The Role of Virulence Proteins in Protection Conferred by Bordetella pertussis Outer Membrane Vesicle Vaccines
Abstract
1. Introduction
2. Results
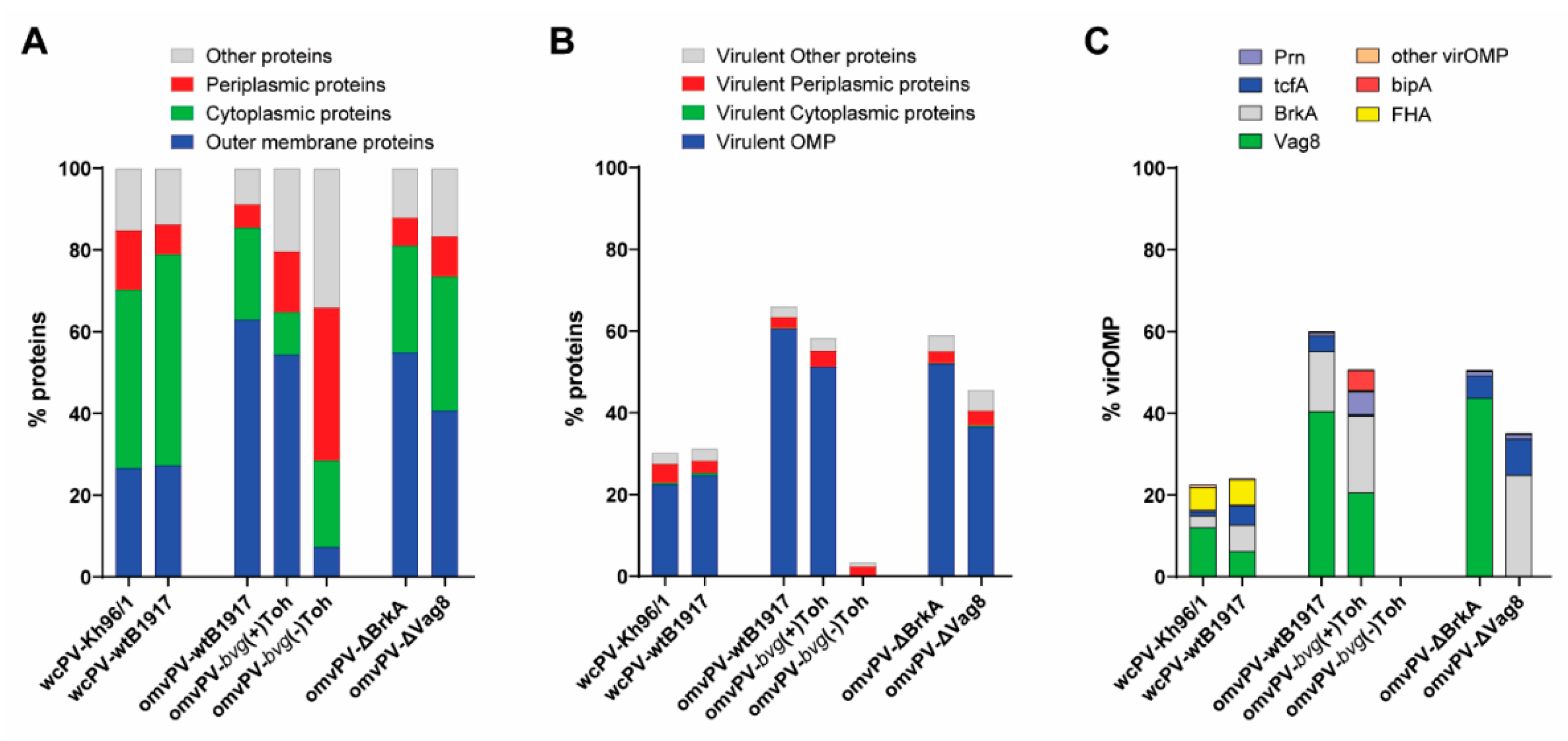
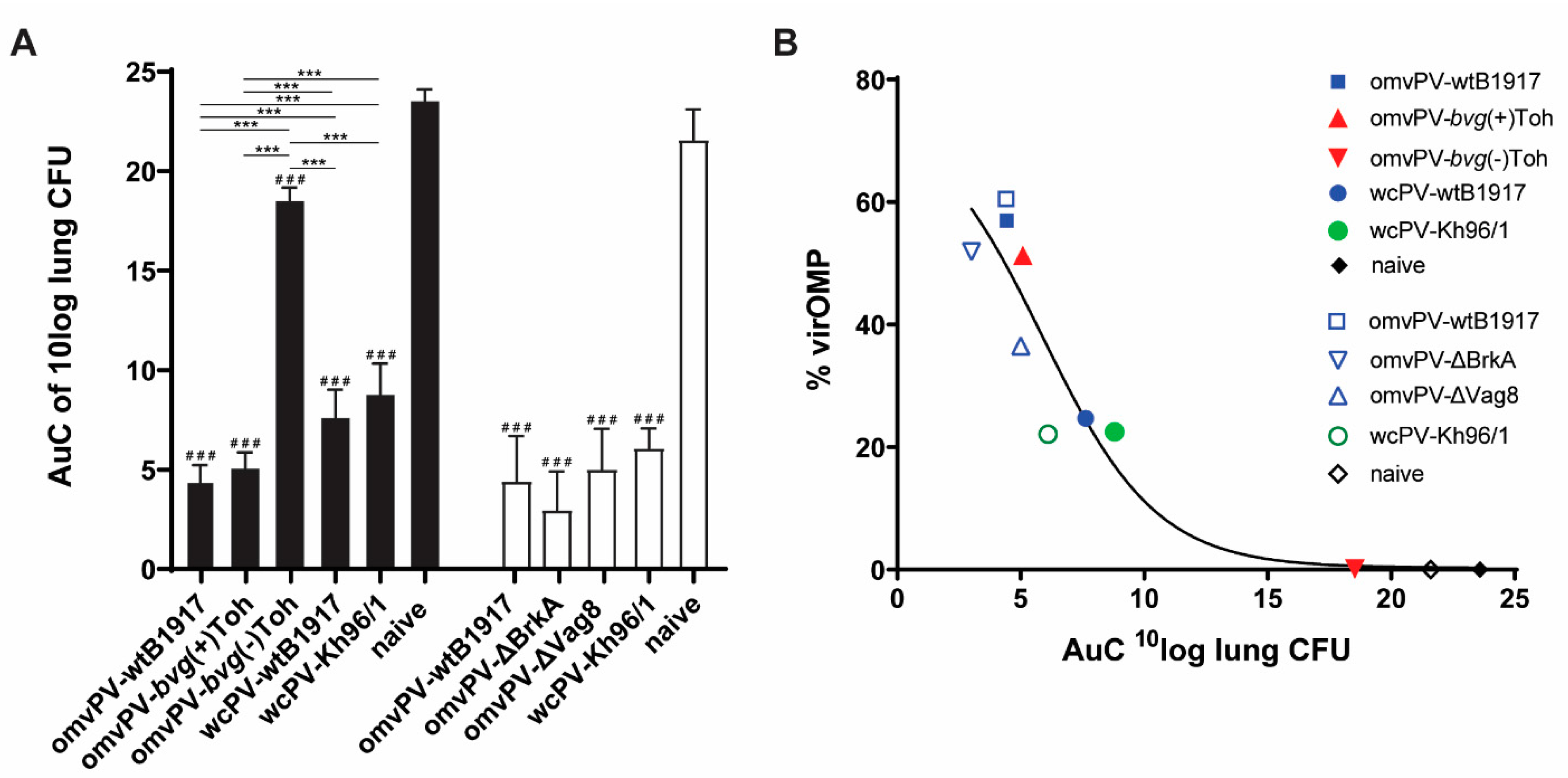
2.1. The Level of VirOMP in omvPV Is Positively Related to Protection against B. Pertussis
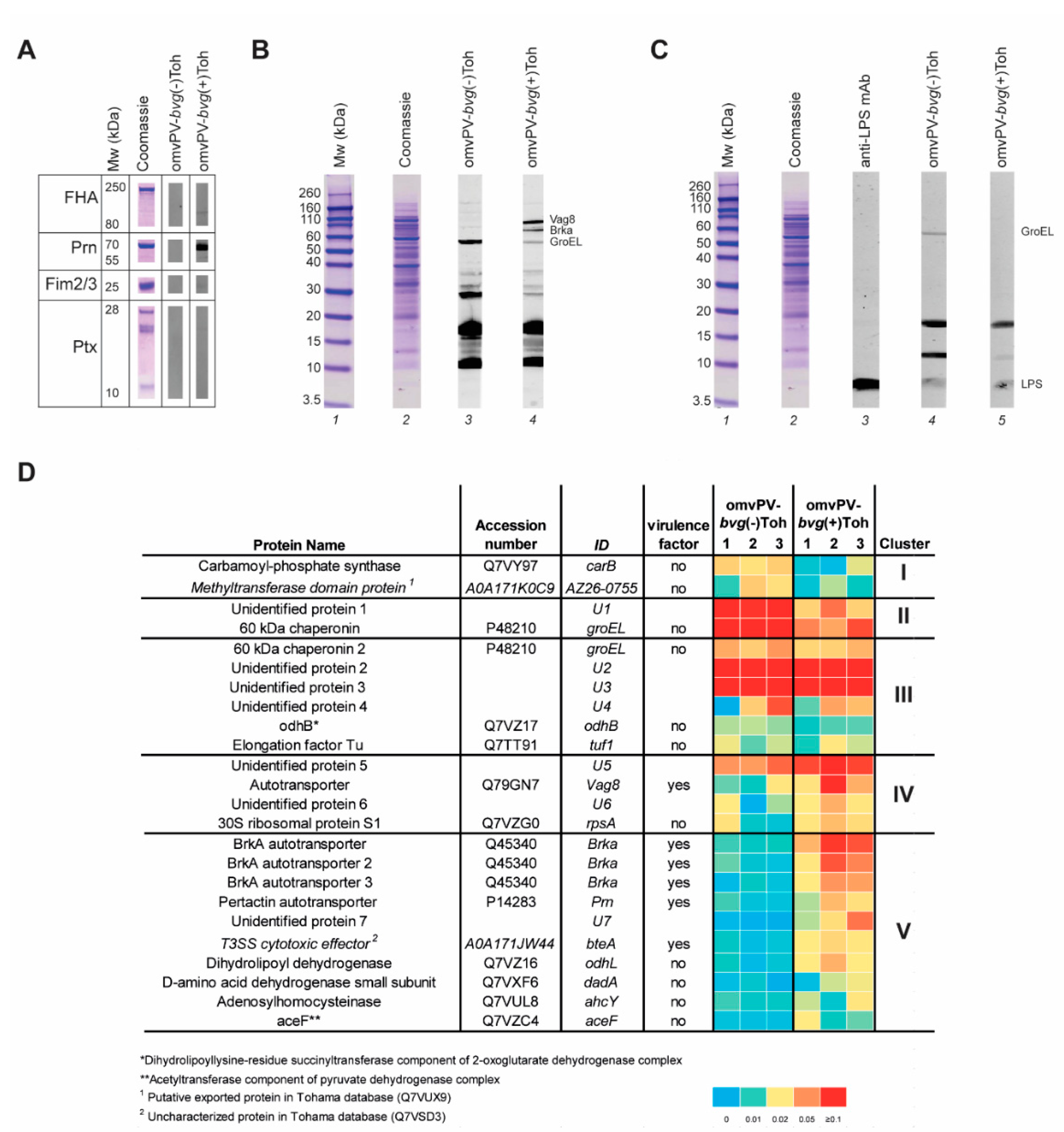
2.2. Immunoproteomic Profiling of High Antibody Responses Induced by omvPV-bvg(-)Toh and omvPV-bvg(+)Toh Reveals Partial Distinct Antigen Specificity
2.3. Deletion of Either Vag8 or BrkA in Outer Membrane Vesicles Does Not Affect Its Protective Capacity and Immunity Profiles
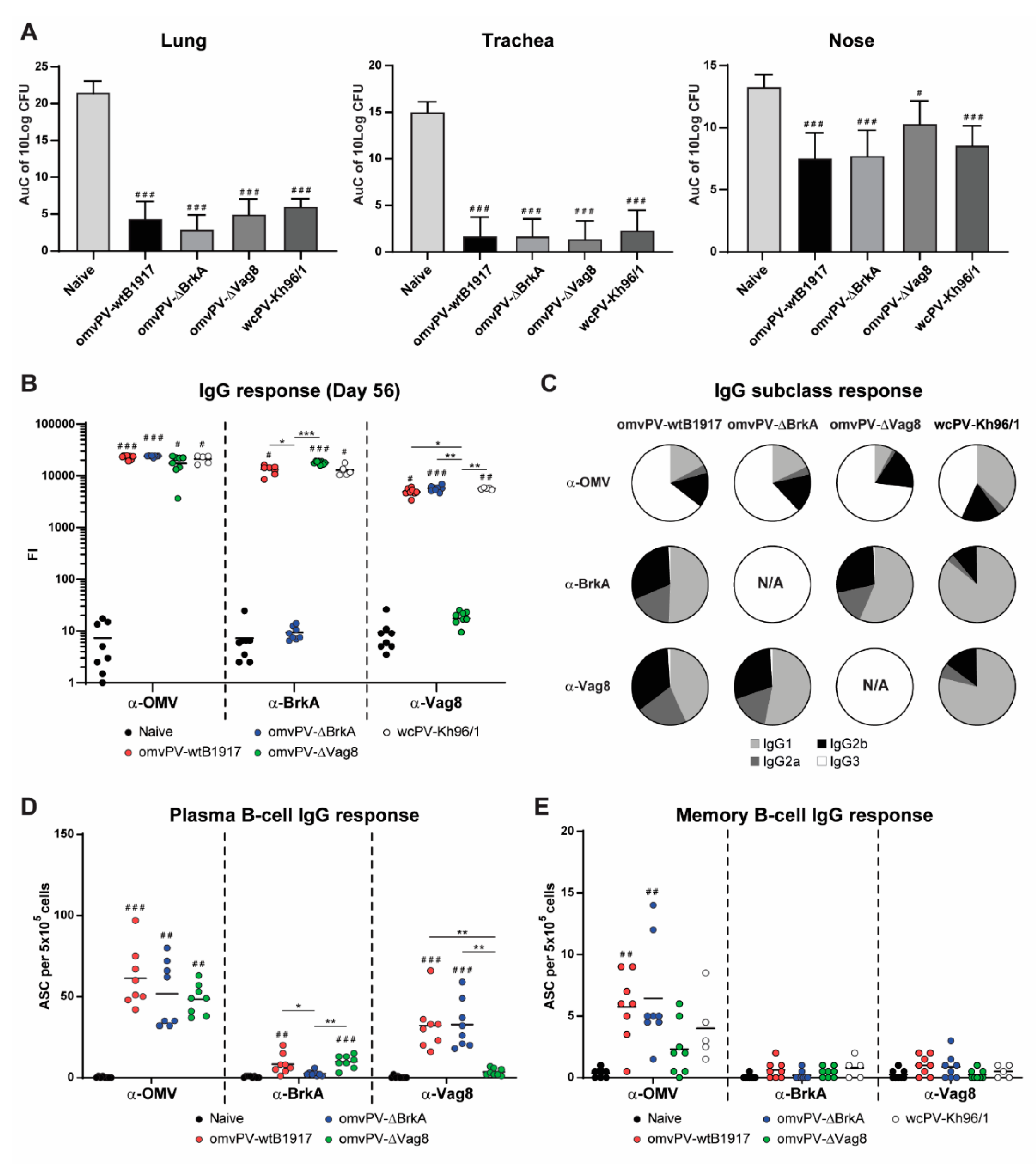
2.4. Immunization with omvPV-∆BrkA and omvPV-∆Vag8 Reveals Equal Protective Capacity as omvPV-wtB1917
2.5. Deletion of BrkA or Vag8 Has Limited Effect on Magnitude, Specificity and Subclass Distribution of Antibody Responses against Other Antigens
2.6. Number of omvPV-Induced Plasma and Memory B-Cells Are Not Influenced by Deletion of BrkA or Vag8
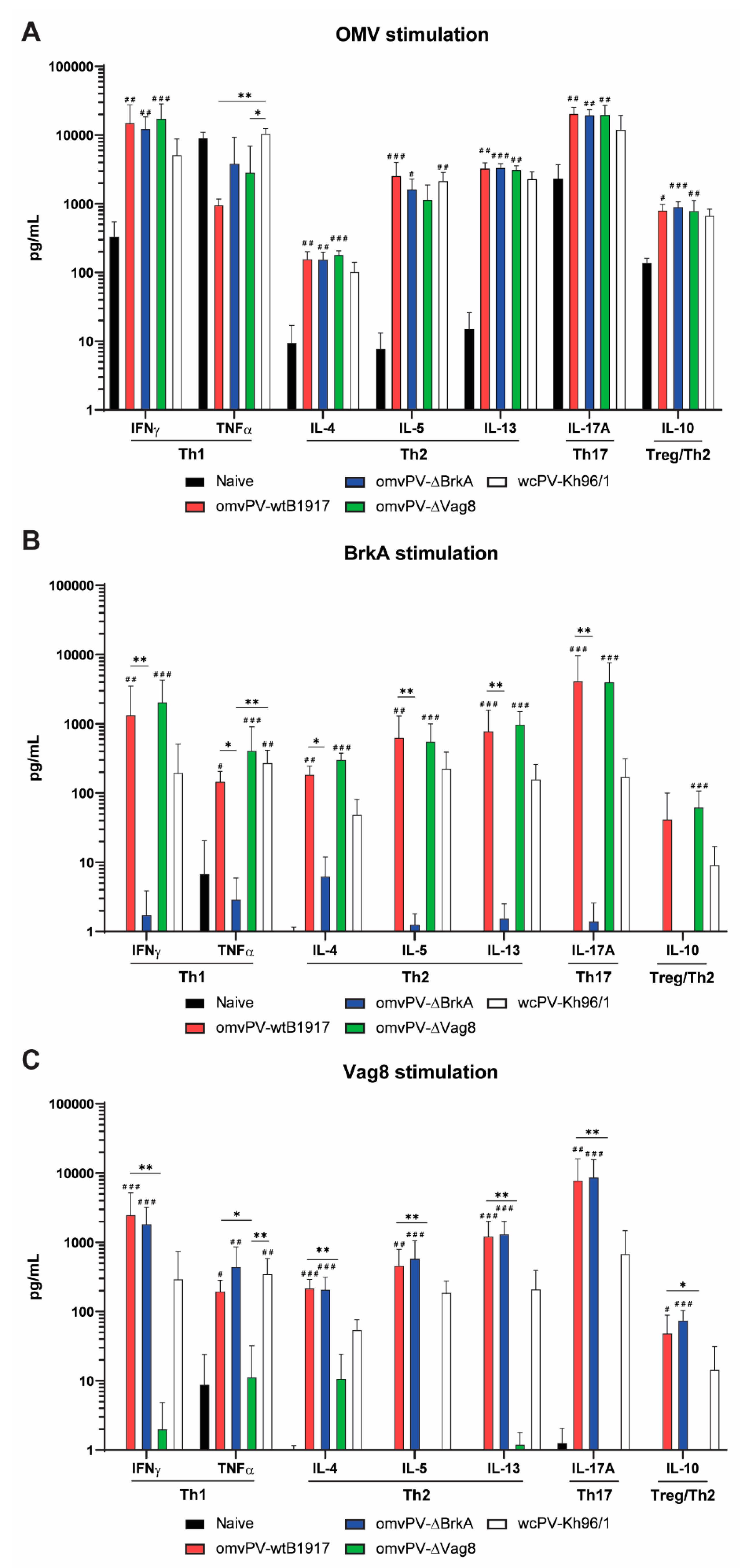
2.7. The omvPV-Induced Mixed Th1/Th2/Th17 Response Is Not Affected by the Deletion of Either Vag8 or BrkA
3. Discussion
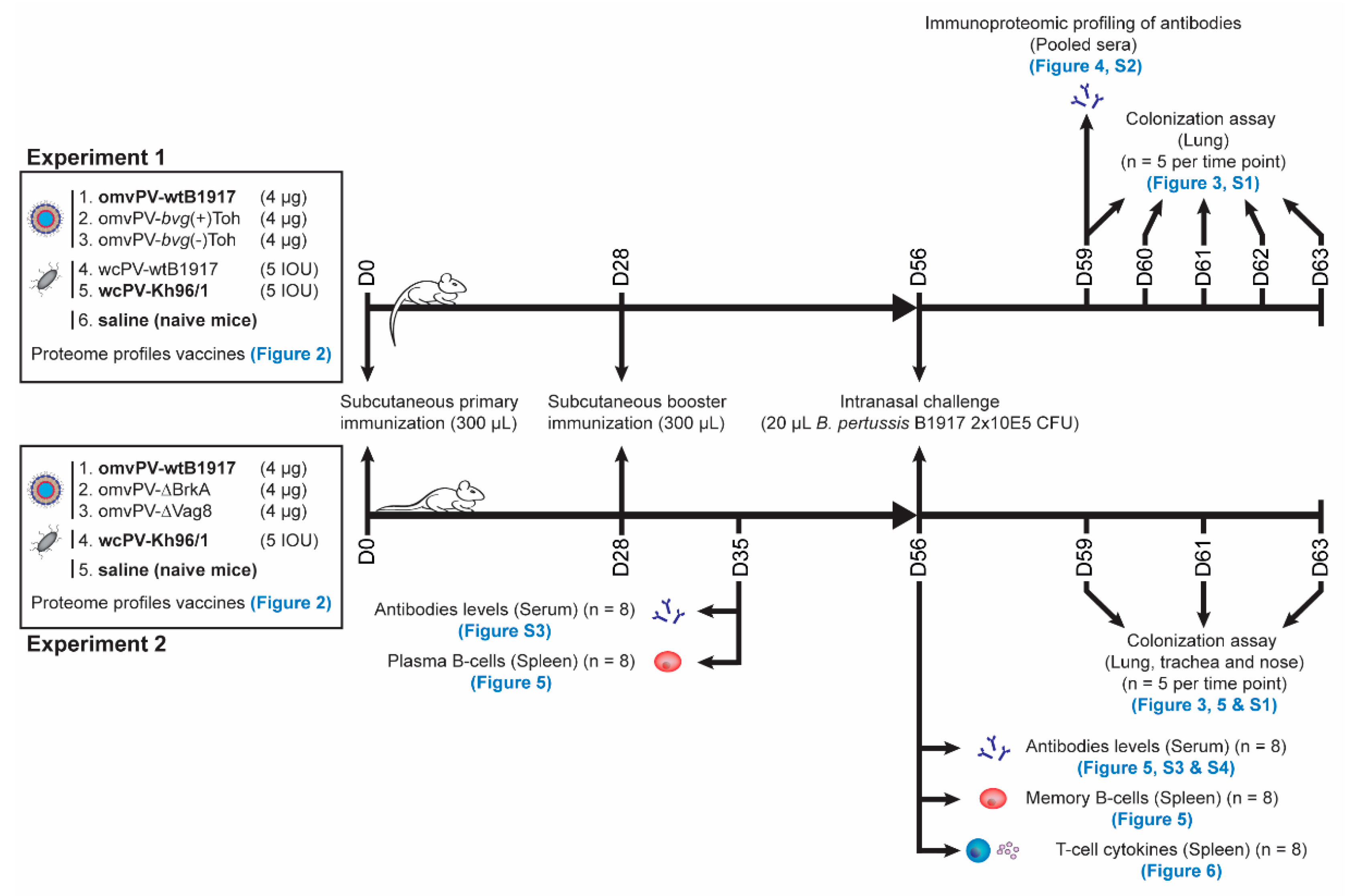
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Construction of BrkA and Vag8 Knockout Mutants
4.3. Whole Cell and Outer Membrane Vesicle Preparations and Analysis
4.4. LC–MS/MS Analysis for Antigen Composition Vaccines
4.4.1. Protein Digestion
4.4.2. Peptide Identification
4.4.3. Data Handling for Protein Digests
4.5. Ethics Statement
4.6. Immunization and Challenge of Mice
4.7. Sample Collection
4.8. Colonization Assays
4.9. Multiplex Immunoassay (MIA) for Antibody Measurements
4.10. B-cell ELISpot for Plasma and Memory B-cells
4.11. Cell Stimulation and MIA for T-Helper (Th) Cytokine Analysis
4.12. Immunoproteomic Antibody Profiling
4.13. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Esposito, S.; Stefanelli, P.; Fry, N.K.; Fedele, G.; He, Q.; Paterson, P.; Tan, T.; Knuf, M.; Rodrigo, C.; Weil, O.C.; et al. Pertussis Prevention: Reasons for Resurgence, and Differences in the Current Acellular Pertussis Vaccines. Front. Immunol. 2019, 10, 1344. [Google Scholar] [CrossRef] [PubMed]
- Diavatopoulos, D.A.; Edwards, K.M. What Is Wrong with Pertussis Vaccine Immunity? Why Immunological Memory to Pertussis Is Failing. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef]
- Pawloski, L.C.; Queenan, A.M.; Cassiday, P.K.; Lynch, A.S.; Harrison, M.J.; Shang, W.; Williams, M.M.; Bowden, K.E.; Burgos-Rivera, B.; Qin, X.; et al. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin. Vaccine Immunol. Cvi. 2014, 21, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Octavia, S.; Luu, L.D.W.; Payne, M.; Timms, V.; Tay, C.Y.; Keil, A.D.; Sintchenko, V.; Guiso, N.; Lan, R. Pertactin-Negative and Filamentous Hemagglutinin-Negative Bordetella pertussis, Australia, 2013–2017. Emerg. Infect. Dis. 2019, 25, 1196–1199. [Google Scholar] [CrossRef]
