1. Introduction
African swine fever (ASF) is a devastating disease of domestic pigs and wild boar, which can result in the death of almost all infected animals. ASF has spread in many countries in sub-Saharan Africa, Russian Federation, Europe and most recently to China and other S. E. Asian countries. Information on disease outbreaks is updated daily (OIE WAHIS
https://www.oie.int/wahis_2/public/), and the situation in Asia is summarised weekly by the United Nations Food and Agriculture Organisation (FAO) (
http://www.fao.org/ag/againfo/programmes/en/empres/ASF/situation_update.html).
African swine fever virus (ASFV) is a large cytoplasmic DNA virus and is the only member of the
Asfarviridae family [
1]. The genome of 170–193 kbp contains about 150–170 genes. These include many that are not essential for virus replication in cells but have roles that include the evasion of host defences [
1]. Several inhibitors of type I interferon (IFN) responses have been identified, including members of the virus multigene families (MGF) 360 and 505/530 and the DP96R/UK protein. The deletion of multiple members of MGF 360 and 505 results in the attenuation of virulent isolates, including genotype I Benin 97/1, genotype II Georgia, and Pr4 [
2,
3,
4]. The deletion of the DP96R/UK gene also resulted in the attenuation of the E70 isolate, although it did not reduce virulence of the genotype II Georgia isolate [
5,
6]. Previously, in cultured macrophages infected with virulent ASFV, the induction of type I IFN and activation of IFN responses was inhibited, whereas, in those infected with virulent ASFV, from which multiple copies of MGF360 or 505/530 were deleted, varying levels of IFN-
β or interferon stimulated genes were expressed [
2,
4]. Increased type I IFN-
β mRNA transcripts were also observed in macrophages infected with the naturally attenuated ASFV isolate OURT88/3 isolate [
4]. This has a deletion of similar numbers of MGF360 and MGF505/530 genes as the ASFV gene deletion mutants BeninΔMGF and Pr4. Thus, there is a good correlation between the increased induction of type I IFN and the attenuation of ASFV.
The ASFV I329L protein is a predicted type I transmembrane protein that contains motifs typical of Toll-like receptors [
7,
8]. These include four leucine-rich repeats (LRRs) in the extracellular domain and a weak homology with the cytoplasmic Toll-interleukin-1 receptor (TIR) domain of Toll-like receptor 3 (TLR3). This domain mediates interactions between TLRs and cytoplasmic adaptor proteins. These similarities suggested that the I329L protein may act as a TLR antagonist by inhibiting the activation of signalling pathways downstream of TLR3 and possibly other TLRs. Transiently expressed I329L inhibited the activation of IFN-β promoter and NF-κB-dependent luciferase reporters following the activation of TLR3 by the double-stranded RNA mimic polyinosinic:polycytidylic acid (poly IC) or of the downstream pathway by overexpression of the TIR-domain-containing adapter-inducing interferon-β (TRIF) adaptor protein. Protein structure modelling suggested that I329L may function as a TLR3 decoy by formation of I329L-TLR3 heterodimers, thus inhibiting the downstream type I IFN induction pathway [
9]. The transient expression of I329L inhibited the secretion of IFN-β into cell supernatants, confirming that the expression of the protein inhibits type I IFN induction [
8].
Although exogenously expressed I329L protein has been shown to reduce type I IFN production from cells, its role during the virus infection of cells or pigs has not previously been investigated. In the current study, we deleted the I329L gene from the genome of the natural attenuated genotype I isolate OURT88/3 (OURT88/3ΔI329L) and from the genotype II virulent Georgia 2007/1 isolate (GeorgiaΔI329L). We hypothesized that the I329L deletion would result in increased amounts of type I IFN being secreted by infected cells, resulting in the inhibition of viral replication in vivo and, importantly, the promotion of the adaptive immune response.
