The African Swine Fever Virus (ASFV) Topoisomerase II as a Target for Viral Prevention and Control
Abstract
1. Introduction
2. The Role of pP1192R during ASFV Infection
3. Designing a Vaccine by Using a P1192R-Defective Strain
4. Controlling ASF through Inhibition of the Viral Type II Topoisomerase pP1192R
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M. ICTV virus taxonomy profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Costard, S.; Mur, L.; Lubroth, J.; Sanchez-Vizcaino, J.M.; Pfeiffer, D.U. Epidemiology of African swine fever virus. Virus Res. 2013, 173, 191–197. [Google Scholar] [CrossRef]
- Blome, S.; Gabriel, C.; Beer, M. Pathogenesis of African swine fever in domestic pigs and European wild boar. Virus Res. 2013, 173, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Panel, E.A. African swine fever. EFSA J. 2015, 13. [Google Scholar] [CrossRef]
- Sánchez-Vizcaíno, J.M.; Mur, L.; Martínez-López, B. African Swine Fever: An Epidemiological Update. Transbound. Emerg. Dis. 2012, 59, 27–35. [Google Scholar] [CrossRef]
- Sánchez-Vizcaíno, J.M.; Mur, L.; Gomez-Villamandos, J.C.; Carrasco, L. An Update on the Epidemiology and Pathology of African Swine Fever. J. Comp. Pathol. 2015, 152, 9–21. [Google Scholar] [CrossRef]
- Cappai, S.; Loi, F.; Rolesu, S.; Coccollone, A.; Laddomada, A.; Sgarangella, F.; Masala, S.; Bitti, G.; Floris, V.; Desini, P. Evaluation of the cost-effectiveness of ASF detection with or without the use of on-field tests in different scenarios, in Sardinia. J. Vet. Sci. 2020, 21, e14. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular characterization of African swine fever virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef] [PubMed]
- Yoo, D.; Kim, H.; Lee, J.Y.; Yoo, H.S. African swine fever: Etiology, epidemiological status in Korea, and perspective on control. J. Vet. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Landreth, S.; Lu, Y.; Pandey, K.; Zhou, Y. A Replication-Defective Influenza Virus Vaccine Confers Complete Protection against H7N9 Viral Infection in Mice. Vaccines 2020, 8, 207. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, E.; Celma, C.C.P.; Boyce, M.; Viarouge, C.; Sailleau, C.; Dubois, E.; Breard, E.; Thiery, R.; Zientara, S.; Roy, P. Generation of Replication-Defective Virus-Based Vaccines That Confer Full Protection in Sheep against Virulent Bluetongue Virus Challenge. J. Virol. 2011, 85, 10213–10221. [Google Scholar] [CrossRef] [PubMed]
- Celma, C.C.; Stewart, M.; Wernike, K.; Eschbaumer, M.; Gonzalez-Molleda, L.; Breard, E.; Schulz, C.; Hoffmann, B.; Haegeman, A.; De Clercq, K.; et al. Replication-Deficient Particles: New Insights into the Next Generation of Bluetongue Virus Vaccines. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly, S.; Brühl, P.; Mayrhofer, J.; Schmid, K.; Gerencer, M.; Falkner, F.G. The nonreplicating smallpox candidate vaccines defective vaccinia Lister (dVV-L) and modified vaccinia Ankara (MVA) elicit robust long-term protection. Virology 2005, 341, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Giel-Moloney, M.; Esteban, M.; Oakes, B.H.; Vaine, M.; Asbach, B.; Wagner, R.; Mize, G.J.; Spies, A.G.; McElrath, J.; Perreau, M.; et al. Recombinant HIV-1 vaccine candidates based on replication-defective flavivirus vector. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Da Costa, X.J.; Jones, C.A.; Knipe, D.M. Immunization against genital herpes with a vaccine virus that has defects in productive and latent infection. Proc. Natl. Acad. Sci. USA 1999, 96, 6994–6998. [Google Scholar] [CrossRef]
