Recombinant Baculovirus-Produced Grass Carp Reovirus Virus-Like Particles as Vaccine Candidate That Provides Protective Immunity against GCRV Genotype II Infection in Grass Carp
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus and Cells
2.2. Construction of Recombinant Baculoviruses
2.3. Acquisition of Recombinant Baculoviruses
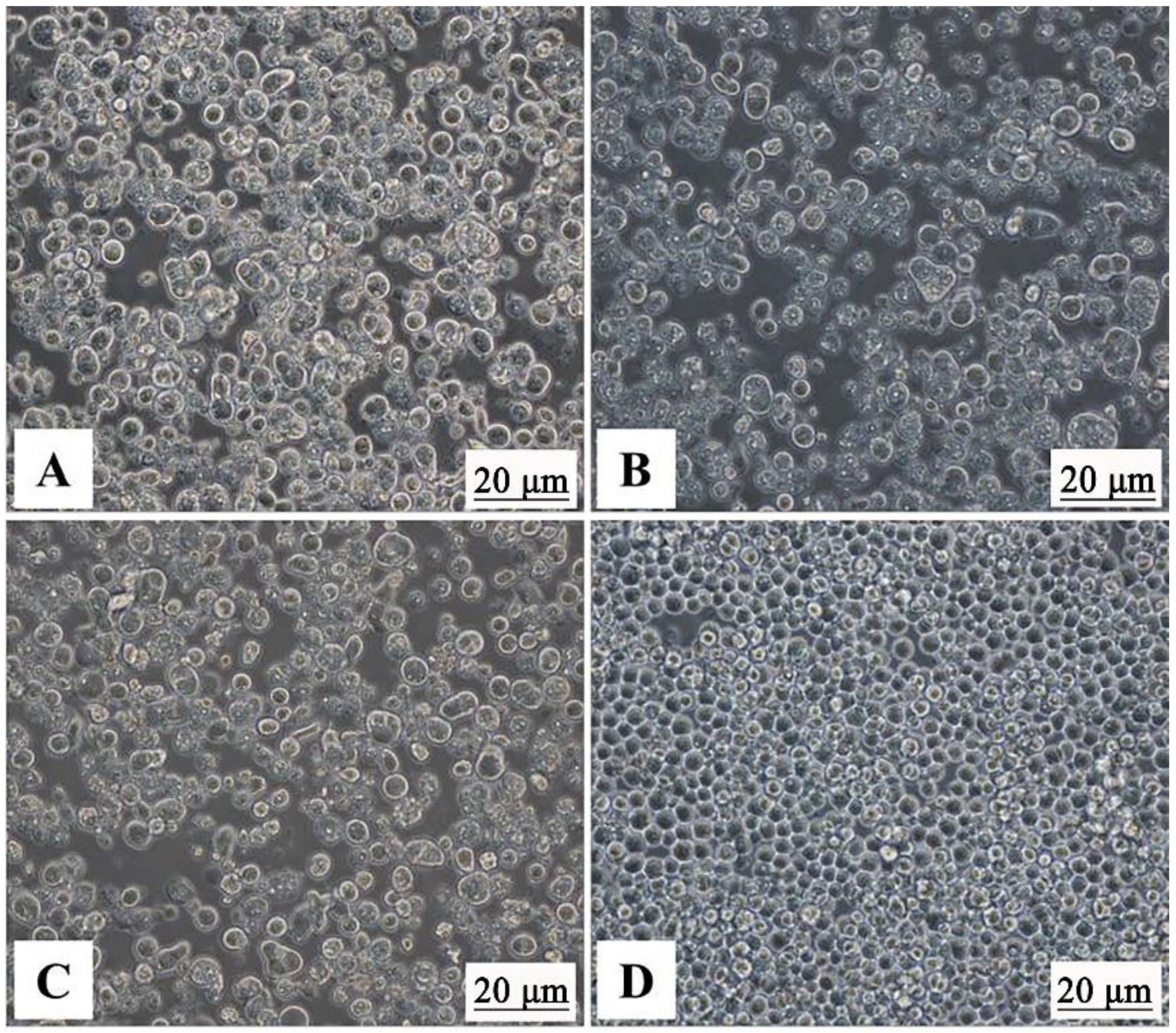
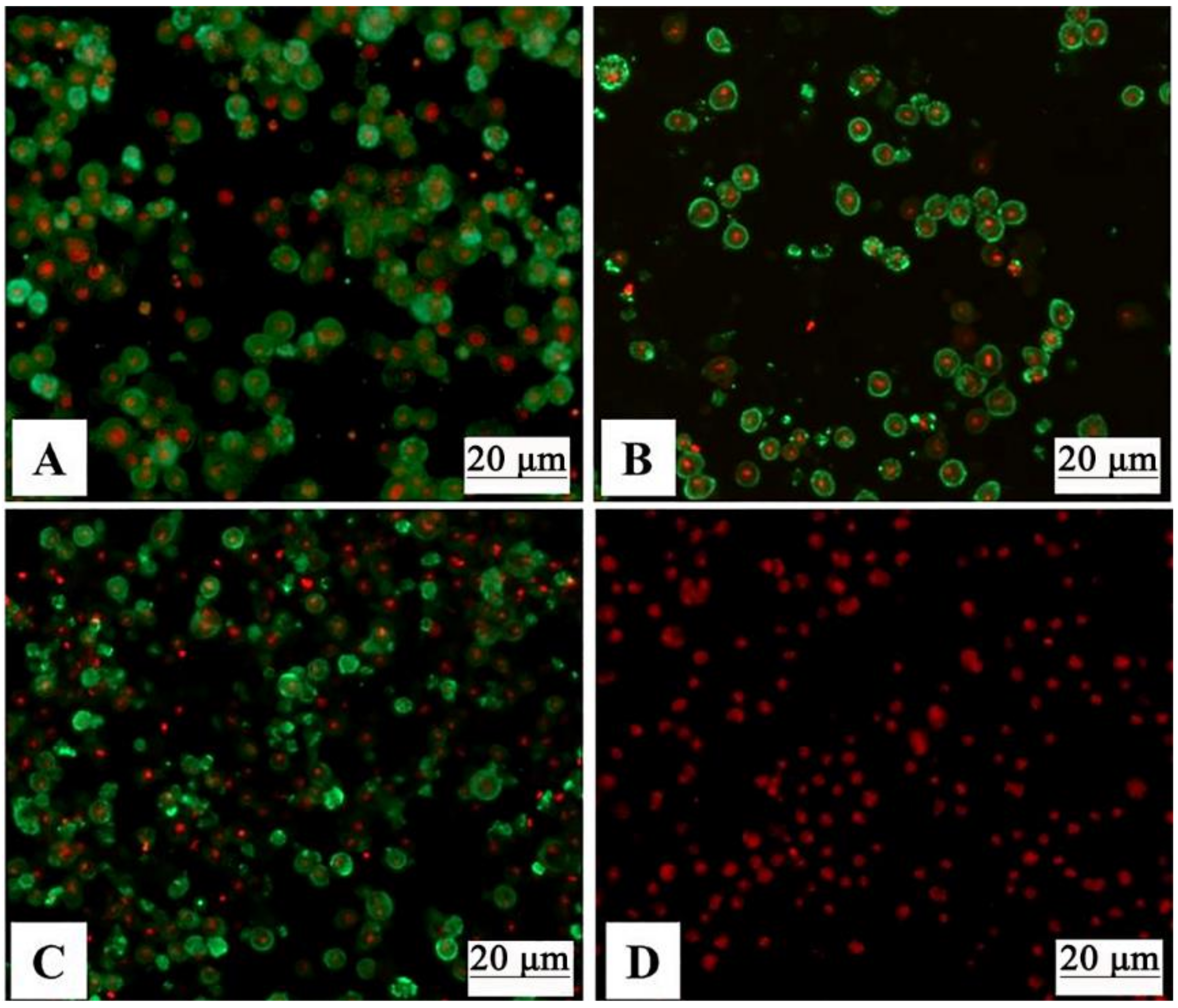
2.4. Detection of Recombinant Baculoviruses by Indirect Fluorescence Assay (IFA)
2.5. Generation and Purification of GCRV-VLPs
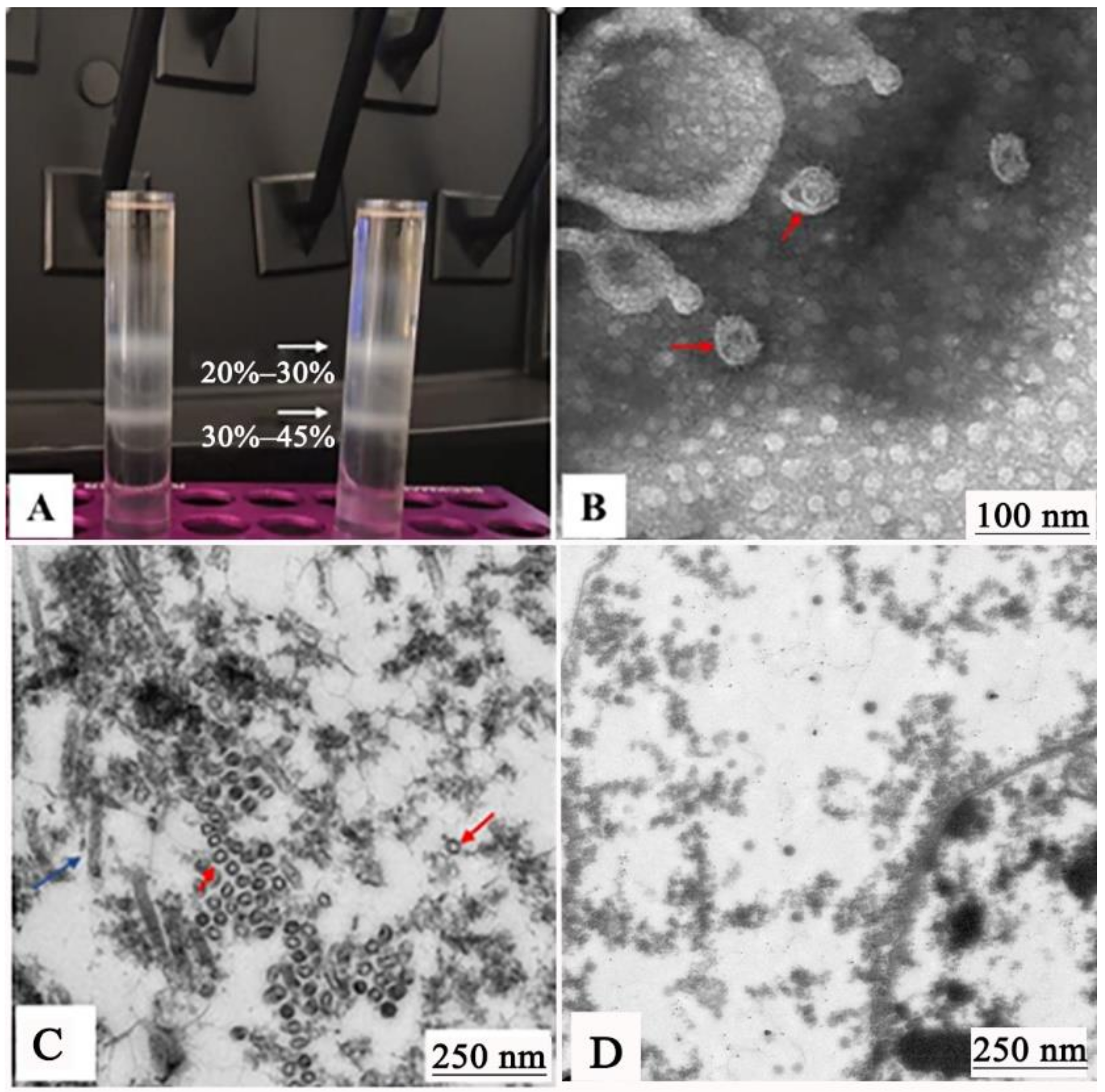
2.6. Identification and Transmission Electron Microscopy (TEM) Analysis of GCRV-VLPs
2.7. Immunization of Grass Carp with GCRV-VLPs
2.8. Detection of Serum-Specific IgM Antibody by Enzyme-linked Immunosorbent Assay (ELISA)
2.9. Detection of Expression of Immune-Related Genes by Quantitative Real-Time PCR (qRT-PCR)
2.10. GCRV Challenge Tests
2.11. Statistical Analysis
3. Results
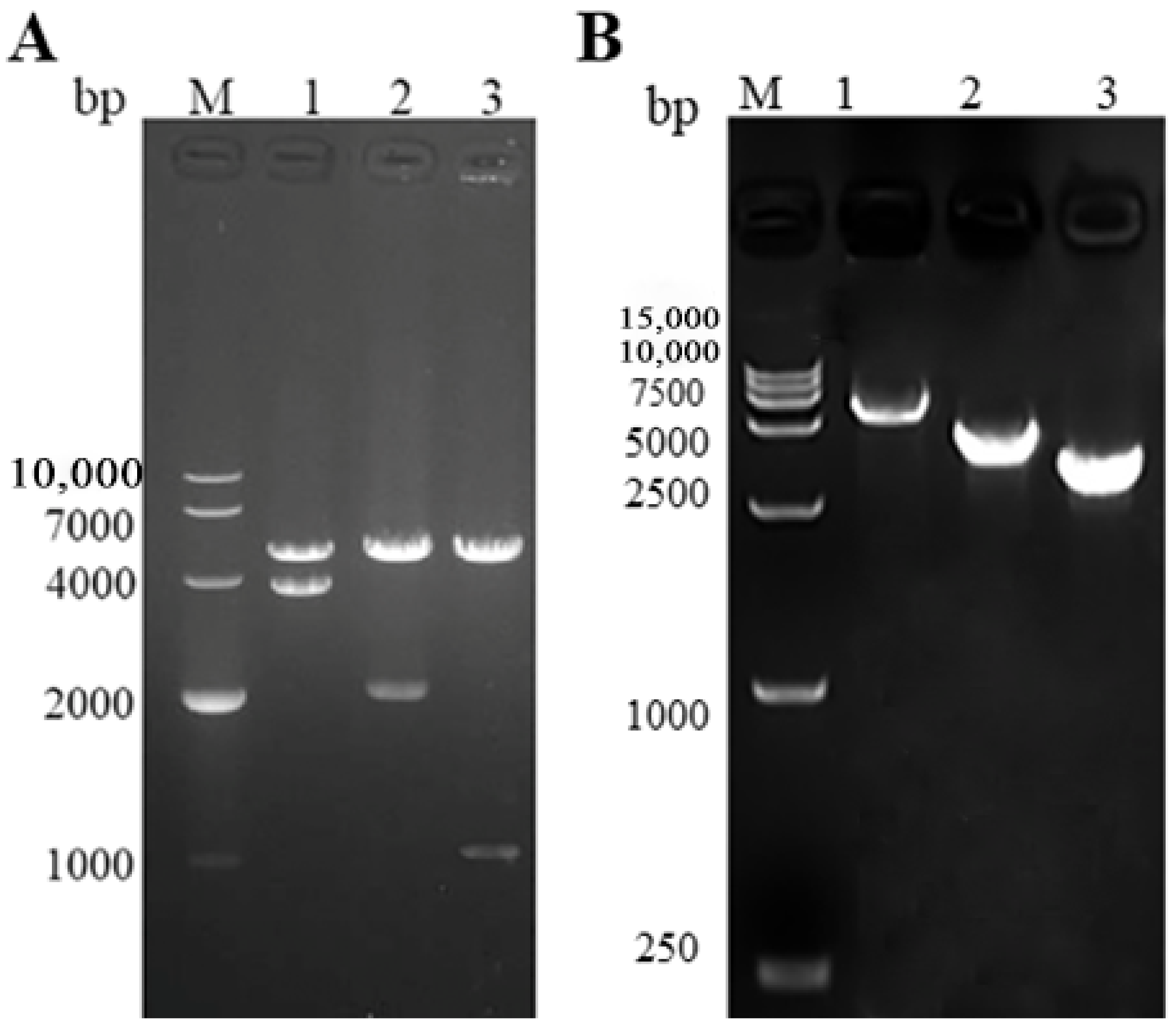
3.1. Construction of Recombinant Baculoviruses
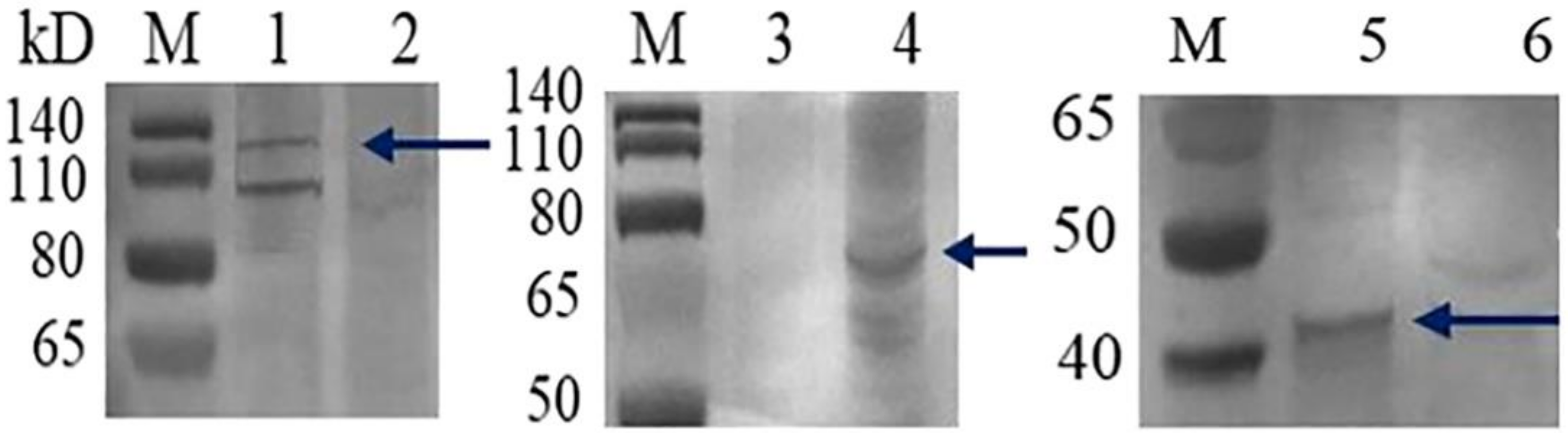
3.2. Verification of Baculovirus-Expressed GCRV VP3, VP4, and VP38 Proteins
3.3. Production and Identification of GCRV-VLPs
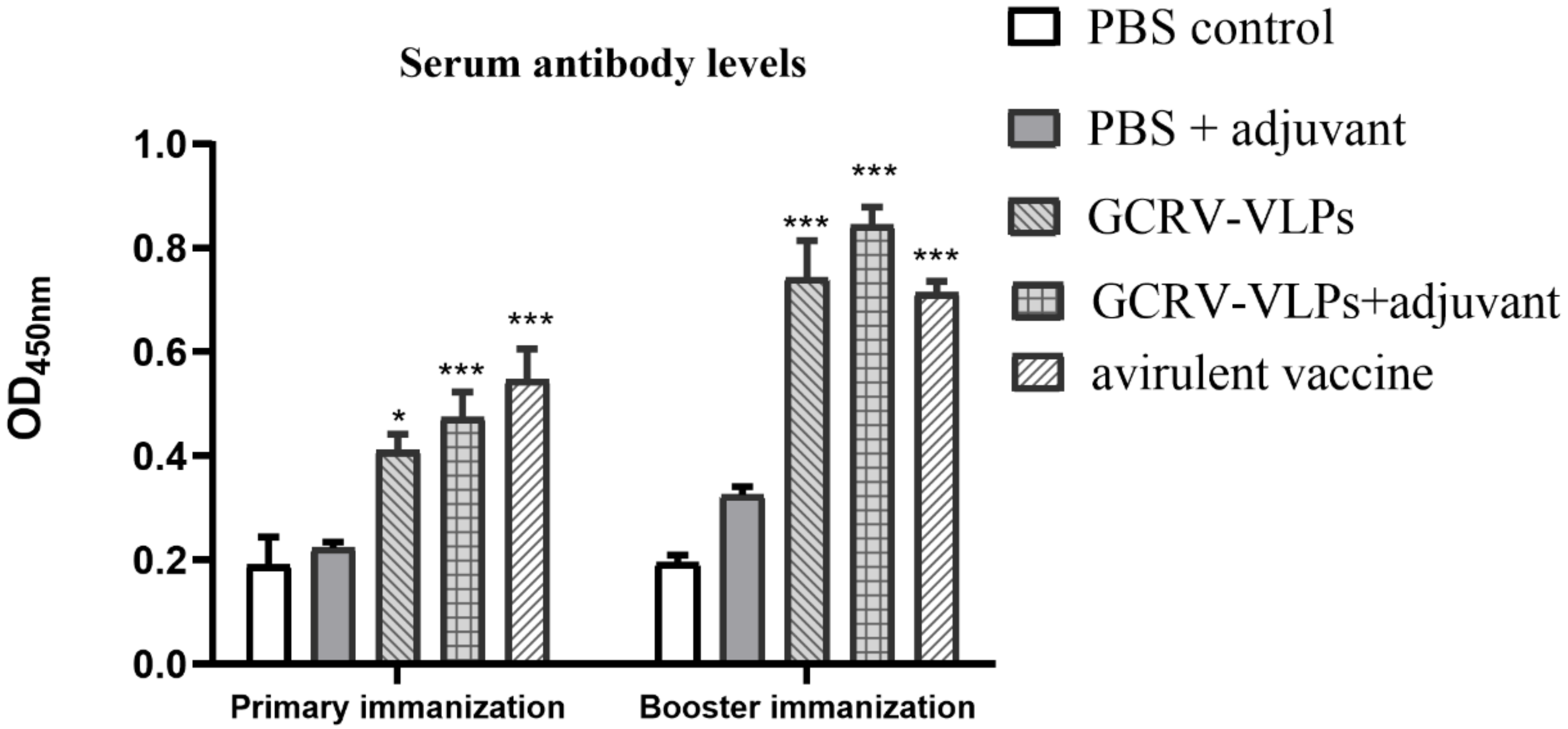
3.4. Detection of Serum IgM Antibody in Immunized Fish
3.5. Expression of Immune-Related Genes after the GCRV VLPs-Based Vaccine
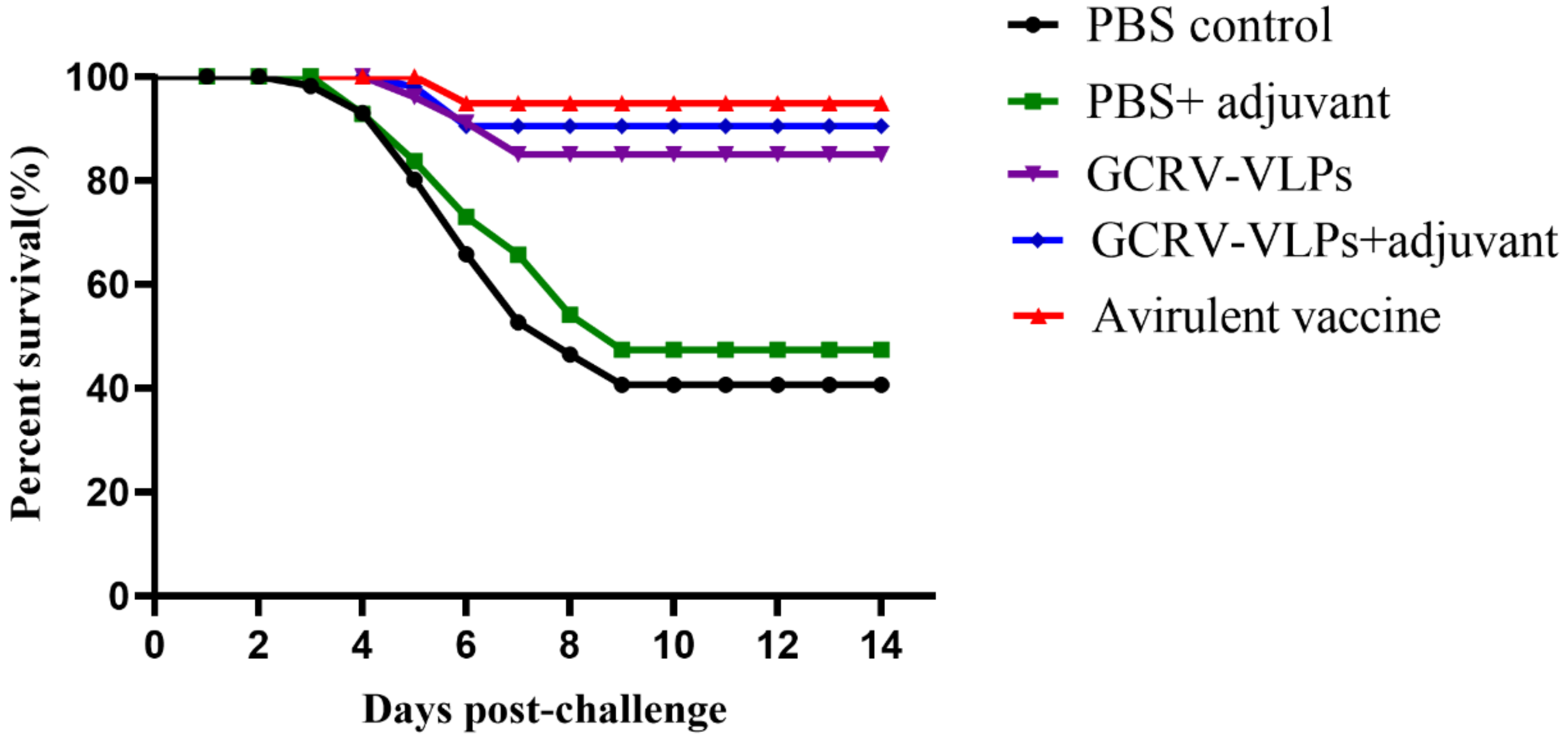
3.6. Protection of GCRV VLPs-Based Vaccine
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Cao, L.; Naylor, R.; Henriksson, P.; Leadbitter, D.; Metian, M.; Troell, M.; Zhang, W. Global food supply. China’s aquaculture and the world’s wild fisheries. Science 2015, 347, 133–135. [Google Scholar] [CrossRef]
- Su, H.; Su, J. Cyprinid viral diseases and vaccine development. Fish Shellfish. Immun. 2018, 83, 84–95. [Google Scholar] [CrossRef]
