Transcriptomic and Metabolic Responses to a Live-Attenuated Francisella tularensis Vaccine
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Study
2.2. Transcriptomics
2.2.1. RNA-Seq Experiments
2.2.2. RNA-Seq Data Processing and Analysis
2.2.3. RNA-Seq Data Availability
2.3. Metabolomics
2.3.1. Amino Acid Sample and Data Processing
2.3.2. Organic Acid Sample Experiment
2.3.3. High-Resolution LC–MS Metabolomics Experiment
2.3.4. Metabolomics Analysis
Filtering and Normalization of Metabolomics Data
Missing Value Imputation
Identification of Differentially Abundant Metabolites
Identification of Metabolomics Responses that Best Predict Adaptive Immune Response
Integration of Metabolomics and Transcriptomics Results
Data Availability
2.4. Ethical Statement
3. Results
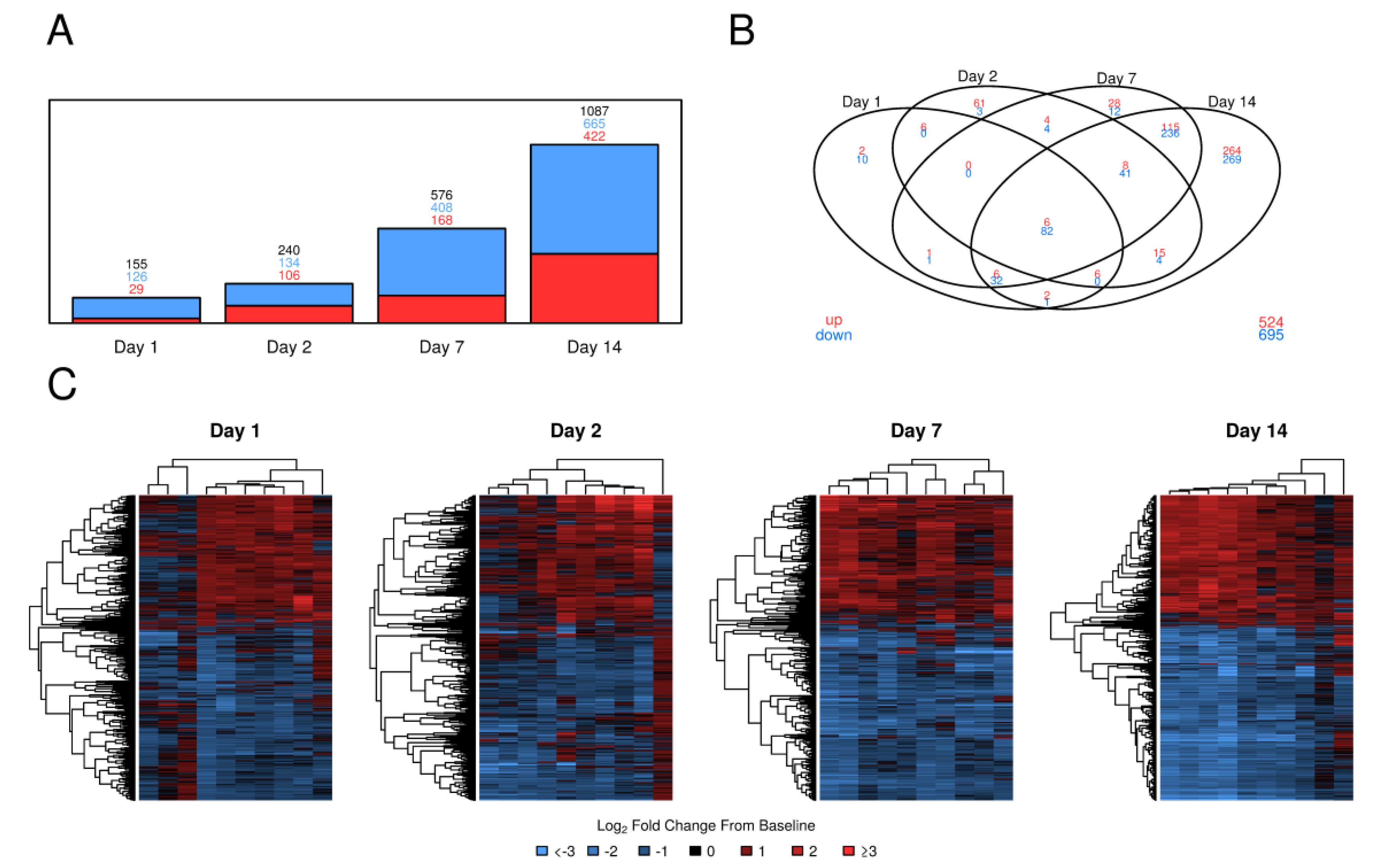
3.1. PBMC Gene Expression Following Tularemia Vaccination Showed Increases over Time in Magnitude and Complexity with the Majority of Genes Being Down-Regulated
3.2. Tularemia Vaccine Activated PBMC Gene Expression Patterns Related to Interferon Signaling But Repressed Other Cytokine and Innate Immune Signaling Pathways Involved in Inflammation
3.3. Tularemia Vaccine Induced Gene Expression Signals Related to the Adaptive Immune Response at Day 7 and 14
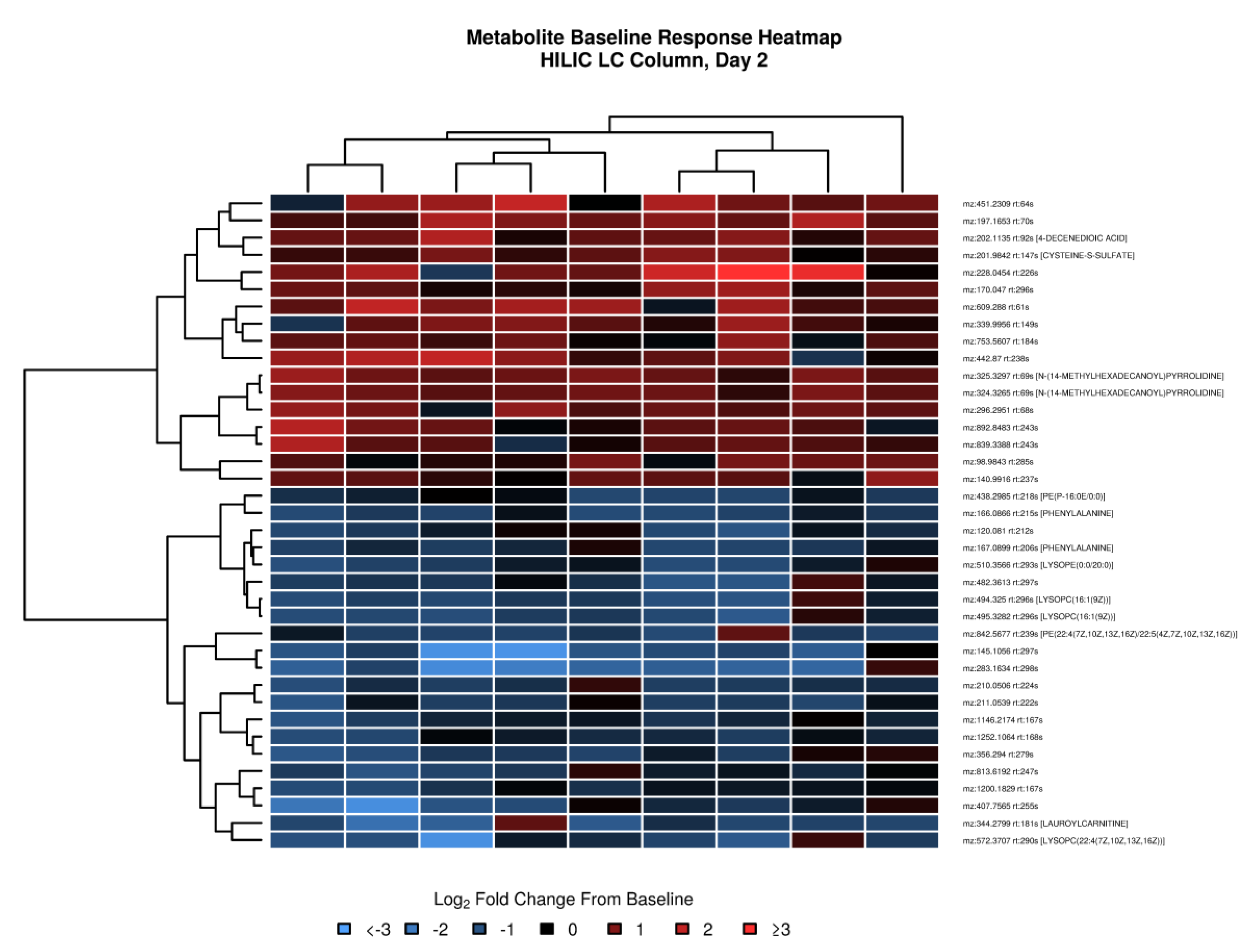