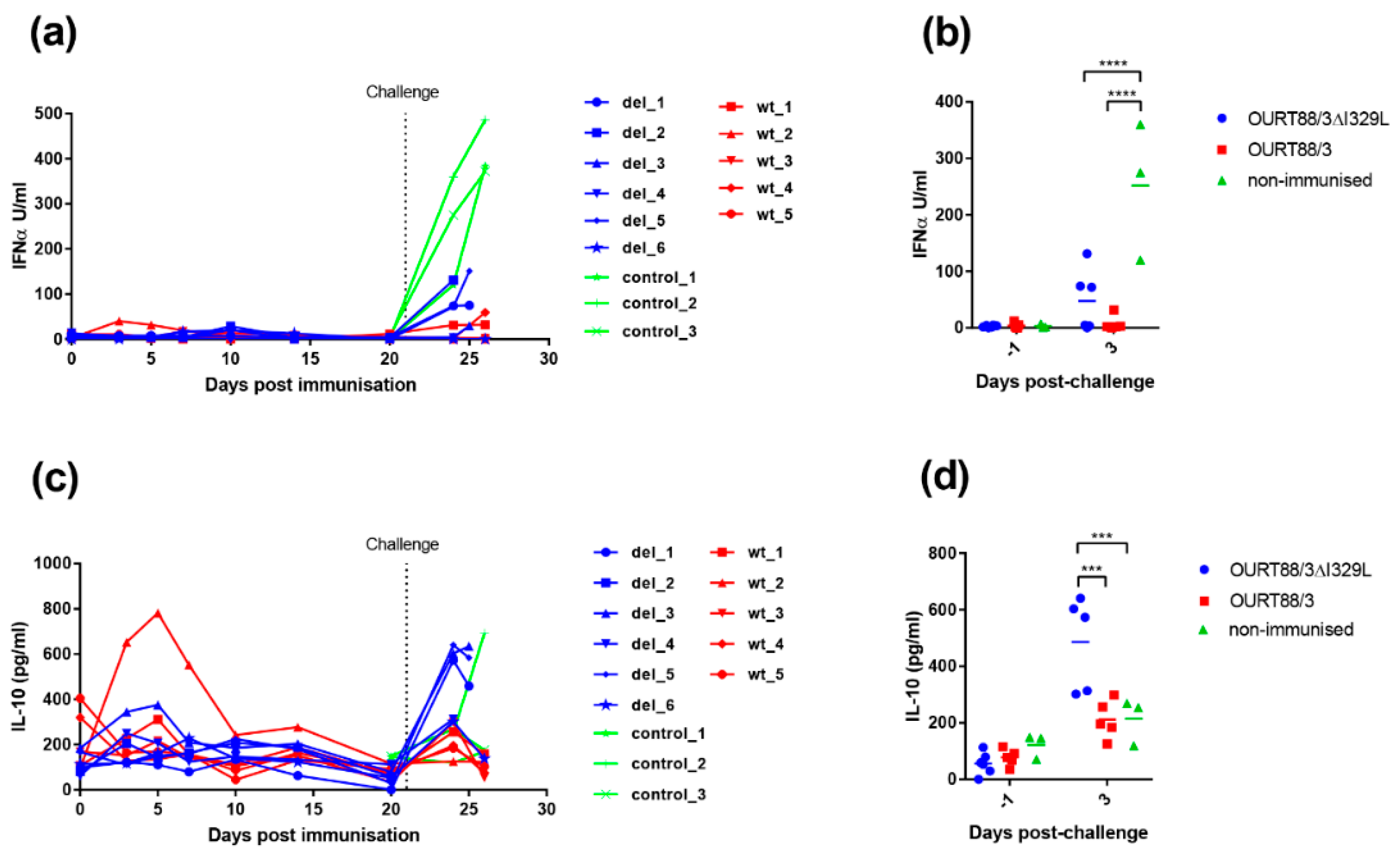
The results show that this gene deletion did not have any significant effect on replication of the viruses in cells. However, porcine macrophages infected with OURT88/3ΔI329L expressed significant higher amounts of type I IFN than the ones infected with wild-type (wt) OURT88/3.