- Langsam, D.; Anis, E.; Haas, E.J.; Gosinov, R.; Yechezkel, M.; Grotto, I.; Shmueli, E.; Yamin, D. Tdap vaccination during pregnancy interrupts a twenty-year increase in the incidence of pertussis. Vaccine 2020, 38, 2700–2706. [Google Scholar] [CrossRef]
- Pinto, M.V.; Merkel, T.J. Pertussis disease and transmission and host responses: Insights from the baboon model of pertussis. J. Infect. 2017, 74 (Suppl. 1), S114–S119. [Google Scholar] [CrossRef]
- Di Mattia, G.; Nicolai, A.; Frassanito, A.; Petrarca, L.; Nenna, R.; Midulla, F. Pertussis: New preventive strategies for an old disease. Paediatr. Respir. Rev. 2019, 29, 68–73. [Google Scholar] [CrossRef]
- Tong, J.; Buikema, A.; Horstman, T. Epidemiology and disease burden of pertussis in the United States among individuals aged 0–64 over a 10-year period (2006–2015). Curr. Med. Res. Opin. 2020, 36, 127–137. [Google Scholar] [CrossRef]
- Hozbor, D.F. Outer membrane vesicles: An attractive candidate for pertussis vaccines. Expert Rev. Vaccines 2017, 16, 193–196. [Google Scholar] [CrossRef]
- Hozbor, D. New Pertussis Vaccines: A Need and a Challenge. Adv. Exp. Med. Biol. 2019, 1183, 115–126. [Google Scholar] [CrossRef]
- Hozbor, D.; Rodriguez, M.E.; Fernandez, J.; Lagares, A.; Guiso, N.; Yantorno, O. Release of outer membrane vesicles from Bordetella pertussis. Curr. Microbiol. 1999, 38, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, M.J.; Kesty, N.C. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005, 19, 2645–2655. [Google Scholar] [CrossRef] [PubMed]
- Ellis, T.N.; Kuehn, M.J. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. MMBR 2010, 74, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Van der Pol, L.; Stork, M.; van der Ley, P. Outer membrane vesicles as platform vaccine technology. Biotechnol. J. 2015, 10, 1689–1706. [Google Scholar] [CrossRef]
- Tan, K.; Li, R.; Huang, X.; Liu, Q. Outer Membrane Vesicles: Current Status and Future Direction of These Novel Vaccine Adjuvants. Front. Microbiol. 2018, 9, 783. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef]
- Yu, Y.J.; Wang, X.H.; Fan, G.C. Versatile effects of bacterium-released membrane vesicles on mammalian cells and infectious/inflammatory diseases. Acta Pharm. Sin. 2018, 39, 514–533. [Google Scholar] [CrossRef]
- Roberts, R.; Moreno, G.; Bottero, D.; Gaillard, M.E.; Fingermann, M.; Graieb, A.; Rumbo, M.; Hozbor, D. Outer membrane vesicles as acellular vaccine against pertussis. Vaccine 2008, 26, 4639–4646. [Google Scholar] [CrossRef]