3.4. Changes in Plasma Amino and Organic Acids within 2 Days after Tularemia Vaccination
3.5. Untargeted Metabolomics Revealed Changes in Energy Metabolism and Nucleotide Metabolism
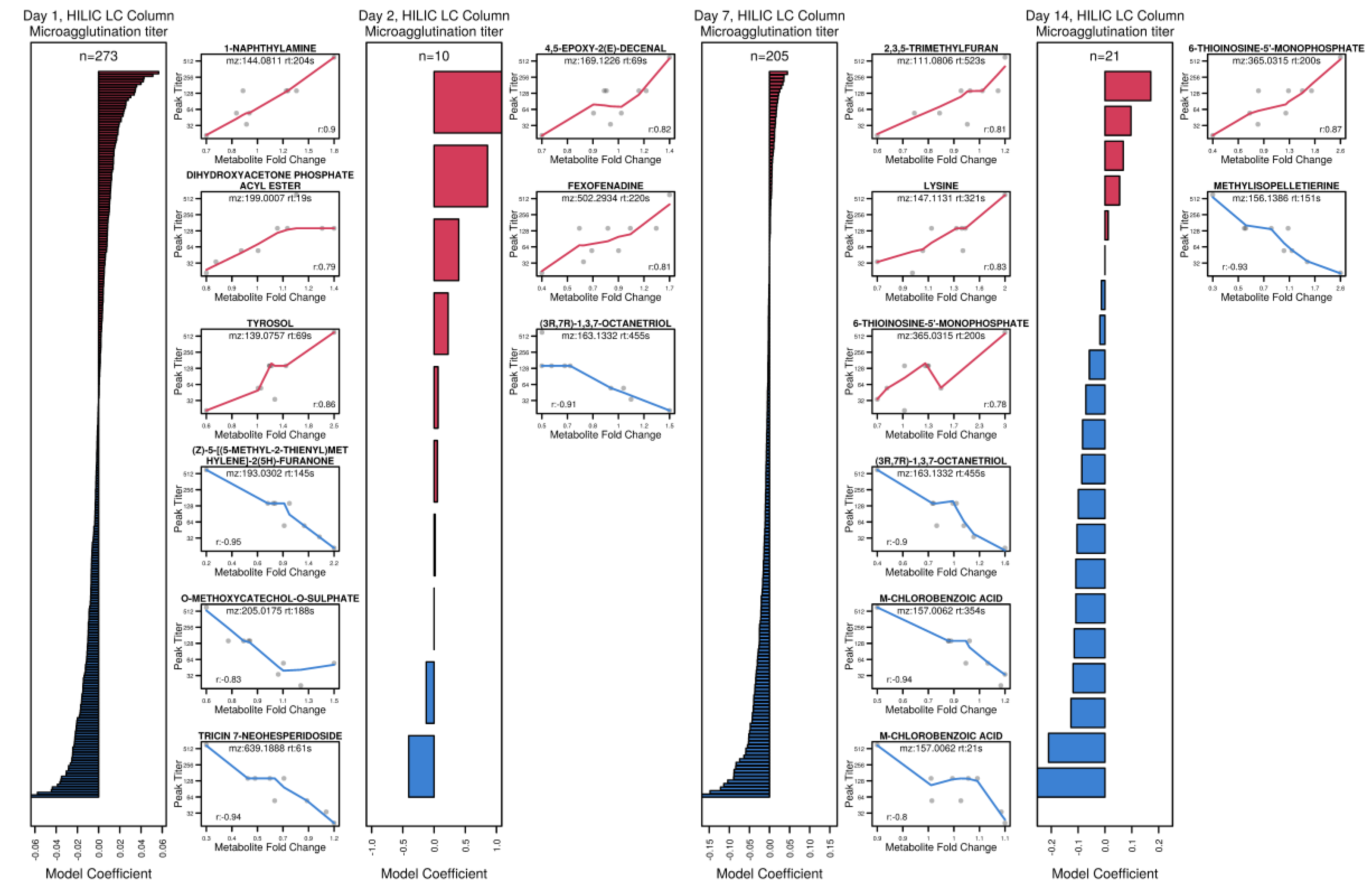
3.6. Exploring Metabolite Predictors of Antibody and T Cell Responses
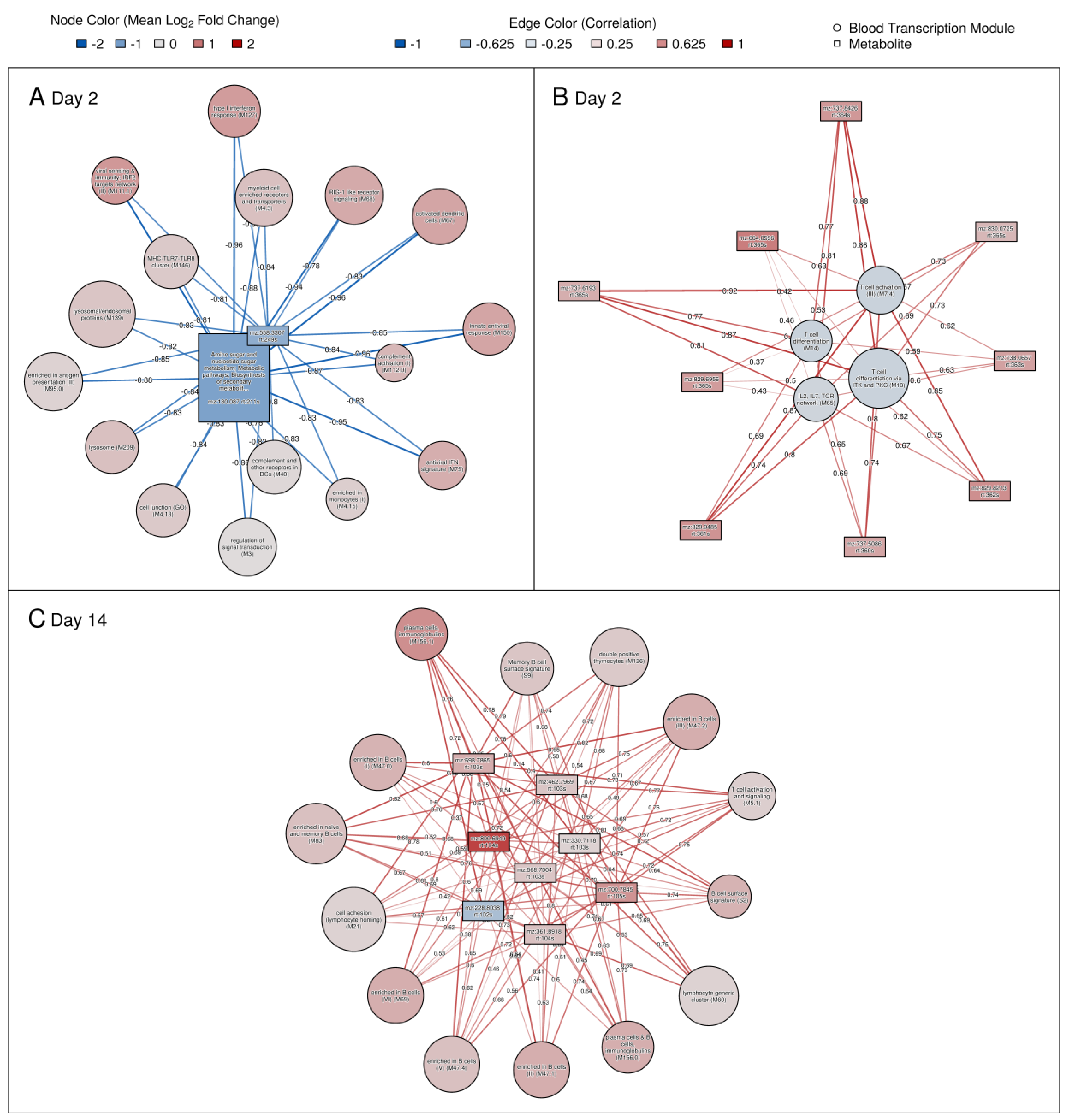
3.7. Significant Correlations between Genes and Metabolites and Association with Immune Responses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mulligan, M.J.; Stapleton, J.T.; Keitel, W.A.; Frey, S.E.; Chen, W.H.; Rouphael, N.; Edupuganti, S.; Beck, A.; Winokur, P.L.; El Sahly, H.M.; et al. Tularemia vaccine: Safety, reactogenicity, “Take” skin reactions, and antibody responses following vaccination with a new lot of the Francisella tularensis live vaccine strain—A phase 2 randomized clinical Trial. Vaccine 2017, 35, 4730–4737. [Google Scholar] [CrossRef] [PubMed]
- Bosio, C. The Subversion of the Immune System by Francisella Tularensis. Front. Microbiol. 2011, 2, 9. [Google Scholar] [CrossRef] [PubMed]
- Querec, T.; Akondy, R.; Lee, E.K.; Cao, W.; Nakaya, H.I.; Teuwen, D.; Pirani, A.; Gernert, K.; Deng, J.; Marzolf, B.; et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2008, 10, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, H.I.; Wrammert, J.; Lee, E.K.; Racioppi, L.; Marie-Kunze, S.; Haining, W.N.; Means, A.R.; Kasturi, S.P.; Khan, N.; Li, G.