- Da Costa, X.; Kramer, M.F.; Zhu, J.; Brockman, M.A.; Knipe, D.M. Construction, Phenotypic Analysis, and Immunogenicity of a UL5/UL29 Double Deletion Mutant of Herpes Simplex Virus 2. J. Virol. 2000, 74, 7963–7971. [Google Scholar] [CrossRef]
- Dropulic, L.K.; Oestreich, M.C.; Pietz, H.L.; Laing, K.J.; Hunsberger, S.; Lumbard, K.; Garabedian, D.; Turk, S.P.; Chen, A.; Hornung, R.L.; et al. A Randomized, Double-Blinded, Placebo-Controlled, Phase 1 Study of a Replication-Defective Herpes Simplex Virus (HSV) Type 2 Vaccine, HSV529, in Adults With or Without HSV Infection. J. Infect. Dis. 2019, 220, 990–1000. [Google Scholar] [CrossRef]
- Goatley, L.C.; Reis, A.L.; Portugal, R.; Goldswain, H.; Shimmon, G.L.; Hargreaves, Z.; Ho, C.-S.; Montoya, M.; Sánchez-Cordón, P.J.; Taylor, G.; et al. A Pool of Eight Virally Vectored African Swine Fever Antigens Protect Pigs Against Fatal Disease. Vaccines 2020, 8, 234. [Google Scholar] [CrossRef]
- Dixon, L.K.; Chapman, D.A.G.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Wang, J.C. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 2002, 3, 430–440. [Google Scholar] [CrossRef]
- Nitiss, J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 2009, 9, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Schoeffler, A.J.; Berger, J.M. DNA topoisomerases: Harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008, 41, 41–101. [Google Scholar] [CrossRef]
- Crisona, N.J.; Strick, T.R.; Bensimon, D.; Croquette, V.; Cozzarelli, N.R. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000, 14, 2881–2892. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.D.; Bryant, Z.; Crisona, N.J.; Smith, S.B.; Vologodskii, A.; Bustamante, C.; Cozzarelli, N.R. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl. Acad. Sci. USA 2003, 100, 8654–8659. [Google Scholar] [CrossRef] [PubMed]
- McClendon, A.K.; Rodriguez, A.C.; Osheroff, N. Human Topoisomerase IIα Rapidly Relaxes Positively Supercoiled DNA. J. Biol. Chem. 2005, 280, 39337–39345. [Google Scholar] [CrossRef]
- McClendon, A.K.; Dickey, J.S.; Osheroff, N. Ability of Viral Topoisomerase II To Discern the Handedness of Supercoiled DNA: Bimodal Recognition of DNA Geometry by Type II Enzymes. Biochemistry 2006, 45, 11674–11680. [Google Scholar] [CrossRef][Green Version]
- Earnshaw, W.C.; Halligan, B.; Cooke, C.A.; Heck, M.M.S.; Liu, L.F. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol. 1985, 100, 1706–1715. [Google Scholar] [CrossRef]
- Holm, C.; Goto, T.; Wang, J.C.; Botstein, D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell 1985, 41, 553–563. [Google Scholar] [CrossRef]
- Baxter, J.; Diffley, J.F.X. Topoisomerase II Inactivation Prevents the Completion of DNA Replication in Budding Yeast. Mol. Cell 2008, 30, 790–802. [Google Scholar] [CrossRef]
- Baylis, S.A.; Dixon, L.K.; Vydelingum, S.; Smith, G.L. African swine fever virus encodes a gene with extensive homology to type II DNA topoisomerases. J. Mol. Biol. 1992, 228, 1003–1010. [Google Scholar] [CrossRef]
- Garcia-Beato, R.; Freije, J.M.P.; López-Otín, C.; Blasco, R.; Viñuela, E.; Salas, M.L. A gene homologous to topoisomerase II in African swine fever virus. Virology 1992, 188, 938–947. [Google Scholar] [CrossRef]
- Coelho, J.; Martins, C.; Ferreira, F.; Leitão, A. African swine fever virus ORF P1192R codes for a functional type II DNA topoisomerase. Virology 2015, 474, 82–93. [Google Scholar] [CrossRef] [PubMed]
- McAuslan, B.R.; Armentrout, R.W. The biochemistry of icosahedral cytoplasmic deoxyviruses. Curr. Top. Microbiol. Immunol. 1974, 77–105. [Google Scholar] [CrossRef]