- Attoui, H.; Fang, Q.; Jaafar, F.M.; Cantaloube, J.F.; Biagini, P.; De Micco, P.; De Lamballerie, X. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of Golden shiner reovirus, Grass carp reovirus, Striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae). J. Gen. Virol. 2002, 83, 1941–1951. [Google Scholar] [CrossRef]
- Subramanian, K.; Hetrick, F.M.; Samal, S.K. Identification of a new genogroup of aquareovirus by RNA-RNA hybridization. J. Gen. Virol. 1997, 78 Pt 6, 1385–1388. [Google Scholar] [CrossRef][Green Version]
- Wang, Q.; Zeng, W.; Liu, C.; Zhang, C.; Wang, Y.; Shi, C.; Wu, S. Complete genome sequence of a reovirus isolated from grass carp, indicating different genotypes of GCRV in China. J. Virol. 2012, 86, 12466. [Google Scholar] [CrossRef]
- Zhang, Q.; Gui, J.F. Virus genomes and virus-host interactions in aquaculture animals. Sci. China Life Sci. 2015, 58, 156–169. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Chen, J.; Wang, Q.; Chang, O.; Zeng, W.; Bergmann, S.M.; Li, Y.; Yin, J.; Wen, H. Establishment of a cell line from egg of rare minnow Gobiocypris rarus for propagation of grass carp reovirus genotype II. Microb. Pathog. 2019, 136, 103715. [Google Scholar] [CrossRef]
- Zeng, W.; Wang, Q.; Wang, Y.; Zhao, C.; Li, Y.; Shi, C.; Wu, S.; Song, X.; Huang, Q.; Li, S. Immunogenicity of a cell culture-derived inactivated vaccine against a common virulent isolate of grass carp reovirus. Fish Shellfish. Immun. 2016, 54, 473–480. [Google Scholar] [CrossRef]
- Collins, C.; Lorenzen, N.; Collet, B. DNA vaccination for finfish aquaculture. Fish Shellfish. Immun. 2019, 85, 106–125. [Google Scholar] [CrossRef]
- Rao, Y.; Su, J. Insights into the antiviral immunity against grass carp (Ctenopharyngodon idella) reovirus (GCRV) in grass carp. J. Immunol. Res. 2015, 2015, 670437. [Google Scholar] [CrossRef]