-M.; et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011, 12, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Nakaya, H.I.; Kazmin, D.A.; Oh, J.Z.; Pulendran, B. Systems biological approaches to measure and understand vaccine immunity in humans. Semin. Immunol. 2013, 25, 209–218. [Google Scholar] [CrossRef][Green Version]
- Davis, M.M.; Tato, C.M.; Furman, D. Systems immunology: Just getting started. Nat. Immunol. 2017, 18, 725–732. [Google Scholar] [CrossRef]
- Howard, L.M.; Hoek, K.L.; Goll, J.B.; Samir, P.; Galassie, A.; Allos, T.M.; Niu, X.; Gordy, L.E.; Creech, C.B.; Prasad, N.; et al. Cell-Based Systems Biology Analysis of Human AS03-Adjuvanted H5N1 Avian Influenza Vaccine Responses: A Phase I Randomized Controlled Trial. PLoS ONE 2017, 12, e0167488. [Google Scholar] [CrossRef]
- Howard, L.M.; Goll, J.B.; Jensen, T.; Hoek, K.L.; Prasad, N.; Gelber, C.E.; Levy, S.E.; Joyce, S.; Link, A.J.; Creech, C.B.; et al. AS03-Adjuvanted H5N1 Avian Influenza Vaccine Modulates Early Innate Immune Signatures in Human Peripheral Blood Mononuclear Cells. J. Infect. Dis. 2018, 219, 1786–1798. [Google Scholar] [CrossRef]
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. [Google Scholar] [CrossRef]