- Ortin, J.; Vińuela, E. Requirement of cell nucleus for African swine fever virus replication in Vero cells. J. Virol. 1977, 21, 902–905. [Google Scholar] [CrossRef] [PubMed]
- Tabares, E.; Botija, C.S. Synthesis of DNA in cells infected with African swine fever virus. Arch. Virol. 1979, 61, 49–59. [Google Scholar] [CrossRef]
- García-Beato, R.; Salas, M.L.; Viñuela, E.; Salas, J. Role of the host cell nucleus in the replication of African swine fever virus DNA. Virology 1992, 188, 637–649. [Google Scholar] [CrossRef]
- Rojo, G.; García-Beato, R.; Viñuela, E.; Salas, M.L.; Salas, J. Replication of African swine fever virus DNA in infected cells. Virology 1999, 257, 524–536. [Google Scholar] [CrossRef]
- Ballester, M.; Galindo-Cardiel, I.; Gallardo, C.; Argilaguet, J.M.; Segalés, J.; Rodríguez, J.M.; Rodríguez, F. Intranuclear detection of African swine fever virus DNA in several cell types from formalin-fixed and paraffin-embedded tissues using a new in situ hybridisation protocol. J. Virol. Methods 2010, 168, 38–43. [Google Scholar] [CrossRef]
- Simões, M.; Martins, C.; Ferreira, F. Host DNA damage response facilitates African swine fever virus infection. Vet. Microbiol. 2013, 165, 140–147. [Google Scholar] [CrossRef]
- Goatley, L.C.; Marron, M.B.; Jacobs, S.C.; Hammond, J.M.; Miskin, J.E.; Abrams, C.C.; Smith, G.L.; Dixon, L.K. Nuclear and nucleolar localization of an African swine fever virus protein, I14L, that is similar to the herpes simplex virus-encoded virulence factor ICP34.5. J. Gen. Virol. 1999, 80, 525–535. [Google Scholar] [CrossRef]
- Eulálio, A.; Nunes-Correia, I.; Salas, J.; Salas, M.L.; Simões, S.; Pedroso de Lima, M.C. African swine fever virus p37 structural protein is localized in nuclear foci containing the viral DNA at early post-infection times. Virus Res. 2007, 130, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Silk, R.N.; Bowick, G.C.; Abrams, C.C.; Dixon, L.K. African swine fever virus A238L inhibitor of NF-κB and of calcineurin phosphatase is imported actively into the nucleus and exported by a CRM1-mediated pathway. J. Gen. Virol. 2007, 88, 411–419. [Google Scholar] [CrossRef]
- Nunes-Correia, I.; Rodríguez, J.M.; Eulálio, A.; Carvalho, A.L.; Citovsky, V.; Simões, S.; Faro, C.; Salas, M.L.; Pedroso de Lima, M.C. African swine fever virus p10 protein exhibits nuclear import capacity and accumulates in the nucleus during viral infection. Vet. Microbiol. 2008, 130, 47–59. [Google Scholar] [CrossRef][Green Version]
- Yáñez, R.J.; Rodríguez, J.M.; Nogal, M.L.; Yuste, L.; Enríquez, C.; Rodriguez, J.F.; Viñuela, E. Analysis of the Complete Nucleotide Sequence of African Swine Fever Virus. Virology 1995, 208, 249–278. [Google Scholar] [CrossRef]
- Portugal, R.; Coelho, J.; Höper, D.; Little, N.S.; Smithson, C.; Upton, C.; Martins, C.; Leitão, A.; Keil, G.M. Related strains of African swine fever virus with different virulence: Genome comparison and analysis. J. Gen. Virol. 2015, 96, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Freitas, F.B.; Frouco, G.; Martins, C.; Leitão, A.; Ferreira, F. In vitro inhibition of African swine fever virus-topoisomerase II disrupts viral replication. Antivir. Res. 2016, 134, 34–41. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosio, C.; Kelly, G.; Shirahige, K.; Uhlmann, F. Condensin-Dependent rDNA Decatenation Introduces a Temporal Pattern to Chromosome Segregation. Curr. Biol. 2008, 18, 1084–1089. [Google Scholar] [CrossRef]
- Piskadlo, E.; Tavares, A.; Oliveira, R.A. Metaphase chromosome structure is dynamically maintained by condensin I-directed DNA (de)catenation. Elife 2017, 6. [Google Scholar] [CrossRef]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P.; Gadelle, D. Phylogenomics of DNA topoisomerases: Their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009, 37, 679–692. [Google Scholar] [CrossRef]