- Tian, Y.; Ye, X.; Zhang, L.; Deng, G.; Bai, Y. Development of a novel candidate subunit vaccine against Grass carp reovirus Guangdong strain (GCRV-GD108). Fish Shellfish. Immun. 2013, 35, 351–356. [Google Scholar] [CrossRef]
- Ji, Q.; Wang, S.; Ma, J.; Liu, Q. A review: Progress in the development of fish Vibrio spp. vaccines. Immunol. Lett. 2020, 226, 46–54. [Google Scholar] [CrossRef]
- Gao, Y.; Pei, C.; Sun, X.; Zhang, C.; Li, L.; Kong, X. Plasmid pcDNA3.1-s11 constructed based on the S11 segment of grass carp reovirus as DNA vaccine provides immune protection. Vaccine 2018, 36, 3613–3621. [Google Scholar] [CrossRef]
- Jeong, K.H.; Kim, H.J.; Kim, H.J. Current status and future directions of fish vaccines employing virus-like particles. Fish Shellfish. Immun. 2020, 100, 49–57. [Google Scholar] [CrossRef]
- Fraenkel-Conrat, H.; Williams, R.C. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl. Acad. Sci. USA 1955, 41, 690–698. [Google Scholar] [CrossRef]
- Ho, J.K.; Jeevan-Raj, B.; Netter, H.J. Hepatitis B Virus (HBV) subviral particles as protective vaccines and vaccine platforms. Viruses 2020, 12, 126. [Google Scholar] [CrossRef]
- Roden, R.B.S.; Stern, P.L. Opportunities and challenges for human papillomavirus vaccination in cancer. Nat. Rev. Cancer 2018, 18, 240–254. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Augusto, G.; Bachmann, M.F. The 3Ds in virus-like particle based-vaccines: “Design, Delivery and Dynamics”. Immunol. Rev. 2020, 296, 155–168. [Google Scholar] [CrossRef]
- Ailor, E.; Betenbaugh, M.J. Modifying secretion and post-translational processing in insect cells. Curr. Opin. Biotechnol. 1999, 10, 142–145. [Google Scholar] [CrossRef]