- Li, S.; Todor, A.; Luo, R. Blood transcriptomics and metabolomics for personalized medicine. Comput. Struct. Biotechnol. J. 2015, 14, 1–7. [Google Scholar] [CrossRef]
- Li, S.; Sullivan, N.L.; Rouphael, N.; Yu, T.; Banton, S.; Maddur, M.S.; McCausland, M.; Chiu, C.; Canniff, J.; Dubey, S.; et al. Metabolic Phenotypes of Response to Vaccination in Humans. Cell 2017, 169, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Medaglini, D.; Santoro, F.; Siegrist, C.-A. Correlates of vaccine-induced protective immunity against Ebola virus disease. Semin. Immunol. 2018, 39, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Tisoncik-Go, J.; Gasper, D.J.; Kyle, J.E.; Eisfeld, A.J.; Selinger, C.; Hatta, M.; Morrison, J.; Korth, M.J.; Zink, E.M.; Kim, Y.-M.; et al. Integrated Omics Analysis of Pathogenic Host Responses during Pandemic H1N1 Influenza Virus Infection: The Crucial Role of Lipid Metabolism. Cell Host Microbe 2016, 19, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479, 609–618. [Google Scholar] [CrossRef]
- Puleston, D.J.; Villa, M.; Pearce, E.L. Ancillary Activity: Beyond Core Metabolism in Immune Cells. Cell Metab. 2017, 26, 131–141. [Google Scholar] [CrossRef]
- Spite, M.; Claria, J.; Serhan, C.N. Resolvins, specialized proresolving lipid mediators, and their potential roles in metabolic diseases. Cell Metab. 2013, 19, 21–36. [Google Scholar] [CrossRef]
