbiosis in Farnesoid X Receptor Knockout Mice Causes Persistent Hepatic Inflammation after Antibiotic Treatment. Am. J. Pathol. 2017, 187, 1800–1813. [Google Scholar] [CrossRef] [Green Version]
- Russell, S.L.; Gold, M.J.; Hartmann, M.; Willing, B.P.; Thorson, L.; Wlodarska, M.; Gill, N.; Blanchet, M.R.; Mohn, W.W.; McNagny, K.M.; et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012, 13, 440–447. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, X.; Zhang, Y.; Zheng, K.; Xiang, Q.; Chen, N.; Chen, Z.; Zhang, N.; Zhu, J.; He, Q. Antibiotic-Induced Disruption of Gut Microbiota Alters Local Metabolomes and Immune Responses. Front. Cell Infect. Microbiol. 2019, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Spinner, C.A.; Lamsoul, I.; Métais, A.; Febrissy, C.; Moog-Lutz, C.; Lutz, P.G. The E3 Ubiquitin Ligase Asb2α in T Helper 2 Cells Negatively Regulates Antitumor Immunity in Colorectal Cancer. Cancer Immunol. Res. 2019, 7, 1332–1344. [Google Scholar] [CrossRef]
- Duchene, J.; Lecomte, F.; Ahmed, S.; Cayla, C.; Pesquero, J.; Bader, M.; Perretti, M.; Ahluwalia, A. A novel inflammatory pathway involved in leukocyte recruitment: Role for the kinin B1 receptor and the chemokine CXCL5. J. Immunol. 2007, 179, 4849–4856. [Google Scholar] [CrossRef] [Green Version]
- Shao, Z.; Schwarz, H. CD137 ligand, a member of the tumor necrosis factor family, regulates immune responses via reverse signal transduction. J. Leukoc. Biol. 2011, 89, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Schenk, R.L.; Tuzlak, S.; Carrington, E.M.; Zhan, Y.; Heinzel, S.; Teh, C.E.; Gray, D.H.; Tai, L.; Lew, A.M.; Villunger, A.; et al. Characterisation of mice lacking all functional isoforms of the pro-survival BCL-2 family member A1 reveals minor defects in the haematopoietic compartment. Cell Death Differ. 2017, 24, 534–545. [Google Scholar] [CrossRef]
- Olivier, J.F.; Fodil, N.; Al Habyan, S.; Gopal, A.; Artusa, P.; Mandl, J.N.; McCaffrey, L.; Gros, P. CCDC88B is required for mobility and inflammatory functions of dendritic cells. J. Leukoc. Biol. 2020, 108, 1787–1802. [Google Scholar] [CrossRef]
- Gilliland, D.G.; Griffin, J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood 2002, 100, 1532–1542. [Google Scholar] [CrossRef] [Green Version]
- Manicone, A.M.; Birkland, T.P.; Lin, M.; Betsuyaku, T.; van Rooijen, N.; Lohi, J.; Keski-Oja, J.; Wang, Y.; Skerrett, S.J.; Parks, W.C. Epilysin (MMP-28) restrains early macrophage recruitment in Pseudomonas aeruginosa pneumonia. J. Immunol. 2009, 182, 3866–3876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharib, S.A.; Johnston, L.K.; Huizar, I.; Birkland, T.P.; Hanson, J.; Wang, Y.; Parks, W.C.; Manicone, A.M. MMP28 promotes macrophage polarization toward M2 cells and augments pulmonary fibrosis. J. Leukoc. Biol. 2014, 95, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Q.; Xing, W.M.; Fan, H.H.; Wang, K.S.; Zhang, H.K.; Wang, Q.W.; Qi, J.; Yang, H.M.; Yang, J.; Ren, Y.N.; et al. The novel lipopolysaccharide-binding protein CRISPLD2 is a critical serum protein to regulate endotoxin function. J. Immunol. 2009, 183, 6646–6656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, I.I.; Frutos Rde, L.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B.; Littman, D.R. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008, 4, 337–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Thackray, L.B.; Handley, S.A.; Gorman, M.J.; Poddar, S.; Bagadia, P.; Briseño, C.G.; Theisen, D.J.; Tan, Q.; Hykes, B.L., Jr.; Lin, H.; et al. Oral Antibiotic Treatment of Mice Exacerbates the Disease Severity of Multiple Flavivirus Infections. Cell Rep. 2018, 22, 3440–3453.e3446. [Google Scholar] [CrossRef] [Green Version]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef] [Green Version]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [Green Version]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Lv, J.; Guo, F.; Li, J.; Jia, Y.; Jiang, D.; Wang, N.; Zhang, C.; Kong, L.; Liu, Y.; et al. Gut Microbiome Influences the Efficacy of PD-1 Antibody Immunotherapy on MSS-Type Colorectal Cancer via Metabolic Pathway. Front. Microbiol. 2020, 11, 814. [Google Scholar] [CrossRef]