- Raeven, R.H.M.; Brummelman, J.; Pennings, J.L.; van der Maas, L.; Tilstra, W.; Helm, K.; van Riet, E.; Jiskoot, W.; van Els, C.A.; Han, W.G.; et al. Bordetella pertussis outer membrane vesicle vaccine confers equal efficacy in mice with milder inflammatory responses compared to a whole-cell vaccine. Sci. Rep. 2016, 6, 38240. [Google Scholar] [CrossRef]
- Raeven, R.H.M.; Brummelman, J.; Pennings, J.; van der Maas, L.; Helm, K.; Tilstra, W.; van der Ark, A.; Sloots, A.; van der Ley, P.; van Eden, W.; et al. Molecular and cellular signatures underlying superior immunity against Bordetella pertussis upon pulmonary vaccination. Mucosal. Immunol. 2018, 11, 979–993. [Google Scholar] [CrossRef]
- Raeven, R.H.M.; Rockx-Brouwer, D.; Kanojia, G.; van der Maas, L.; Bindels, T.H.E.; Ten Have, R.; van Riet, E.; Metz, B.; Kersten, G.F.A. Intranasal immunization with outer membrane vesicle pertussis vaccine confers broad protection through mucosal IgA and Th17 responses. Sci. Rep. 2020, 10, 7396. [Google Scholar] [CrossRef] [PubMed]
- Bottero, D.; Gaillard, M.E.; Zurita, E.; Moreno, G.; Martinez, D.S.; Bartel, E.; Bravo, S.; Carriquiriborde, F.; Errea, A.; Castuma, C.; et al. Characterization of the immune response induced by pertussis OMVs-based vaccine. Vaccine 2016, 34, 3303–3309. [Google Scholar] [CrossRef]
- Lambert, E.E.; Buisman, A.M.; van Els, C.; Superior, B. pertussis Specific CD4+ T-Cell Immunity Imprinted by Natural Infection. Adv. Exp. Med. Biol. 2019, 1183, 81–98. [Google Scholar] [CrossRef]
- Kapil, P.; Merkel, T.J. Pertussis vaccines and protective immunity. Curr. Opin. Immunol. 2019, 59, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Raeven, R.H.M.; van der Maas, L.; Tilstra, W.; Uittenbogaard, J.P.; Bindels, T.H.; Kuipers, B.; van der Ark, A.; Pennings, J.L.; van Riet, E.; Jiskoot, W.; et al. Immunoproteomic Profiling of Bordetella pertussis Outer Membrane Vesicle Vaccine Reveals Broad and Balanced Humoral Immunogenicity. J. Proteome Res. 2015, 14, 2929–2942. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Stibitz, S. The BvgASR virulence regulon of Bordetella pertussis. Curr. Opin. Microbiol. 2019, 47, 74–81. [Google Scholar] [CrossRef]
- Moon, K.; Bonocora, R.P.; Kim, D.D.; Chen, Q.; Wade, J.T.; Stibitz, S.; Hinton, D.M. The BvgAS Regulon of Bordetella pertussis. mBio 2017, 8. [Google Scholar] [CrossRef]
- Metz, B.; Hoonakker, M.; Uittenbogaard, J.P.; Weyts, M.; Mommen, G.P.; Meiring, H.D.; Tilstra, W.; Pennings, J.L.; van der Pol, L.A.; Kuipers, B.; et al. Proteome Analysis Is a Valuable Tool to Monitor Antigen Expression during Upstream Processing of Whole-Cell Pertussis Vaccines. J. Proteome Res. 2017, 16, 528–537. [Google Scholar] [CrossRef]
- Lesne, E.; Coutte, L.; Solans, L.; Slupek, S.; Debrie, A.S.; Dhennin, V.; Froguel, P.; Hot, D.; Locht, C.; Antoine, R.; et al. Distinct virulence ranges for infection of mice by Bordetella pertussis revealed by engineering of the sensor-kinase BvgS. PLoS ONE 2018, 13, e0204861. [Google Scholar] [CrossRef]