- Berger, I.; Fitzgerald, D.J.; Richmond, T.J. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004, 22, 1583–1587. [Google Scholar] [CrossRef]
- Han, X.; Huang, Y.; Hou, Y.; Dang, H.; Li, R. Recombinant expression and functional analysis of antimicrobial Siganus oraminl-amino acid oxidase using the Bac-to-Bac baculovirus expression system. Fish Shellfish. Immun. 2020, 98, 962–970. [Google Scholar] [CrossRef]
- Fang, Q.; Attoui, H.; Cantaloube, J.F.; Biagini, P.; Zhu, Z.; De Micco, P.; De Lamballerie, X. Sequence of genome segments 1, 2, and 3 of the grass carp reovirus (Genus Aquareovirus, family Reoviridae). Biochem. Biophys. Res. Commun. 2000, 274, 762–766. [Google Scholar] [CrossRef]
- Su, H.; Fan, C.; Liao, Z.; Yang, C.; Clarke, J.L.; Zhang, Y.; Su, J. Grass carp reovirus major outer capsid protein VP4 interacts with RNA sensor RIG-I to suppress interferon response. Biomolecules 2020, 10, 560. [Google Scholar] [CrossRef]
- Qiu, T.; Lu, R.H.; Zhang, J.; Zhu, Z.Y. Complete nucleotide sequence of the S10 genome segment of grass carp reovirus (GCRV). Dis. Aquat. Organ. 2001, 44, 69–74. [Google Scholar] [CrossRef]
- Jiang, H.; Bian, Q.; Zeng, W.; Ren, P.; Sun, H.; Lin, Z.; Tang, Z.; Zhou, X.; Wang, Q.; Wang, Y.; et al. Oral delivery of Bacillus subtilis spores expressing grass carp reovirus VP4 protein produces protection against grass carp reovirus infection. Fish Shellfish. Immun. 2019, 84, 768–780. [Google Scholar] [CrossRef]
- Chen, D.D.; Yao, Y.Y.; Cui, Z.W.; Zhang, X.Y.; Peng, K.S.; Guo, X.; Wang, B.; Zhou, Y.Y.; Li, S.; Wu, N.; et al. Comparative study of the immunoprotective effect of two DNA vaccines against grass carp reovirus. Fish Shellfish. Immun. 2018, 75, 66–73. [Google Scholar] [CrossRef]