- Salas, M.L.; Kuznar, J.; Viñuela, E. Effect of rifamycin derivatives and coumermycin A1 on in vitro RNA synthesis by African swine fever virus. Arch. Virol. 1983, 77, 77–80. [Google Scholar] [CrossRef]
- Chelikani, V.; Ranjan, T.; Zade, A.; Shukla, A.; Kondabagil, K. Genome Segregation and Packaging Machinery in Acanthamoeba polyphaga Mimivirus Is Reminiscent of Bacterial Apparatus. J. Virol. 2014, 88, 6069–6075. [Google Scholar] [CrossRef] [PubMed]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Frouco, G.; Freitas, F.B.; Coelho, J.; Leitão, A.; Martins, C.; Ferreira, F. DNA-Binding Properties of African Swine Fever Virus pA104R, a Histone-Like Protein Involved in Viral Replication and Transcription. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Salas, M.L.; Rey-Campos, J.; Almendral, J.M.; Talavera, A.; Viñuela, E. Transcription and translation maps of african swine fever virus. Virology 1986, 152, 228–240. [Google Scholar] [CrossRef]
- Hingamp, P.M.; Leyland, M.L.; Webb, J.; Twigger, S.; Mayer, R.J.; Dixon, L.K. Characterization of a ubiquitinated protein which is externally located in African swine fever virions. J. Virol. 1995, 69, 1785–1793. [Google Scholar] [CrossRef] [PubMed]
- Brookes, S.M.; Sun, H.; Dixon, L.K.; Parkhouse, R.M.E. Characterization of African swine fever virion proteins j5R and j13L: Immune-localization in virus particles and assembly sites. J. Gen. Virol. 1998, 79, 1179–1188. [Google Scholar] [CrossRef][Green Version]
- Montgomery, R.E. On A Form of Swine Fever Occurring in British East Africa (Kenya Colony). J. Comp. Pathol. Ther. 1921, 34, 159–191. [Google Scholar] [CrossRef]
- Hornby, H.E. Trypanosomes and Trypanosomiases of Cattle. J. Comp. Pathol. Ther. 1921, 34, 243–262. [Google Scholar] [CrossRef]
- DeTray, D.E. Persistence of viremia and immunity in African swine fever. Am. J. Vet. Res. 1957, 18, 811–816. [Google Scholar]
- Malmquist, W.A. Serologic and immunologic studies with African swine fever virus. Am. J. Vet. Res. 1963, 24, 450–459. [Google Scholar] [PubMed]
- Mebus, C.A.; Dardiri, A.H. Western hemisphere isolates of African swine fever virus: Asymptomatic carriers and resistance to challenge inoculation. Am. J. Vet. Res. 1980, 41, 1867–1869. [Google Scholar] [PubMed]
- Arias, M.; de la Torre, A.; Dixon, L.; Gallardo, C.; Jori, F.; Laddomada, A.; Martins, C.; Parkhouse, R.M.; Revilla, Y.; Rodriguez, F.; et al. Approaches and Perspectives for Development of African Swine Fever Virus Vaccines. Vaccines 2017, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Sang, H.; Miller, G.; Lokhandwala, S.; Sangewar, N.; Waghela, S.D.; Bishop, R.P.; Mwangi, W. Progress Toward Development of Effective and Safe African Swine Fever Virus Vaccines. Front. Vet. Sci. 2020, 7, 84. [Google Scholar] [CrossRef]
- Pérez-Núñez, D.; Sunwoo, S.Y.; Sánchez, E.G.; Haley, N.; García-Belmonte, R.; Nogal, M.; Morozov, I.; Madden, D.; Gaudreault, N.N.; Mur, L.; et al. Evaluation of a viral DNA-protein immunization strategy against African swine fever in domestic pigs. Vet. Immunol. Immunopathol. 2019, 208, 34–43. [Google Scholar] [CrossRef]
- Sunwoo, S.-Y.; Pérez-Núñez, D.; Morozov, I.; Sánchez, E.; Gaudreault, N.; Trujillo, J.; Mur, L.; Nogal, M.; Madden, D.; Urbaniak, K.; et al. DNA-Protein Vaccination Strategy Does Not Protect from Challenge with African Swine Fever Virus Armenia 2007 Strain. Vaccines 2019, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.S.; DeLay, P.D.; Sharman, E.C. The antibody response in pigs inoculated with attenuated African swine fever virus. Can. J. Comp. Med. Rev. Can. Med. Comp. 1968, 32, 455–460. [Google Scholar]