- Raeven, R.H.M.; van der Maas, L.; Pennings, J.L.A.; Fuursted, K.; Jorgensen, C.S.; van Riet, E.; Metz, B.; Kersten, G.F.A.; Dalby, T. Antibody Specificity Following a Recent Bordetella pertussis Infection in Adolescence Is Correlated With the Pertussis Vaccine Received in Childhood. Front. Immunol. 2019, 10, 1364. [Google Scholar] [CrossRef]
- Hovingh, E.S.; Kuipers, B.; Bonacic Marinovic, A.A.; Jan Hamstra, H.; Hijdra, D.; Mughini Gras, L.; van Twillert, I.; Jongerius, I.; van Els, C.; Pinelli, E. Detection of opsonizing antibodies directed against a recently circulating Bordetella pertussis strain in paired plasma samples from symptomatic and recovered pertussis patients. Sci. Rep. 2018, 8, 12039. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.A.; Mobberley, P.S.; Fernandez, R.C.; Mink, C.M. Characterization of human bactericidal antibodies to Bordetella pertussis. Infect. Immun. 1999, 67, 1424–1431. [Google Scholar] [CrossRef] [PubMed]
- Oliver, D.C.; Fernandez, R.C. Antibodies to BrkA augment killing of Bordetella pertussis. Vaccine 2001, 20, 235–241. [Google Scholar] [CrossRef]
- Gouw, D.; Jonge, M.I.; Hermans, P.W.; Wessels, H.J.; Zomer, A.; Berends, A.; Pratt, C.; Berbers, G.A.; Mooi, F.R.; Diavatopoulos, D.A. Proteomics-Identified Bvg-Activated Autotransporters Protect against Bordetella pertussis in a Mouse Model. PLoS ONE 2014, 9, e105011. [Google Scholar] [CrossRef] [PubMed]
- Marr, N.; Oliver, D.C.; Laurent, V.; Poolman, J.; Denoel, P.; Fernandez, R.C. Protective activity of the Bordetella pertussis BrkA autotransporter in the murine lung colonization model. Vaccine 2008, 26, 4306–4311. [Google Scholar] [CrossRef]
- Cainelli Gebara, V.C.; Risoleo, L.; Lopes, A.P.; Ferreira, V.R.; Quintilio, W.; Lepine, F.; Silva, W.D.; Raw, I. Adjuvant and immunogenic activities of the 73kDa N-terminal alpha-domain of BrkA autotransporter and Cpn60/60kDa chaperonin of Bordetella pertussis. Vaccine 2007, 25, 621–629. [Google Scholar] [CrossRef]
- Gasperini, G.; Biagini, M.; Arato, V.; Gianfaldoni, C.; Vadi, A.; Norais, N.; Bensi, G.; Delany, I.; Pizza, M.; Arico, B.; et al. Outer Membrane Vesicles (OMV)-based and Proteomics-driven Antigen Selection Identifies Novel Factors Contributing to Bordetella pertussis Adhesion to Epithelial Cells. Mol. Cell. Proteom. MCP 2018, 17, 205–215. [Google Scholar] [CrossRef]
- Suzuki, K.; Shinzawa, N.; Ishigaki, K.; Nakamura, K.; Abe, H.; Fukui-Miyazaki, A.; Ikuta, K.; Horiguchi, Y. Protective effects of in vivo-expressed autotransporters against Bordetella pertussis infection. Microbiol. Immunol. 2017, 61, 371–379. [Google Scholar] [CrossRef]