- Woo, P.C.; Tsoi, H.W.; Wong, L.P.; Leung, H.C.; Yuen, K.Y. Antibiotics modulate vaccine-induced humoral immune response. Clin. Diagn. Lab. Immunol. 1999, 6, 832–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchiyama, R.; Chassaing, B.; Zhang, B.; Gewirtz, A.T. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 2014, 210, 171–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zackular, J.P.; Baxter, N.T.; Chen, G.Y.; Schloss, P.D. Manipulation of the Gut Microbiota Reveals Role in Colon Tumorigenesis. mSphere 2016, 1. [Google Scholar] [CrossRef] [Green Version]
- Schubert, A.M.; Sinani, H.; Schloss, P.D. Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile. mBio 2015, 6, e00974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciabattini, A.; Olivieri, R.; Lazzeri, E.; Medaglini, D. Role of the Microbiota in the Modulation of Vaccine Immune Responses. Front. Microbiol. 2019, 10, 1305. [Google Scholar] [CrossRef] [Green Version]
- Levine, M.M. Immunogenicity and efficacy of oral vaccines in developing countries: Lessons from a live cholera vaccine. BMC Biol. 2010, 8, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, P.; Curtis, N. The influence of the intestinal microbiome on vaccine responses. Vaccine 2018, 36, 4433–4439. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.; Kim, M.V.; Rakib, T.; Wong, P.W.; van Zandt, M.; Barry, N.A.; Kaisho, T.; Goodman, A.L.; Iwasaki, A. Topical application of aminoglycoside antibiotics enhances host resistance to viral infections in a microbiota-independent manner. Nat. Microbiol. 2018, 3, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Bhargava, P.; McCloskey, D.; Mao, N.; Palsson, B.O.; Collins, J.J. Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function. Cell Host Microbe 2017, 22, 757–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swaminathan, G.; Citron, M.; Xiao, J.; Norton, J.E., Jr.; Reens, A.L.; Topçuoğlu, B.D.; Maritz, J.M.; Lee, K.-J.; Freed, D.C.; Weber, T.M.; et al. Vaccine Hyporesponse Induced by Individual Antibiotic Treatment in Mice and Non-Human Primates Is Diminished upon Recovery of the Gut Microbiome. Vaccines 2021, 9, 1340. https://doi.org/10.3390/vaccines9111340
Swaminathan G, Citron M, Xiao J, Norton JE Jr., Reens AL, Topçuoğlu BD, Maritz JM, Lee K-J, Freed DC, Weber TM, et al. Vaccine Hyporesponse Induced by Individual Antibiotic Treatment in Mice and Non-Human Primates Is Diminished upon Recovery of the Gut Microbiome. Vaccines. 2021; 9(11):1340. https://doi.org/10.3390/vaccines9111340
Chicago/Turabian StyleSwaminathan, Gokul, Michael Citron, Jianying Xiao, James E. Norton, Jr., Abigail L. Reens, Begüm D. Topçuoğlu, Julia M. Maritz, Keun-Joong Lee, Daniel C. Freed, Teresa M. Weber, and et al. 2021. "Vaccine Hyporesponse Induced by Individual Antibiotic Treatment in Mice and Non-Human Primates Is Diminished upon Recovery of the Gut Microbiome" Vaccines 9, no. 11: 1340. https://doi.org/10.3390/vaccines9111340
APA StyleSwaminathan, G., Citron, M., Xiao, J., Norton, J. E., Jr., Reens, A. L., Topçuoğlu, B. D., Maritz, J. M., Lee, K.-J., Freed, D. C., Weber, T. M., White, C. H., Kadam, M., Spofford, E., Bryant-Hall, E., Salituro, G., Kommineni, S., Liang, X., Danilchanka, O., Fontenot, J. A., ... Hannigan, G. D. (2021). Vaccine Hyporesponse Induced by Individual Antibiotic Treatment in Mice and Non-Human Primates Is Diminished upon Recovery of the Gut Microbiome. Vaccines, 9(11), 1340. https://doi.org/10.3390/vaccines9111340