- Di Gennaro, A.; Haeggström, J.Z. The Leukotrienes: Immune-Modulating Lipid Mediators of Disease. Adv. Immunol. 2012, 116, 51–92. [Google Scholar] [CrossRef]
- Healy, L.M.; Antel, J.P. Sphingosine-1-Phosphate Receptors in the Central Nervous and Immune Systems. Curr. Drug Targets 2015, 17, 1841–1850. [Google Scholar] [CrossRef]
- Frey, B.; Gaipl, U.S. The immune functions of phosphatidylserine in membranes of dying cells and microvesicles. Semin. Immunopathol. 2010, 33, 497–516. [Google Scholar] [CrossRef]
- Man, K.; Kutyavin, V.I.; Chawla, A. Tissue Immunometabolism: Development, Physiology, and Pathobiology. Cell Metab. 2016, 25, 11–26. [Google Scholar] [CrossRef]
- Natrajan, M.S.; Rouphael, N.; Lai, L.; Kazmin, D.; Jensen, T.L.; Weiss, D.S.; Ibegbu, C.; Sztein, M.B.; Hooper, W.F.; Hill, H.; et al. Systems Vaccinology for a Live Attenuated Tularemia Vaccine Reveals Unique Transcriptional Signatures That Predict Humoral and Cellular Immune Responses. Vaccines 2019, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-H.; Duong, D.M.; Goll, J.B.; Wood, D.C.; Jensen, T.L.; Yin, L.; Gelber, C.E.; Seyfried, N.T.; Anderson, E.; Natrajan, M.S.; et al. Proteomic Analysis of Human Immune Responses to Live-Attenuated Tularemia Vaccine. Vaccines. accepted.