- Streefland, M.; van de Waterbeemd, B.; Happe, H.; van der Pol, L.A.; Beuvery, E.C.; Tramper, J.; Martens, D.E. PAT for vaccines: The first stage of PAT implementation for development of a well-defined whole-cell vaccine against whooping cough disease. Vaccine 2007, 25, 2994–3000. [Google Scholar] [CrossRef]
- Fuchslocher, B.; Millar, L.L.; Cotter, P.A. Comparison of bipA alleles within and across Bordetella species. Infect. Immun. 2003, 71, 3043–3052. [Google Scholar] [CrossRef]
- Fedele, G.; Bianco, M.; Ausiello, C.M. The virulence factors of Bordetella pertussis: Talented modulators of host immune response. Archivum Immunologiae et Therapiae Experimentalis 2013, 61, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Luu, L.D.W.; Octavia, S.; Zhong, L.; Raftery, M.; Sintchenko, V.; Lan, R. Characterisation of the Bordetella pertussis secretome under different media. J. Proteom. 2017, 158, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Thalen, M.; van der Ark, A.; van den Ijssel, J.; van Straaten, I.; Jansen, D.; Beuvery, C.; Martens, D.; Tramper, J. Improving the cellular pertussis vaccine: Increased potency and consistency. Vaccine 2008, 26, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Vidakovics, M.L.; Paba, J.; Lamberti, Y.; Ricart, C.A.; de Sousa, M.V.; Rodriguez, M.E. Profiling the Bordetella pertussis proteome during iron starvation. J. Proteome Res. 2007, 6, 2518–2528. [Google Scholar] [CrossRef] [PubMed]
- Streefland, M.; Van Herpen, P.F.; Van de Waterbeemd, B.; Van der Pol, L.A.; Beuvery, E.C.; Tramper, J.; Martens, D.E.; Toft, M. A practical approach for exploration and modeling of the design space of a bacterial vaccine cultivation process. Biotechnol. Bioeng. 2009, 104, 492–504. [Google Scholar] [CrossRef]
- Van de Waterbeemd, B.; Streefland, M.; Pennings, J.; van der Pol, L.; Beuvery, C.; Tramper, J.; Martens, D. Gene-expression-based quality scores indicate optimal harvest point in Bordetella pertussis cultivation for vaccine production. Biotechnol. Bioeng. 2009, 103, 900–908. [Google Scholar] [CrossRef]
- Zollinger, W.D.; Donets, M.A.; Schmiel, D.H.; Pinto, V.B.; Labrie, J.E., III; Moran, E.E.; Brandt, B.L.; Ionin, B.; Marques, R.; Wu, M.; et al. Design and evaluation in mice of a broadly protective meningococcal group B native outer membrane vesicle vaccine. Vaccine 2010, 28, 5057–5067. [Google Scholar] [CrossRef]
- King, A.J.; van der Lee, S.; Mohangoo, A.; van Gent, M.; van der Ark, A.; van de Waterbeemd, B. Genome-wide gene expression analysis of Bordetella pertussis isolates associated with a resurgence in pertussis: Elucidation of factors involved in the increased fitness of epidemic strains. PLoS ONE 2013, 8, e66150. [Google Scholar] [CrossRef]
- Stockbauer, K.E.; Fuchslocher, B.; Miller, J.F.; Cotter, P.A. Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol. Microbiol. 2001, 39, 65–78. [Google Scholar] [CrossRef]