- Leitão, A.; Cartaxeiro, C.; Coelho, R.; Cruz, B.; Parkhouse, R.M.E.; Portugal, F.C.; Vigário, J.D.; Martins, C.L.V. The non-haemadsorbing African swine fever virus isolate ASFV/NH/P68 provides a model for defining the protective anti-virus immune response. J. Gen. Virol. 2001, 82, 513–523. [Google Scholar] [CrossRef]
- Boinas, F.S.; Hutchings, G.H.; Dixon, L.K.; Wilkinson, P.J. Characterization of pathogenic and non-pathogenic African swine fever virus isolates from Ornithodoros erraticus inhabiting pig premises in Portugal. J. Gen. Virol. 2004, 85, 2177–2187. [Google Scholar] [CrossRef]
- Mulumba-Mfumu, L.K.; Goatley, L.C.; Saegerman, C.; Takamatsu, H.-H.; Dixon, L.K. Immunization of African Indigenous Pigs with Attenuated Genotype I African Swine Fever Virus OURT88/3 Induces Protection Against Challenge with Virulent Strains of Genotype I. Transbound. Emerg. Dis. 2016, 63, e323–e327. [Google Scholar] [CrossRef]
- Gallardo, C.; Sánchez, E.G.; Pérez-Núñez, D.; Nogal, M.; de León, P.; Carrascosa, Á.L.; Nieto, R.; Soler, A.; Arias, M.L.; Revilla, Y. African swine fever virus (ASFV) protection mediated by NH/P68 and NH/P68 recombinant live-attenuated viruses. Vaccine 2018, 36, 2694–2704. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Soler, A.; Rodze, I.; Nieto, R.; Cano-Gómez, C.; Fernandez-Pinero, J.; Arias, M. Attenuated and non-haemadsorbing (non-HAD) genotype II African swine fever virus (ASFV) isolated in Europe, Latvia 2017. Transbound. Emerg. Dis. 2019, 66, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Borca, M.V.; Carrillo, C.; Zsak, L.; Laegreid, W.W.; Kutish, G.F.; Neilan, J.G.; Burrage, T.G.; Rock, D.L. Deletion of a CD2-Like Gene, 8-DR, from African Swine Fever Virus Affects Viral Infection in Domestic Swine. J. Virol. 1998, 72, 2881–2889. [Google Scholar] [CrossRef] [PubMed]
- Borca, M.V.; O’Donnell, V.; Holinka, L.G.; Risatti, G.R.; Ramirez-Medina, E.; Vuono, E.A.; Shi, J.; Pruitt, S.; Rai, A.; Silva, E.; et al. Deletion of CD2-like gene from the genome of African swine fever virus strain Georgia does not attenuate virulence in swine. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- O’Donnell, V.; Holinka, L.G.; Krug, P.W.; Gladue, D.P.; Carlson, J.; Sanford, B.; Alfano, M.; Kramer, E.; Lu, Z.; Arzt, J.; et al. African Swine Fever Virus Georgia 2007 with a Deletion of Virulence-Associated Gene 9GL (B119L), when Administered at Low Doses, Leads to Virus Attenuation in Swine and Induces an Effective Protection against Homologous Challenge. J. Virol. 2015, 89, 8556–8566. [Google Scholar] [CrossRef]
- O’Donnell, V.; Holinka, L.G.; Gladue, D.P.; Sanford, B.; Krug, P.W.; Lu, X.; Arzt, J.; Reese, B.; Carrillo, C.; Risatti, G.R.; et al. African Swine Fever Virus Georgia Isolate Harboring Deletions of MGF360 and MGF505 Genes Is Attenuated in Swine and Confers Protection against Challenge with Virulent Parental Virus. J. Virol. 2015, 89, 6048–6056. [Google Scholar] [CrossRef]