- Maner-Smith, K.M.; Goll, J.B.; Khadka, M.; Jensen, T.L.; Colucci, J.K.; Gelber, C.E.; Albert, C.J.; Bosinger, S.; Franke, J.I.; Natrajan, M.; et al. Alterations in the Human Plasma Lipidome in Response to Tularemia Vaccination. Vaccines. accepted.
- Yates, A.; Akanni, W.; Amode, M.R.; Barrell, D.; Billis, K.; Carvalho-Silva, D.; Cummins, C.; Clapham, P.; Fitzgerald, S.; Gil, L.; et al. Ensembl 2016. Nucleic Acids Res. 2015, 44, D710–D716. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Li, S.; Rouphael, N.; Duraisingham, S.; Romero-Steiner, S.; Presnell, S.; Davis, C.; Schmidt, D.S.; Johnson, S.E.; Milton, A.; Rajam, G.; et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 2013, 15, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Boil. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Uppal, K.; Walker, D.I.; Jones, D.P. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal. Chem. 2017, 89, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Troyanskaya, O.; Cantor, M.; Sherlock, G.; Brown, P.; Hastie, T.; Tibshirani, R.; Botstein, D.; Altman, R.B. Missing value estimation methods for DNA microarrays. Bioinformatics 2001, 17, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.H.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Gardinassi, L.; Arévalo-Herrera, M.; Herrera, S.; Cordy, R.J.; Tran, V.; Smith, M.R.; Johnson, M.S.; Chacko, B.; Liu, K.H.; Darley-Usmar, V.; et al. Integrative metabolomics and transcriptomics signatures of clinical tolerance to Plasmodium vivax reveal activation of innate cell immunity and T cell signaling. Redox Boil. 2018, 17, 158–170. [Google Scholar] [CrossRef]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef]
- Szanto, A.; Bálint, B.L.; Nagy, Z.S.; Barta, E.; Dezso, B.; Pap, A.; Szeles, L.; Poliska, S.; Oros, M.; Evans, R.M.; et al. STAT6 Transcription Factor Is a Facilitator of the Nuclear Receptor PPARγ-Regulated Gene Expression in Macrophages and Dendritic Cells. Immunity 2010, 33, 699–712. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.H.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Boil. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Krocova, Z.; Macela, A.; Kubelkova, K. Innate Immune Recognition: Implications for the Interaction of Francisella tularensis with the Host Immune System. Front. Microbiol. 2017, 7, 446. [Google Scholar] [CrossRef]
- Gillette, D.D.; Curry, H.M.; Cremer, T.; Ravneberg, D.; Fatehchand, K.; Shah, P.A.; Wewers, M.D.; Schlesinger, L.S.; Butchar, J.P.; Tridandapani, S.; et al. Virulent Type A Francisella tularensis actively suppresses cytokine responses in human monocytes. Front. Microbiol. 2014, 4, 45. [Google Scholar] [CrossRef]
- Telepnev, M.; Golovliov, I.; Sjöstedt, A. Francisella tularensis LVS initially activates but subsequently down-regulates intracellular signaling and cytokine secretion in mouse monocytic and human peripheral blood mononuclear cells. Microb. Pathog. 2005, 38, 239–247. [Google Scholar] [CrossRef]
- Telepnev, M.; Golovliov, I.; Grundström, T.; Tärnvik, A.; Sjöstedt, A. Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-alpha and IL-1 from murine macrophages. Cell. Microbiol. 2003, 5, 41–51. [Google Scholar] [CrossRef]