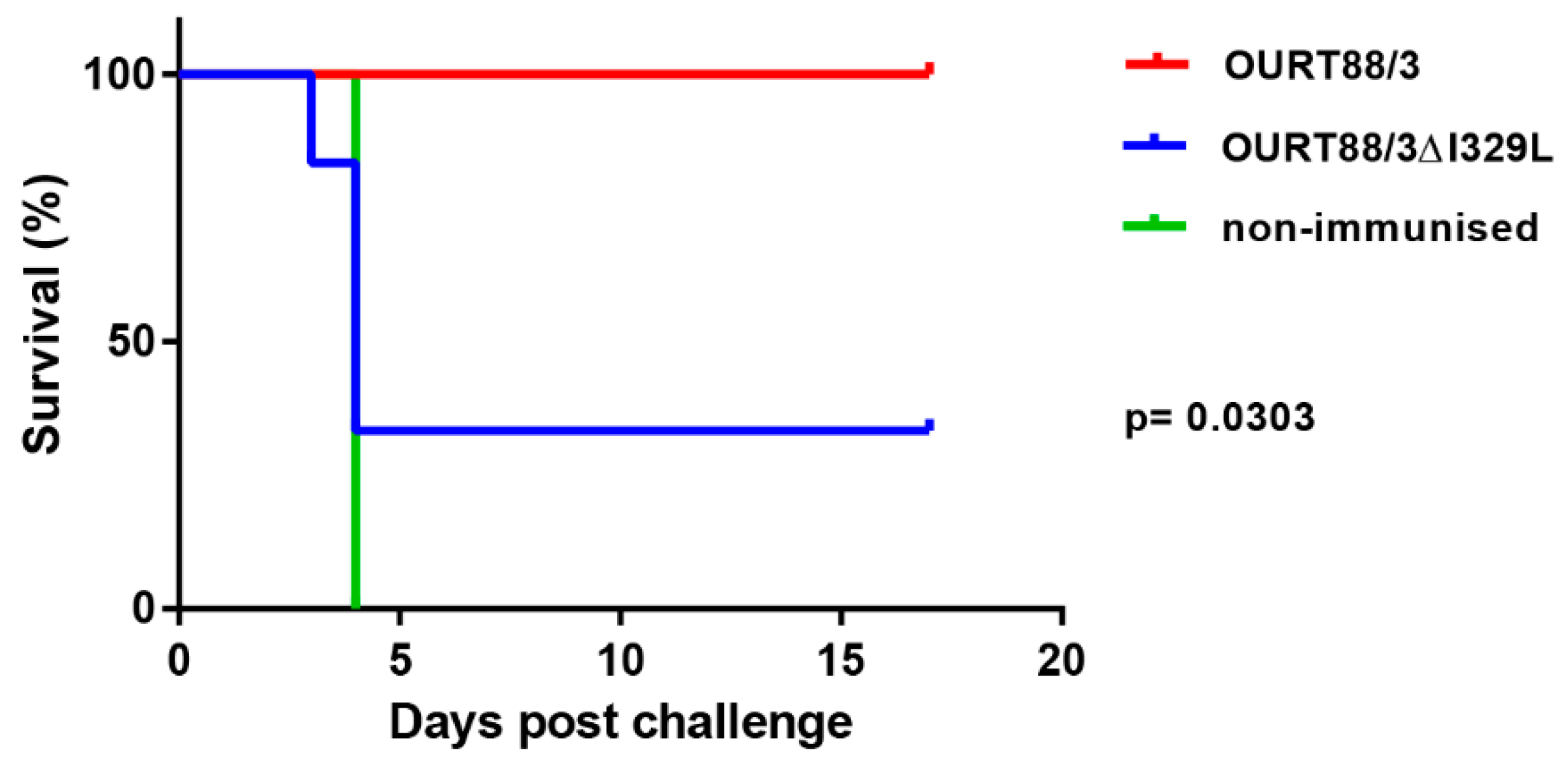
Pigs infected with the GeorgiaΔI329L virus developed high viremia as well as clinical and pathological signs typical of acute ASFV. The deletion of I329L from the OURT88/3 strain did not result in a reduction in clinical signs but unexpectedly reduced the level of protection against challenge. Thus, an effect of deleting I329L was observed when it was deleted in combination with other type I interferon inhibitors from an attenuated strain but not singly from a highly virulent strain.
2. Materials and Methods
2.1. Viruses and Cells
The OURT88/3 low virulence non-haemabsorbing genotype I ASFV isolate and the virulent Georgia 2007/1 genotype II isolate have been described previously [
10,
11]. These viruses were grown in primary pig macrophage cultures from bone marrow (PBMs). Virus titres were determined by end-point dilution in PBM cultures. The virus was detected by immunofluorescence using a monoclonal antibody against virus protein p30/CP204L (mouse monoclonal IgG1 antibody clone C18, Pirbright, UK) and an appropriate secondary antibody. Virus titres were calculated as the amount of virus infecting 50% of the PBM cultures (TCID
50/mL).