- Mu, C.; Vakharia, V.N.; Zhou, Y.; Jiang, N.; Liu, W.; Meng, Y.; Li, Y.; Xue, M.; Zhang, J.; Zeng, L.; et al. A novel subunit vaccine based on outer capsid proteins of Grass Carp Reovirus (GCRV) provides protective immunity against GCRV Infection in rare minnow (Gobiocypris rarus). Pathogens 2020, 9, 945. [Google Scholar] [CrossRef]
- Zeng, W.; Wang, Y.; Guo, Y.; Bergmann, S.M.; Yin, J.; Li, Y.; Ren, Y.; Shi, C.; Wang, Q. Development of a VP38 recombinant protein-based indirect ELISA for detection of antibodies against grass carp reovirus genotype II (iELISA for detection of antibodies against GCRV II). J. Fish Dis. 2018, 41, 1811–1819. [Google Scholar] [CrossRef]
- Li, G.; Zhao, Y.; Wang, J.; Liu, B.; Sun, X.; Guo, S.; Feng, J. Transcriptome profiling of developing spleen tissue and discovery of immune-related genes in grass carp (Ctenopharyngodon idella). Fish Shellfish. Immun. 2017, 60, 400–410. [Google Scholar] [CrossRef]
- Chang, L.J.; Dowd, K.A.; Mendoza, F.H.; Saunders, J.G.; Sitar, S.; Plummer, S.H.; Yamshchikov, G.; Sarwar, U.N.; Hu, Z.; Enama, M.E.; et al. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: A phase 1 dose-escalation trial. Lancet 2014, 384, 2046–2052. [Google Scholar] [CrossRef]
- Roldao, A.; Mellado, M.C.; Castilho, L.R.; Carrondo, M.J.; Alves, P.M. Virus-like particles in vaccine development. Expert Rev. Vaccines 2010, 9, 1149–1176. [Google Scholar] [CrossRef]
- Chan, H.L.; Thompson, A.; Martinot-Peignoux, M.; Piratvisuth, T.; Cornberg, M.; Brunetto, M.R.; Tillmann, H.L.; Kao, J.H.; Jia, J.D.; Wedemeyer, H.; et al. Hepatitis B surface antigen quantification: Why and how to use it in 2011—A core group report. J. Hepatol. 2011, 55, 1121–1131. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Zha, L.; Cabral-Miranda, G.; Bachmann, M.F. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin. Immunol. 2017, 34, 123–132. [Google Scholar] [CrossRef]