- Brummelman, J.; Wilk, M.M.; Han, W.G.; van Els, C.A.; Mills, K.H. Roads to the development of improved pertussis vaccines paved by immunology. Pathog. Dis. 2015, 73, ftv067. [Google Scholar] [CrossRef]
- Ross, P.J.; Sutton, C.E.; Higgins, S.; Allen, A.C.; Walsh, K.; Misiak, A.; Lavelle, E.C.; McLoughlin, R.M.; Mills, K.H. Relative contribution of Th1 and Th17 cells in adaptive immunity to Bordetella pertussis: Towards the rational design of an improved acellular pertussis vaccine. PLoS Pathog. 2013, 9, e1003264. [Google Scholar] [CrossRef] [PubMed]
- Warfel, J.M.; Merkel, T.J. Bordetella pertussis infection induces a mucosal IL-17 response and long-lived Th17 and Th1 immune memory cells in nonhuman primates. Mucosal. Immunol. 2013, 6, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Raeven, R.H.M.; Brummelman, J.; van der Maas, L.; Tilstra, W.; Pennings, J.L.; Han, W.G.; van Els, C.A.; van Riet, E.; Kersten, G.F.; Metz, B. Immunological Signatures after Bordetella pertussis Infection Demonstrate Importance of Pulmonary Innate Immune Cells. PLoS ONE 2016, 11, e0164027. [Google Scholar] [CrossRef] [PubMed]
- Brummelman, J.; Helm, K.; Hamstra, H.J.; van der Ley, P.; Boog, C.J.; Han, W.G.; van Els, C.A. Modulation of the CD4(+) T cell response after acellular pertussis vaccination in the presence of TLR4 ligation. Vaccine 2015, 33, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Brookes, C.; Freire-Martin, I.; Cavell, B.; Alexander, F.; Taylor, S.; Persaud, R.; Fry, N.; Preston, A.; Diavatopoulos, D.; Gorringe, A. Bordetella pertussis isolates vary in their interactions with human complement components. Emerg. Microbes Infect. 2018, 7, 81. [Google Scholar] [CrossRef]
- Tuomanen, E.I.; Zapiain, L.A.; Galvan, P.; Hewlett, E.L. Characterization of antibody inhibiting adherence of Bordetella pertussis to human respiratory epithelial cells. J. Clin. Microbiol. 1984, 20, 167–170. [Google Scholar] [CrossRef]
- Burns, D.L.; Gould-Kostka, J.L.; Kessel, M.; Arciniega, J.L. Purification and immunological characterization of a GroEL-like protein from Bordetella pertussis. Infect. Immun. 1991, 59, 1417–1422. [Google Scholar] [CrossRef]
- Zurita, M.E.; Wilk, M.M.; Carriquiriborde, F.; Bartel, E.; Moreno, G.; Misiak, A.; Mills, K.H.G.; Hozbor, D. A Pertussis Outer Membrane Vesicle-Based Vaccine Induces Lung-Resident Memory CD4 T Cells and Protection Against Bordetella pertussis, Including Pertactin Deficient Strains. Front. Cell. Infect. Microbiol. 2019, 9, 125. [Google Scholar] [CrossRef]
- Jongerius, I.; Schuijt, T.J.; Mooi, F.R.; Pinelli, E. Complement evasion by Bordetella pertussis: Implications for improving current vaccines. J. Mol. Med. 2015, 93, 395–402. [Google Scholar] [CrossRef]
- Marr, N.; Shah, N.R.; Lee, R.; Kim, E.J.; Fernandez, R.C. Bordetella pertussis autotransporter Vag8 binds human C1 esterase inhibitor and confers serum resistance. PLoS ONE 2011, 6, e20585. [Google Scholar] [CrossRef]