2.2. Recombinant Viruses
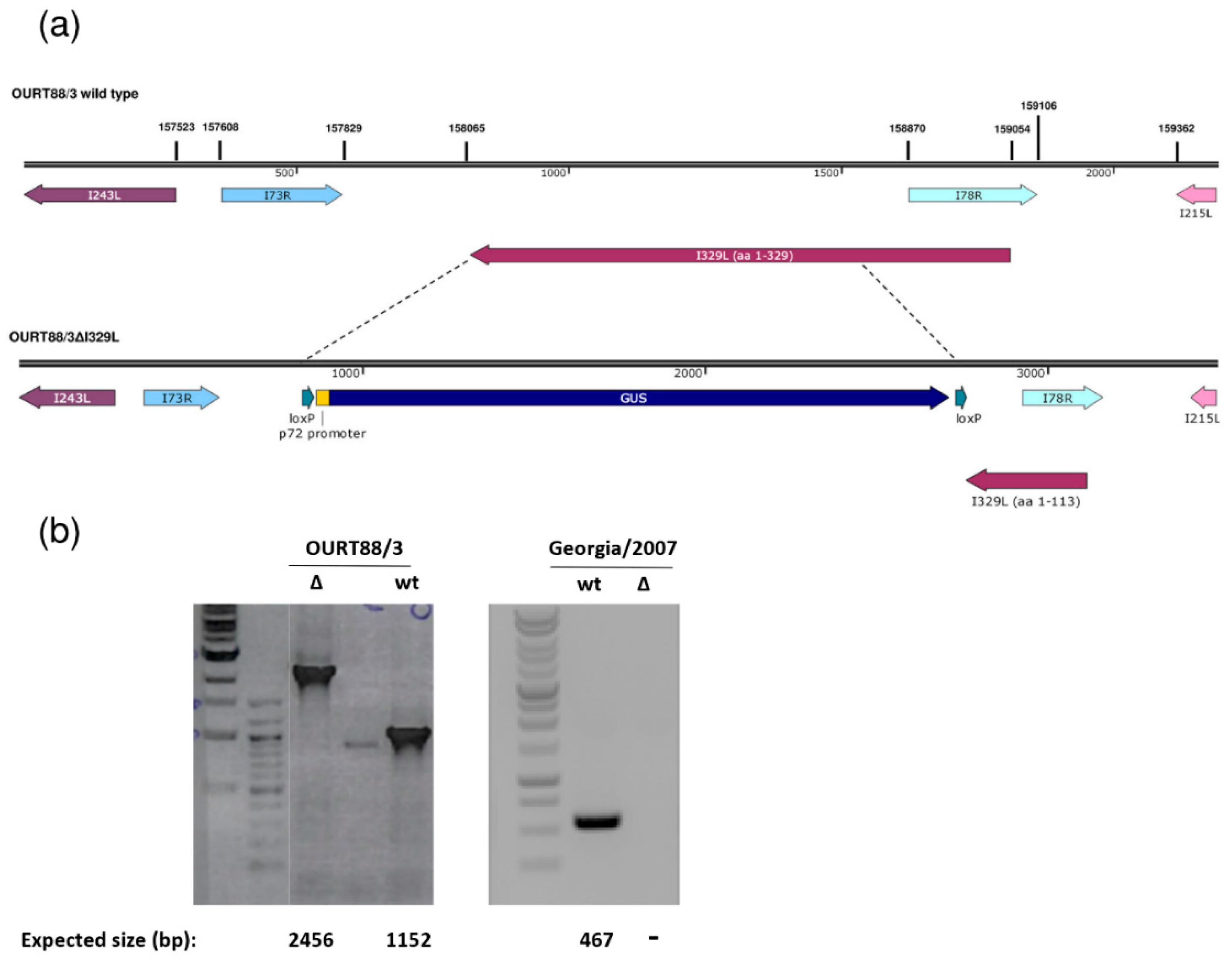
Right and left genome fragments of approximately 700 to 800 bp flanking the I329L gene were amplified by PCR. The following primers were used for the left flanking fragment encompassing genome positions 157245 to 158064: CATATGTTTTTGAAGCGTTCTAAAAAACATC and ATGTGTGGTTTATTTTAGTATG. The right flanking fragment was amplified with the GAGTTCTTTACCAAAGCC and GGAGGATGACACATATATCTTAACC primers comprising genome positions 158716 to 159434 of the OURT88/3 isolate. Similar fragments were amplified from the Georgia 2007/1 isolate. The obtained DNA fragments were cloned into the pLoxPVP72GUSLoxP vector to construct the pΔI329LGUS plasmids. Pig alveolar macrophages (PAMs) were infected with OURT88/3 or Georgia 2007/1 isolates and transfected with the pΔI329LGUS plasmids using TransIT-LT1 (Mirus Bio, Madison, WI, USA). Recombinant viruses expressing the β-GUS gene were identified by incubation with 5-bromo-4-chloro-1H-indol-3-yl β-D-glucopyranosiduronic acid and purified by limiting dilution. Viral genomic DNA was purified from supernatants from infected porcine macrophages using MagVet™ Universal Isolation Kit (Life Technologies). The analysis of viral DNA was carried out by PCR amplification using primers binding within the I329L deletion (GGACTGTTTGCTGAGGTTGTATG and CCCTTATACTACTTCCTACTGAAACAGG) or flanking regions (GGTTCTATAAATAGCATACTGTACAG and CTGCTGGCATTTCATGCACTTG).