- Dai, S.; Rajaram, M.V.S.; Curry, H.M.; Leander, R.; Schlesinger, L.S. Fine Tuning Inflammation at the Front Door: Macrophage Complement Receptor 3-mediates Phagocytosis and Immune Suppression for Francisella tularensis. PLoS Pathog. 2013, 9, e1003114. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Dueñas, A.I.; Aceves, M.; Orduña, A.; Diaz, R.; Crespo, M.S.; Garcia-Rodriguez, C. Francisella tularensis LPS induces the production of cytokines in human monocytes and signals via Toll-like receptor 4 with much lower potency than E. coli LPS. Int. Immunol. 2006, 18, 785–795. [Google Scholar] [CrossRef]
- Gunn, J.S.; Ernst, R.K. The structure and function of Francisella lipopolysaccharide. Ann. N. Y. Acad. Sci. 2007, 1105, 202–218. [Google Scholar] [CrossRef]
- Thakran, S.; Li, H.; Lavine, C.L.; Miller, M.A.; Bina, J.E.; Bina, X.R.; Re, F. Identification of Francisella tularensis lipoproteins that stimulate the toll-like receptor (TLR) 2/TLR1 heterodimer. J. Biol. Chem. 2008, 283, 3751–3760. [Google Scholar] [CrossRef]
- Malik, M.; Bakshi, C.S.; Sahay, B.; Shah, A.; Lotz, S.A.; Sellati, T.J. Toll-Like Receptor 2 Is Required for Control of Pulmonary Infection with Francisella tularensis. Infect. Immun. 2006, 74, 3657–3662. [Google Scholar] [CrossRef]
- Cole, L.E.; Shirey, K.A.; Barry, E.; Santiago, A.; Rallabhandi, P.; Elkins, K.L.; Puche, A.C.; Michalek, S.M.; Vogel, S.N. Toll-Like Receptor 2-Mediated Signaling Requirements for Francisella tularensis Live Vaccine Strain Infection of Murine Macrophages. Infect. Immun. 2007, 75, 4127–4137. [Google Scholar] [CrossRef] [PubMed]
- Katz, J.; Zhang, P.; Martin, M.; Vogel, S.N.; Michalek, S.M. Toll-Like Receptor 2 Is Required for Inflammatory Responses to Francisella tularensis LVS. Infect. Immun. 2006, 74, 2809–2816. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef]
- Rahman, M.M.; McFadden, G. Modulation of NF-κB signalling by microbial pathogens. Nat. Rev. Genet. 2011, 9, 291–306. [Google Scholar] [CrossRef] [PubMed]
- Le Negrate, G. Subversion of innate immune responses by bacterial hindrance of NF-κB pathway. Cell. Microbiol. 2011, 14, 155–167. [Google Scholar] [CrossRef]
- Mahawar, M.; Atianand, M.K.; Dotson, R.J.; Mora, V.; Rabadi, S.M.; Metzger, D.W.; Huntley, J.; Harton, J.; Malik, M.; Bakshi, C.S. Identification of a Novel Francisella tularensis Factor Required for Intramacrophage Survival and Subversion of Innate Immune Response. J. Boil. Chem. 2012, 287, 25216–25229. [Google Scholar] [CrossRef]
- Thomson, S.J.; Askari, A.; Bishop-Bailey, D. Anti-Inflammatory Effects of Epoxyeicosatrienoic Acids. Int. J. Vasc. Med. 2012, 2012, 1–7. [Google Scholar] [CrossRef]
- Torres, A.; Luke, J.D.; Kullas, A.L.; Kapilashrami, K.; Botbol, Y.; Koller, A.; Tonge, P.J.; Chen, E.I.; Macian, F.; Van Der Velden, A.W.M. Asparagine deprivation mediated by Salmonella asparaginase causes suppression of activation-induced T cell metabolic reprogramming. J. Leukoc. Boil. 2015, 99, 387–398. [Google Scholar] [CrossRef]
- Newsholme, P.; Calder, P.C. The proposed role of glutamine in some cells of the immune system and speculative consequences for the whole animal. Nutrition 1997, 13, 728–730. [Google Scholar] [CrossRef]
- Assmann, N.; Finlay, D.K. Metabolic regulation of immune responses: Therapeutic opportunities. J. Clin. Investig. 2016, 126, 2031–2039. [Google Scholar] [CrossRef]