- Luu, V.T.; Moon, H.Y.; Hwang, J.Y.; Kang, B.K.; Kang, H.A. Development of recombinant Yarrowia lipolytica producing virus-like particles of a fish nervous necrosis virus. J. Microbiol. 2017, 55, 655–664. [Google Scholar] [CrossRef]
- Marsian, J.; Hurdiss, D.L.; Ranson, N.A.; Ritala, A.; Paley, R.; Cano, I.; Lomonossoff, G.P. Plant-made nervous necrosis virus-like particles protect fish against disease. Front. Plant Sci. 2019, 10, 880. [Google Scholar] [CrossRef]
- Lin, C.F.; Jiang, H.K.; Chen, N.C.; Wang, T.Y.; Chen, T.Y. Novel subunit vaccine with linear array epitope protect giant grouper against nervous necrosis virus infection. Fish Shellfish. Immun. 2018, 74, 551–558. [Google Scholar] [CrossRef]
- Magadan, S.; Sunyer, O.J.; Boudinot, P. Unique Features of Fish Immune Repertoires: Particularities of Adaptive Immunity within the Largest Group of Vertebrates; Hsu, E., Du Pasquier, L., Eds.; Springer: Berlin, Germany, 2015; pp. 235–264. [Google Scholar] [CrossRef]
- Fillatreau, S.; Six, A.; Magadan, S.; Castro, R.; Sunyer, J.O.; Boudinot, P. The astonishing diversity of Ig classes and B cell repertoires in teleost fish. Front. Immunol. 2013, 4, 28. [Google Scholar] [CrossRef]
- Stefan, K.L.; Kim, M.V.; Iwasaki, A.; Kasper, D.L. Commensal microbiota modulation of natural resistance to virus infection. Cell 2020, 183, 1312–1324.e10. [Google Scholar] [CrossRef]
- Lu, L.F.; Li, Z.C.; Zhang, C.; Zhou, X.Y.; Zhou, Y.; Jiang, J.Y.; Chen, D.D.; Li, S.; Zhang, Y.A. Grass Carp Reovirus (GCRV) giving its all to suppress IFN production by countering MAVS signaling transduction. Front. Immunol. 2020, 11, 545302. [Google Scholar] [CrossRef]
- Feng, X.; Su, J.; Yang, C.; Yan, N.; Rao, Y.; Chen, X. Molecular characterizations of grass carp (Ctenopharyngodon idella) TBK1 gene and its roles in regulating IFN-I pathway. Dev. Comp. Immunol. 2014, 45, 278–290. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, Y.B.; Liu, T.K.; Shi, J.; Sun, F.; Gui, J.F. Fish viperin exerts a conserved antiviral function through RLR-triggered IFN signaling pathway. Dev. Comp. Immunol. 2014, 47, 140–149. [Google Scholar] [CrossRef]