- Reis, A.L.; Goatley, L.C.; Jabbar, T.; Sanchez-Cordon, P.J.; Netherton, C.L.; Chapman, D.A.G.; Dixon, L.K. Deletion of the African Swine Fever Virus Gene DP148R Does Not Reduce Virus Replication in Culture but Reduces Virus Virulence in Pigs and Induces High Levels of Protection against Challenge. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Monteagudo, P.L.; Lacasta, A.; López, E.; Bosch, L.; Collado, J.; Pina-Pedrero, S.; Correa-Fiz, F.; Accensi, F.; Navas, M.J.; Vidal, E.; et al. BA71ΔCD2: A New Recombinant Live Attenuated African Swine Fever Virus with Cross-Protective Capabilities. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Sánchez-Cordón, P.J.; Jabbar, T.; Chapman, D.; Dixon, L.K.; Montoya, M. Absence of long-term protection in domestic pigs immunized with attenuated African swine fever virus isolate OURT88/3 or BeninΔMFG correlates with increased levels of regulatory T cells and IL-10. J. Virol. 2020. [Google Scholar] [CrossRef]
- Takamatsu, H.H.; Denyer, M.S.; Lacasta, A.; Stirling, C.M.A.; Argilaguet, J.M.; Netherton, C.L.; Oura, C.A.L.; Martins, C.; Rodríguez, F. Cellular immunity in ASFV responses. Virus Res. 2013, 173, 110–121. [Google Scholar] [CrossRef]
- Correia, S.; Ventura, S.; Parkhouse, R.M. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013, 173, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Islam, M.; Nash, R.; Reis, A.L. African swine fever virus evasion of host defences. Virus Res. 2019, 266, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Reis, A.L.; Netherton, C.; Dixon, L.K. Unraveling the Armor of a Killer: Evasion of Host Defenses by African Swine Fever Virus. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- García-Belmonte, R.; Pérez-Núñez, D.; Pittau, M.; Richt, J.A.; Revilla, Y. African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Abrams, C.C.; Goatley, L.; Fishbourne, E.; Chapman, D.; Cooke, L.; Oura, C.A.; Netherton, C.L.; Takamatsu, H.H.; Dixon, L.K. Deletion of virulence associated genes from attenuated African swine fever virus isolate OUR T88/3 decreases its ability to protect against challenge with virulent virus. Virology 2013, 443, 99–105. [Google Scholar] [CrossRef]
- Chen, W.; Zhao, D.; He, X.; Liu, R.; Wang, Z.; Zhang, X.; Li, F.; Shan, D.; Chen, H.; Zhang, J.; et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020, 1–12. [Google Scholar] [CrossRef]
- Chapman, D.A.G.; Tcherepanov, V.; Upton, C.; Dixon, L.K. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 2008, 89, 397–408. [Google Scholar] [CrossRef]
- Sereda, A.D.; Balyshev, V.M.; Kazakova, A.S.; Imatdinov, A.R.; Kolbasov, D.V. Protective Properties of Attenuated Strains of African Swine Fever Virus Belonging to Seroimmunotypes I–VIII. Pathogens 2020, 9, 274. [Google Scholar] [CrossRef]
- Karger, A.; Pérez-Núñez, D.; Urquiza, J.; Hinojar, P.; Alonso, C.; Freitas, F.B.; Revilla, Y.; Le Potier, M.-F.; Montoya, M. An Update on African Swine Fever Virology. Viruses 2019, 11, 864. [Google Scholar] [CrossRef]
- Sánchez, E.G.; Pérez-Núñez, D.; Revilla, Y. Development of vaccines against African swine fever virus. Virus Res. 2019, 265, 150–155. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Salas, M.L. African swine fever virus transcription. Virus Res. 2013, 173, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Dudek, T.; Knipe, D.M. Replication-defective viruses as vaccines and vaccine vectors. Virology 2006, 344, 230–239. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, C.M.; Jasperse, B.; Hagen, C.J.; Titong, A.; Verardi, P.H. Replication-inducible vaccinia virus vectors with enhanced safety in vivo. PLoS ONE 2020, 15, e0230711. [Google Scholar] [CrossRef]
- Nguyen, L.H.; Knipe, D.M.; Finberg, R.W. Replication-defective mutants of herpes simplex virus (HSV) induce cellular immunity and protect against lethal HSV infection. J. Virol. 1992, 66, 7067–7072. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Meng, G. ASFVdb: An integrative resource for genomic and proteomic analyses of African swine fever virus. Database 2020, 2020. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lokhandwala, S.; Waghela, S.D.; Bray, J.; Martin, C.L.; Sangewar, N.; Charendoff, C.; Shetti, R.; Ashley, C.; Chen, C.H.; Berghman, L.R.; et al. Induction of robust immune responses in swine by using a cocktail of adenovirus-vectored African swine fever virus antigens. Clin. Vaccine Immunol. 2016, 23, 888–900. [Google Scholar] [CrossRef]