- Hovingh, E.S.; van den Broek, B.; Kuipers, B.; Pinelli, E.; Rooijakkers, S.H.M.; Jongerius, I. Acquisition of C1 inhibitor by Bordetella pertussis virulence associated gene 8 results in C2 and C4 consumption away from the bacterial surface. PLoS Pathog. 2017, 13, e1006531. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.G.; Weiss, A.A. BrkA protein of Bordetella pertussis inhibits the classical pathway of complement after C1 deposition. Infect. Immun. 2001, 69, 3067–3072. [Google Scholar] [CrossRef]
- Finn, T.M.; Amsbaugh, D.F. Vag8, a Bordetella pertussis bvg-regulated protein. Infect. Immun. 1998, 66, 3985–3989. [Google Scholar] [CrossRef] [PubMed]
- Kanojia, G.; Raeven, R.H.M.; van der Maas, L.; Bindels, T.H.E.; van Riet, E.; Metz, B.; Soema, P.C.; Ten Have, R.; Frijlink, H.W.; Amorij, J.P.; et al. Development of a thermostable spray dried outer membrane vesicle pertussis vaccine for pulmonary immunization. J. Control. Release 2018, 286, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Bio/Technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Thalen, M.; van den, I.J.; Jiskoot, W.; Zomer, B.; Roholl, P.; de Gooijer, C.; Beuvery, C.; Tramper, J. Rational medium design for Bordetella pertussis: Basic metabolism. J. Biotechnol. 1999, 75, 147–159. [Google Scholar] [CrossRef]
- Stibitz, S. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzym. 1994, 235, 458–465. [Google Scholar] [CrossRef]
- Stibitz, S.; Black, W.; Falkow, S. The construction of a cloning vector designed for gene replacement in Bordetella pertussis. Gene 1986, 50, 133–140. [Google Scholar] [CrossRef]
- Horton, R.M.; Hunt, H.D.; Ho, S.N.; Pullen, J.K.; Pease, L.R. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 1989, 77, 61–68. [Google Scholar] [CrossRef]
- Verwey, W.F.; Thiele, E.H.; Sage, D.N.; Schuchardt, L.F. A simplified liquid culture medium for the growth of Hemophilus pertussis. J. Bacteriol. 1949, 58, 127–134. [Google Scholar] [CrossRef]
- Meiring, H.D.; Van Der Heeft, E.; Ten Hove, G.J.; De Jong, A.P.J.M. Nanoscale LC-MS(n): Technical design and applications to peptide and protein analysis. J. Sep. Sci. 2002, 25, 557–568. [Google Scholar] [CrossRef]
- Silva, J.C.; Gorenstein, M.V.; Li, G.Z.; Vissers, J.P.; Geromanos, S.J. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteom. MCP 2006, 5, 144–156. [Google Scholar] [CrossRef] [PubMed]
| Accession Number | Name | ID |
|---|---|---|
| P12255 | Filamentous hemagglutinin | fhaB |
| P14283 | Pertactin autotransporter | Prn |
| P33410 | Outer membrane usher protein FimC | fimC |
| P81549 | Probable TonB-dependent receptor BfrD | bfrD |
| Q45340 | BrkA autotransporter | BrkA |
| Q79GG1 | Outer membrane porin protein OmpQ | ompQ |