- Chen, L.; Li, Q.; Su, J.; Yang, C.; Li, Y.; Rao, Y. Trunk kidney of grass carp (Ctenopharyngodon idella) mediates immune responses against GCRV and viral/bacterial PAMPs in vivo and in vitro. Fish Shellfish. Immun. 2013, 34, 909–919. [Google Scholar] [CrossRef]
- Fan, Y.; Zhou, Y.; Zeng, L.; Jiang, N.; Liu, W.; Zhao, J.; Zhong, Q. Identification, structural characterization, and expression analysis of toll-like receptors 2 and 3 from gibel carp (Carassius auratus gibelio). Fish Shellfish. Immun. 2018, 72, 629–638. [Google Scholar] [CrossRef]

| Gene | Primer | Primer Sequence (5′→3′) 1 | Restriction Site | Amplicon Size(bp) |
|---|---|---|---|---|
| S3-VP3 | VP3-F VP3-R | 5′-ATAGCGGCCGCCCAATGCATCGTCATAACAGA-3′ 5′-ATAGGTACCATGTGCGTGGTCCAGAACGTG-3′ | Not I Kpn I | 3700 |
| S6-VP4 | VP4-F VP4-R | 5′-ATCGGATCCCTGACTTACGGCCACTATCAT-3′ 5′-ATACTCGAGATGCAAAGCGGGGGTCGGTGT-3′ | BamH I Xho I | 1953 |
| S10-VP38 | VP38-F VP38-R | 5′-ATAGCGGCCGCTCAACATGGCGGGTGTGTCTC-3′ 5′-ATACTCGAGGTACCGTTATCGCCGACCACA-3′ | Not I Xho I | 1038 |
| M13 | M13-F M13-R | 5′-GTTTTCCCAGTCACGAC-3′ 5′-CAGGAAACAGCTATGAC-3′ | 2430 bp + the inset |
| Gene | Primer Sequence (5′→3′) | GenBank Accession |
|---|---|---|
| β-actin-F | 5′-GGATGATGAAATTGCCGCACTGG-3′ | M25013.1 |
| β-actin-R | 5′-ACCGACCATGACGCCCTGATGT-3′ | |
| IFN-F | 5′-GACACATACAGTAGGATATTCACTCGC-3′ | DQ357216.1 |
| IFN-R | 5′-TTGCCTGGGAAGTAGTTTTCTTG-3′ | |
| TLR3-F | 5′-GAGAACAATCGTGACTCCCTGA-3′ | DQ885910.1 |
| TLR3-R | 5′-CCAGTAGAGAACACAGCGAGGT-3′ | |
| IgM-F | 5′-TCTACCTCCAACTCCACCACC-3′ | DQ417927.1 |
| IgM-R | 5′-TGTTTATTGTATTTGCCACCTGAT-3′ | |