- Lokhandwala, S.; Waghela, S.D.; Bray, J.; Sangewar, N.; Charendoff, C.; Martin, C.L.; Hassan, W.S.; Koynarski, T.; Gabbert, L.; Burrage, T.G.; et al. Adenovirus-vectored novel African Swine Fever Virus antigens elicit robust immune responses in swine. PLoS ONE 2017, 12, e0177007. [Google Scholar] [CrossRef]
- Lokhandwala, S.; Petrovan, V.; Popescu, L.; Sangewar, N.; Elijah, C.; Stoian, A.; Olcha, M.; Ennen, L.; Bray, J.; Bishop, R.P.; et al. Adenovirus-vectored African Swine Fever Virus antigen cocktails are immunogenic but not protective against intranasal challenge with Georgia 2007/1 isolate. Vet. Microbiol. 2019, 235, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Netherton, C.L.; Goatley, L.C.; Reis, A.L.; Portugal, R.; Nash, R.H.; Morgan, S.B.; Gault, L.; Nieto, R.; Norlin, V.; Gallardo, C.; et al. Identification and Immunogenicity of African Swine Fever Virus Antigens. Front. Immunol. 2019, 10, 1318. [Google Scholar] [CrossRef] [PubMed]
- Maschera, B.; Ferrazzi, E.; Rassu, M.; Toni, M.; Palù, G. Evaluation of Topoisomerase Inhibitors as Potential Antiviral Agents. Antivir. Chem. Chemother. 1993, 4, 85–91. [Google Scholar] [CrossRef]
- González-Molleda, L.; Wang, Y.; Yuan, Y. Potent Antiviral Activity of Topoisomerase I and II Inhibitors against Kaposi’s Sarcoma-Associated Herpesvirus. Antimicrob. Agents Chemother. 2012, 56, 893–902. [Google Scholar] [CrossRef]
- Tachedjian, G.; Tyssen, D.; Locarnini, S.; Gust, I.; Birch, C. Investigation of Topoisomerase Inhibitors for Activity against Human Immunodeficiency Virus: Inhibition by Coumermycin A1. Antivir. Chem. Chemother. 1990, 1, 131–138. [Google Scholar] [CrossRef]
- Coelho, J.; Ferreira, F.; Martins, C.; Leitão, A. Functional characterization and inhibition of the type II DNA topoisomerase coded by African swine fever virus. Virology 2016, 493, 209–216. [Google Scholar] [CrossRef]
- Drlica, K.; Hiasa, H.; Kerns, R.; Malik, M.; Mustaev, A.; Zhao, X. Quinolones: Action and Resistance Updated. Curr. Top. Med. Chem. 2009, 9, 981–998. [Google Scholar] [CrossRef]
- Laponogov, I.; Sohi, M.K.; Veselkov, D.A.; Pan, X.S.; Sawhney, R.; Thompson, A.W.; McAuley, K.E.; Fisher, L.M.; Sanderson, M.R. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009, 16, 667–669. [Google Scholar] [CrossRef]
- Heddle, J.G.; Barnard, F.M.; Wentzell, L.M.; Maxwell, A. The Interaction of Drugs with DNA Gyrase: A Model for the Molecular Basis of Quinolone Action. Nucleosides Nucleotides Nucleic Acids 2000, 19, 1249–1264. [Google Scholar] [CrossRef]
- Mottola, C.; Freitas, F.B.; Simões, M.; Martins, C.; Leitão, A.; Ferreira, F. In vitro antiviral activity of fluoroquinolones against African swine fever virus. Vet. Microbiol. 2013, 165, 86–94. [Google Scholar] [CrossRef]
- Wu, C.-C.; Li, T.-K.; Farh, L.; Lin, L.-Y.; Lin, T.-S.; Yu, Y.-J.; Yen, T.-J.; Chiang, C.-W.; Chan, N.-L. Structural Basis of Type II Topoisomerase Inhibition by the Anticancer Drug Etoposide. Science (80-) 2011, 333, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Hakobyan, A.; Arabyan, E.; Avetisyan, A.; Abroyan, L.; Hakobyan, L.; Zakaryan, H. Apigenin inhibits African swine fever virus infection in vitro. Arch. Virol. 2016, 161, 3445–3453. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.; Fachin, A.; Beleboni, R.; França, S.; Marins, M. In Vitro Action of Flavonoids in the Canine Malignant Histiocytic Cell Line DH82. Molecules 2013, 18, 15448–15463. [Google Scholar] [CrossRef] [PubMed]