| Q79GN7 | Autotransporter | Vag8 |
| Q79GQ6 | Putative type III secretion protein | bscJ |
| Q79GR8 | Putative type III secretion protein | bscC |
| Q79GX8 | Tracheal colonization factor | TcfA |
| Q7VSD3 | Type III secretion system cytotoxic effector | BteA |
| Q7VSG8 | Putative heme receptor | hemC |
| Q7VVD6 | Autotransporter | bapC |
| Q7VVJ2 | Adhesin | fhaS |
| Q7VZ27 | Putative outer membrane ligand binding protein | bipA |
| Q7VZP0 | Probable TonB-dependent receptor for iron transport | bfrE |
| Primer Name | Primer Sequence (5′→3′) | PCR |
|---|---|---|
| BrkA U-F | gcctcttcgccaaagaagg | Fusion Ia, fusion II |
| BrkA U-R2 | GGAGCTCGCTCAGAAGCTGTgagaagttgaacaaaccgac | Fusion Ia |
| BrkA D-F2 | GTCGGTTTGTTCAACTTCTCacagcttctgagcgagctcc | Fusion Ib |
| BrkA D-R | agaaggcgtggttcctgg | Fusion Ib, II |
| BrkA check-F2 | ttcaggaaagctcttgttgg | Deletion check |
| BrkA check-R2 | cggcatggacttctaagtcc | Deletion check |
| Vag8 U-F | ctgcgtcaaccgcttagc | Fusion Ia, fusion II |
| Vag8U-R2 | GGTCACCAGCTGTAGCGATACctcaacacctcttggctag | Fusion Ia |
| Vag8D-F2 | CTAGCCAAGAGGTGTTGAGgtatcgctacagctggtgacc | Fusion Ib |
| Vag8-R | aagccgcgtgcgactacgtc | Fusion Ib, II |
| Vag8 check-F2 | ggcgttttctgtcaatcgtc | Deletion check |
| Vag8 check-R2 | gccgaactgcaacgctactg | Deletion check |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raeven, R.H.M.; van Vlies, N.; Salverda, M.L.M.; van der Maas, L.; Uittenbogaard, J.P.; Bindels, T.H.E.; Rigters, J.; Verhagen, L.M.; Kruijer, S.; van Riet, E.; et al. The Role of Virulence Proteins in Protection Conferred by Bordetella pertussis Outer Membrane Vesicle Vaccines. Vaccines 2020, 8, 429. https://doi.org/10.3390/vaccines8030429
Raeven RHM, van Vlies N, Salverda MLM, van der Maas L, Uittenbogaard JP, Bindels THE, Rigters J, Verhagen LM, Kruijer S, van Riet E, et al. The Role of Virulence Proteins in Protection Conferred by Bordetella pertussis Outer Membrane Vesicle Vaccines. Vaccines. 2020; 8(3):429. https://doi.org/10.3390/vaccines8030429
Chicago/Turabian StyleRaeven, René H. M., Naomi van Vlies, Merijn L. M. Salverda, Larissa van der Maas, Joost P. Uittenbogaard, Tim H. E. Bindels, Jolanda Rigters, Lisa M. Verhagen, Sabine Kruijer, Elly van Riet, and et al. 2020. "The Role of Virulence Proteins in Protection Conferred by Bordetella pertussis Outer Membrane Vesicle Vaccines" Vaccines 8, no. 3: 429. https://doi.org/10.3390/vaccines8030429
APA StyleRaeven, R. H. M., van Vlies, N., Salverda, M. L. M., van der Maas, L., Uittenbogaard, J. P., Bindels, T. H. E., Rigters, J., Verhagen, L. M., Kruijer, S., van Riet, E., Metz, B., & van der Ark, A. A. J. (2020). The Role of Virulence Proteins in Protection Conferred by Bordetella pertussis Outer Membrane Vesicle Vaccines. Vaccines, 8(3), 429. https://doi.org/10.3390/vaccines8030429