- Ganeshan, K.; Chawla, A. Metabolic regulation of immune responses. Annu. Rev. Immunol. 2014, 32, 609–634. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.; O’Neill, L.A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015, 25, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, C.M.; Holowka, T.; Sun, J.; Blagih, J.; Amiel, E.; DeBerardinis, R.J.; Cross, J.; Jung, E.; Thompson, C.B.; Jones, R.G.; et al. Toll-like receptor–induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 2010, 115, 4742–4749. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, E.V.; Diaz, K.; Griffin, A.J.; Rassmussen, J.A.; Crane, D.D.; Jones, B.D.; Bosio, C. Metabolic reprogramming of host cells by virulent Francisella tularensis for optimal replication and modulation of inflammation. J. Immunol. 2016, 196, 4227–4236. [Google Scholar] [CrossRef] [PubMed]
- Gieger, C.; Geistlinger, L.; Altmaier, E.; De Angelis, M.H.; Kronenberg, F.; Meitinger, T.; Mewes, H.-W.; Wichmann, H.-E.; Weinberger, K.M.; Adamski, J.; et al. Genetics Meets Metabolomics: A Genome-Wide Association Study of Metabolite Profiles in Human Serum. PLoS Genet. 2008, 4, e1000282. [Google Scholar] [CrossRef] [PubMed]
- Bartel, J.; Krumsiek, J.; Schramm, K.; Adamski, J.; Gieger, C.; Herder, C.; Carstensen, M.; Peters, A.; Rathmann, W.; Roden, M.; et al. The Human Blood Metabolome-Transcriptome Interface. PLoS Genet. 2015, 11, e1005274. [Google Scholar] [CrossRef]
- Tsang, J.S.; Schwartzberg, P.L.; Kotliarov, Y.; Biancotto, A.; Xie, Z.; Germain, R.N.; Wang, E.; Olnes, M.J.; Narayanan, M.; Golding, H.; et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell 2014, 157, 499–513. [Google Scholar] [CrossRef]
- HIPC-I Consortium. Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci. Immunol. 2017, 2, eaal4656. [Google Scholar] [CrossRef]
- Fuller, C.; Brittingham, K.C.; Porter, M.W.; Hepburn, M.J.; Petitt, P.L.; Pittman, P.; Bavari, S. Transcriptome analysis of human immune responses following live vaccine strain (LVS) Francisella tularensis vaccination. Mol. Immunol. 2007, 44, 3173–3184. [Google Scholar] [CrossRef][Green Version]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Boil. 2019, 20, 353–367. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goll, J.B.; Li, S.; Edwards, J.L.; Bosinger, S.E.; Jensen, T.L.; Wang, Y.; Hooper, W.F.; Gelber, C.E.; Sanders, K.L.; Anderson, E.J.; et al. Transcriptomic and Metabolic Responses to a Live-Attenuated Francisella tularensis Vaccine. Vaccines 2020, 8, 412. https://doi.org/10.3390/vaccines8030412
Goll JB, Li S, Edwards JL, Bosinger SE, Jensen TL, Wang Y, Hooper WF, Gelber CE, Sanders KL, Anderson EJ, et al. Transcriptomic and Metabolic Responses to a Live-Attenuated Francisella tularensis Vaccine. Vaccines. 2020; 8(3):412. https://doi.org/10.3390/vaccines8030412
Chicago/Turabian StyleGoll, Johannes B., Shuzhao Li, James L. Edwards, Steven E. Bosinger, Travis L. Jensen, Yating Wang, William F. Hooper, Casey E. Gelber, Katherine L. Sanders, Evan J. Anderson, and et al. 2020. "Transcriptomic and Metabolic Responses to a Live-Attenuated Francisella tularensis Vaccine" Vaccines 8, no. 3: 412. https://doi.org/10.3390/vaccines8030412
APA StyleGoll, J. B., Li, S., Edwards, J. L., Bosinger, S. E., Jensen, T. L., Wang, Y., Hooper, W. F., Gelber, C. E., Sanders, K. L., Anderson, E. J., Rouphael, N., Natrajan, M. S., Johnson, R. A., Sanz, P., Hoft, D., & Mulligan, M. J. (2020). Transcriptomic and Metabolic Responses to a Live-Attenuated Francisella tularensis Vaccine. Vaccines, 8(3), 412. https://doi.org/10.3390/vaccines8030412