- Constantinou, A.; Mehta, R.; Runyan, C.; Rao, K.; Vaughan, A.; Moon, R. Flavonoids as DNA topoisomerase antagonists and poisons: Structure-activity relationships. J. Nat. Prod. 1995, 58, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Arabyan, E.; Hakobyan, A.; Kotsinyan, A.; Karalyan, Z.; Arakelov, V.; Arakelov, G.; Nazaryan, K.; Simonyan, A.; Aroutiounian, R.; Ferreira, F.; et al. Genistein inhibits African swine fever virus replication in vitro by disrupting viral DNA synthesis. Antivir. Res. 2018, 156, 128–137. [Google Scholar] [CrossRef]
- Hsieh, T.; Brutlag, D. ATP-dependent DNA topoisomerase from D. melanogaster reversibly catenates duplex DNA rings. Cell 1980, 21, 115–125. [Google Scholar] [CrossRef]
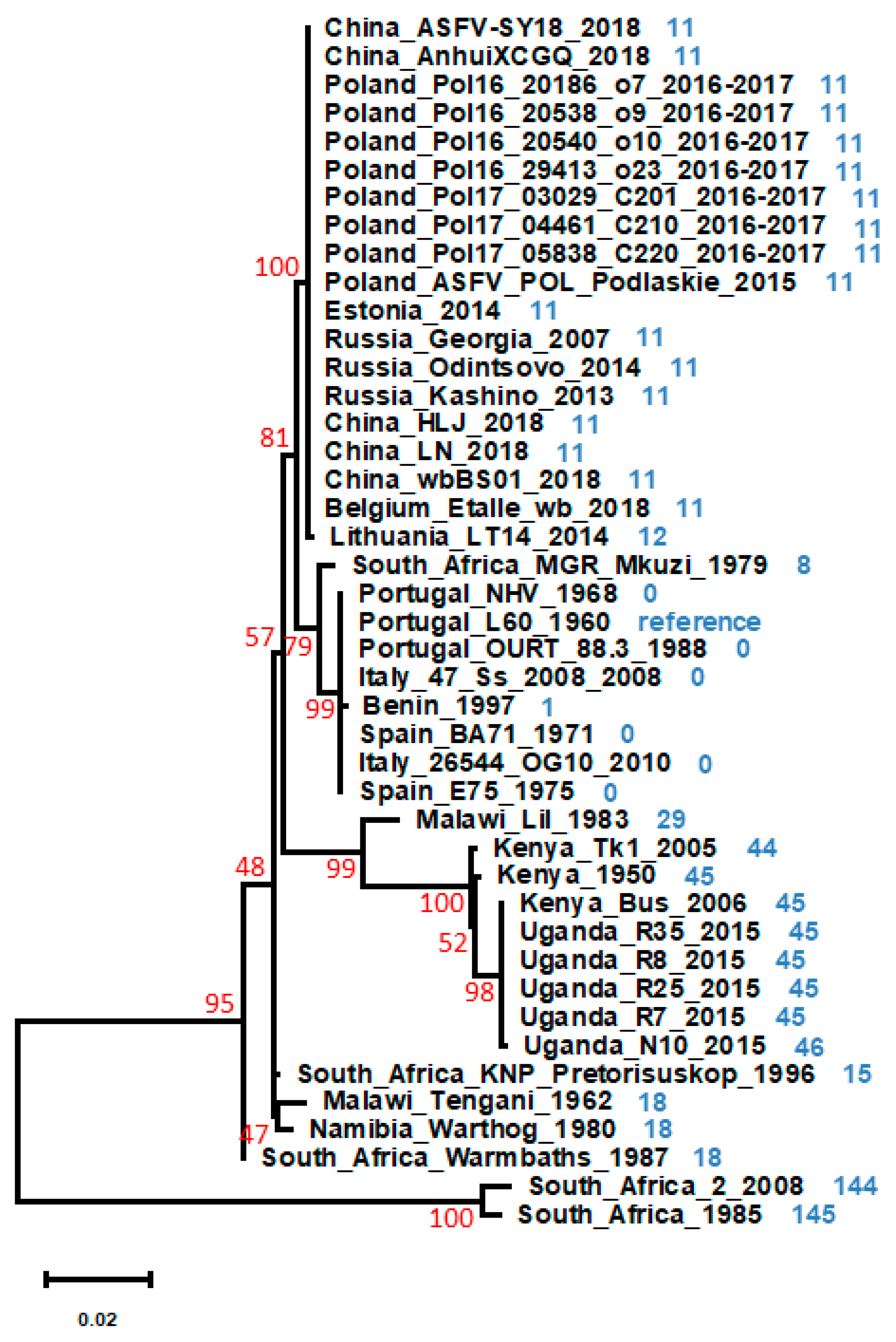
| L60 | HLJ | Tk1 | RSA_2 | ||
|---|---|---|---|---|---|
| pP1192R topoisomerase II | Portugal_L60_1960 (L60) | - | 99.08% | 96.31% | 87.92% |
| China_HLJ_2018 (HLJ) | 99.08% | - | 96.31% | 88.42% | |
| Kenya_Tk1_2005 (Tk1) | 96.31% | 96.31% | - | 86.07% | |
| South_Africa_2_2008 (RSA_2) | 87.92% | 88.42% | 86.07% | - | |
| pA104R histone-like protein | Portugal_L60_1960 | - | 100% | 100% | 80.77% |
| China_HLJ_2018 | 100% | - | 100% | 80.77% | |
| Kenya_Tk1_2005 | 100% | 100% | - | 80.77% | |
| South_Africa_2_2008 | 80.77% | 80.77% | 80.77% | - | |
| pG1211R DNA polymerase alpha-like protein | Portugal_L60_1960 | - | 98.35% | 96.19% | 86.37% |
| China_HLJ_2018 | 98.35% | - | 96.51% | 86.65% | |
| Kenya_Tk1_2005 | 96.19% | 96.51% | - | 85.20% | |
| South_Africa_2_2008 | 86.37% | 86.65% | 85.20% | - | |
| pA224L IAP-like protein | Portugal_L60_1960 | - | 98.21% | 90.18% | 97.32% |
| China_HLJ_2018 | 98.21% | - | 90.18% | 98.21% | |
| Kenya_Tk1_2005 | 90.18% | 90.18% | - | 90.18% | |
| South_Africa_2_2008 | 97.32% | 98.21% | 90.18% | - | |
| pEP402R CD2 homolog | Portugal_L60_1960 | - | 54.55% | 50.83% | 26.02% |
| China_HLJ_2018 | 54.55% | - | 68.75% | 26.79% | |
| Kenya_Tk1_2005 | 50.83% | 68.75% | - | 23.21% | |
| South_Africa_2_2008 | 26.02% | 26.79% | 23.21% | - | |
| pDP96R uncharacterized protein | Portugal_L60_1960 | - | 95.83% | 76.04% | 81.25% |
| China_HLJ_2018 | 95.83% | - | 77.08% | 82.29% | |
| Kenya_Tk1_2005 | 76.04% | 77.08% | - | 68.42% | |
| South_Africa_2_2008 | 81.25% | 82.29% | 68.42% | - | |
| pEP153R C-type lectin-like protein | Portugal_L60_1960 | - | 51.83% | 65.66% | 56.21% |
| China_HLJ_2018 | 51.83% | - | 57.32% | 37.20% | |
| Kenya_Tk1_2005 | 65.66% | 57.32% | - | 46.99% | |
| South_Africa_2_2008 | 56.21% | 37.20% | 46.99% | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coelho, J.; Leitão, A. The African Swine Fever Virus (ASFV) Topoisomerase II as a Target for Viral Prevention and Control. Vaccines 2020, 8, 312. https://doi.org/10.3390/vaccines8020312
Coelho J, Leitão A. The African Swine Fever Virus (ASFV) Topoisomerase II as a Target for Viral Prevention and Control. Vaccines. 2020; 8(2):312. https://doi.org/10.3390/vaccines8020312
Chicago/Turabian StyleCoelho, João, and Alexandre Leitão. 2020. "The African Swine Fever Virus (ASFV) Topoisomerase II as a Target for Viral Prevention and Control" Vaccines 8, no. 2: 312. https://doi.org/10.3390/vaccines8020312
APA StyleCoelho, J., & Leitão, A. (2020). The African Swine Fever Virus (ASFV) Topoisomerase II as a Target for Viral Prevention and Control. Vaccines, 8(2), 312. https://doi.org/10.3390/vaccines8020312