| TLR7-F | 5′-GAGCATACAGTTGAGTAAACGCAC-3′ | AB553573.1 |
| TLR7-R | 5′-TCTCCAAGAATATCAGGACGATAA-3 |
| Project | Mortality Rate (%) | Survival Rate (%) | RPS 2 (%) |
|---|---|---|---|
| PBS control | 60 | 40.00 | -- |
| PBS + adjuvant | 52.5 | 47.50 | 12.50 |
| Attenuated vaccine | 5.00 | 95.00 | 91.67 |
| GCRV-VLPs | 15.00 | 85.00 | 75.00 |
| GCRV-VLPs + adjuvant | 10.00 | 90.00 | 83.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, T.; Gao, C.; Wu, S.; Wang, Y.; Yin, J.; Li, Y.; Zeng, W.; Bergmann, S.M.; Wang, Q. Recombinant Baculovirus-Produced Grass Carp Reovirus Virus-Like Particles as Vaccine Candidate That Provides Protective Immunity against GCRV Genotype II Infection in Grass Carp. Vaccines 2021, 9, 53. https://doi.org/10.3390/vaccines9010053
Gao T, Gao C, Wu S, Wang Y, Yin J, Li Y, Zeng W, Bergmann SM, Wang Q. Recombinant Baculovirus-Produced Grass Carp Reovirus Virus-Like Particles as Vaccine Candidate That Provides Protective Immunity against GCRV Genotype II Infection in Grass Carp. Vaccines. 2021; 9(1):53. https://doi.org/10.3390/vaccines9010053
Chicago/Turabian StyleGao, Ting, Caixia Gao, Siyu Wu, Yingying Wang, Jiyuan Yin, Yingying Li, Weiwei Zeng, Sven M. Bergmann, and Qing Wang. 2021. "Recombinant Baculovirus-Produced Grass Carp Reovirus Virus-Like Particles as Vaccine Candidate That Provides Protective Immunity against GCRV Genotype II Infection in Grass Carp" Vaccines 9, no. 1: 53. https://doi.org/10.3390/vaccines9010053
APA StyleGao, T., Gao, C., Wu, S., Wang, Y., Yin, J., Li, Y., Zeng, W., Bergmann, S. M., & Wang, Q. (2021). Recombinant Baculovirus-Produced Grass Carp Reovirus Virus-Like Particles as Vaccine Candidate That Provides Protective Immunity against GCRV Genotype II Infection in Grass Carp. Vaccines, 9(1), 53. https://doi.org/10.3390/vaccines9010053