2.3. Quantification of IFN-β Transcripts
The expression of IFN-
β was quantified as described previously [
4]. Briefly, PAMs (5 × 10
5 cells) were infected with ASFV or mock infected. At selected times, RNA was extracted (RNeasy mini kit, Qiagen, Hilden, Germany) and equal amounts were used as a template to synthesise cDNA (Superscript III reverse transcriptase kit, Invitrogen, San Diego, CA, USA). IFN-
β transcripts were measured by quantitative real time PCR (qPCR) using a power SYBR Green Master Mix (Thermo Fisher Scientific, Hemel Hempstead, UK). IFN-
β and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) copy numbers were calculated by the standard curve method, and the results are presented as the IFN-
β/GAPDH ratio. Assays were carried out in duplicate.
2.4. Quantification of IFN-α in Supernatants and Pig Sera
PBMs were purified from bone marrow suspensions by gradient centrifugation and seeded at 1 × 106 cells per well (24 well plate). Cells were infected with the different ASFV isolates at a multiplicity of infection (MOI) of 1 (1.45 × 106 TCID50 per well). After 1 h of incubation at 37 °C, the inoculum was removed and fresh medium was added. The infected cells were further incubated for 16 h and the supernatants were collected. The amount of IFN-α in supernatants or pig sera was quantified by an in-house ELISA. Briefly, Maxisorp plates (Nunc, Roskilde, Denmark) were coated overnight at room temperature with anti-pig IFN-α antibody (clone K9) at 0.5 µg/mL in 0.5 M carbonate-bicarbonate coating buffer. Plates were washed with wash buffer (0.05% Tween 20 in PBS) and blocked with 1% BSA in PBS. Samples and standards were then added and the plates were incubated at room temperature for 2 h. After washing, detection antibody (biotinylated anti-pig IFN-α antibody—clone F17) diluted 1:5000 in blocking buffer was added and the plates were incubated at room temperature for 2 h. The plates were then washed, incubated with Streptavidin horseradish peroxidase (HRP) and finally developed with 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (R and D Systems, DY999). After stopping the reaction with 2 N H2SO4, the absorbances were read at 450 nm.
2.5. Virus Growth Analysis
PBMs were infected at an MOI of 0.3 with the OURT88/3, OURT88/3ΔI329L, Georgia 2007/1 or GeorgiaΔI329L. Cells and supernatants were collected at different times post-infection and subjected to 3 freeze-thaw cycles. Cellular debris was removed by centrifugation, and virus titres were determined as above.
2.6. Pig Immunisation and Challenge
The experiments were conducted in the SAPO4 high containment large animal isolation units at the Pirbright Institute under Home Office License PPL70/8852. One group of 5 and one group of 6 Large white/Landrace pigs of 15–20 kg weight, in separate rooms, were immunised by the intramuscular route with 10
4 TCID
50 in 1 mL of the wild-type OURT88/3 virus (OURT88/3_wt 1–5) or the OURT88/3 virus with the I329L gene deleted (OURT88/3_del 1–6). After 21 days, all pigs were challenged by the intramuscular route with 10
4 TCID
50 of virulent strain OURT88/1, in parallel with 3 control non-immune pigs in a separate room. Pigs were observed and scored for the development of clinical signs, including fever, loss of appetite, lethargy and external signs of haemorrhage [
12]. At defined humane endpoints, pigs were euthanized by an overdose of barbiturates. Those pigs that survived challenge were terminated at the end of the experiment 21 days post-challenge. Macroscopic lesions were scored at post-mortem examination [
13]. In a second experiment, a group of 6 pigs were infected with 10
4 TCID
50 of the GeorgiaΔI329L virus by the intramuscular route and clinical signs were scored as above. In both experiments, blood and serum samples were collected at defined time points during the experiment and tissues at post-mortem. These samples were stored at −80 °C.
2.7. Measurement of Virus Genome Copy Numbers by Quantitative PCR
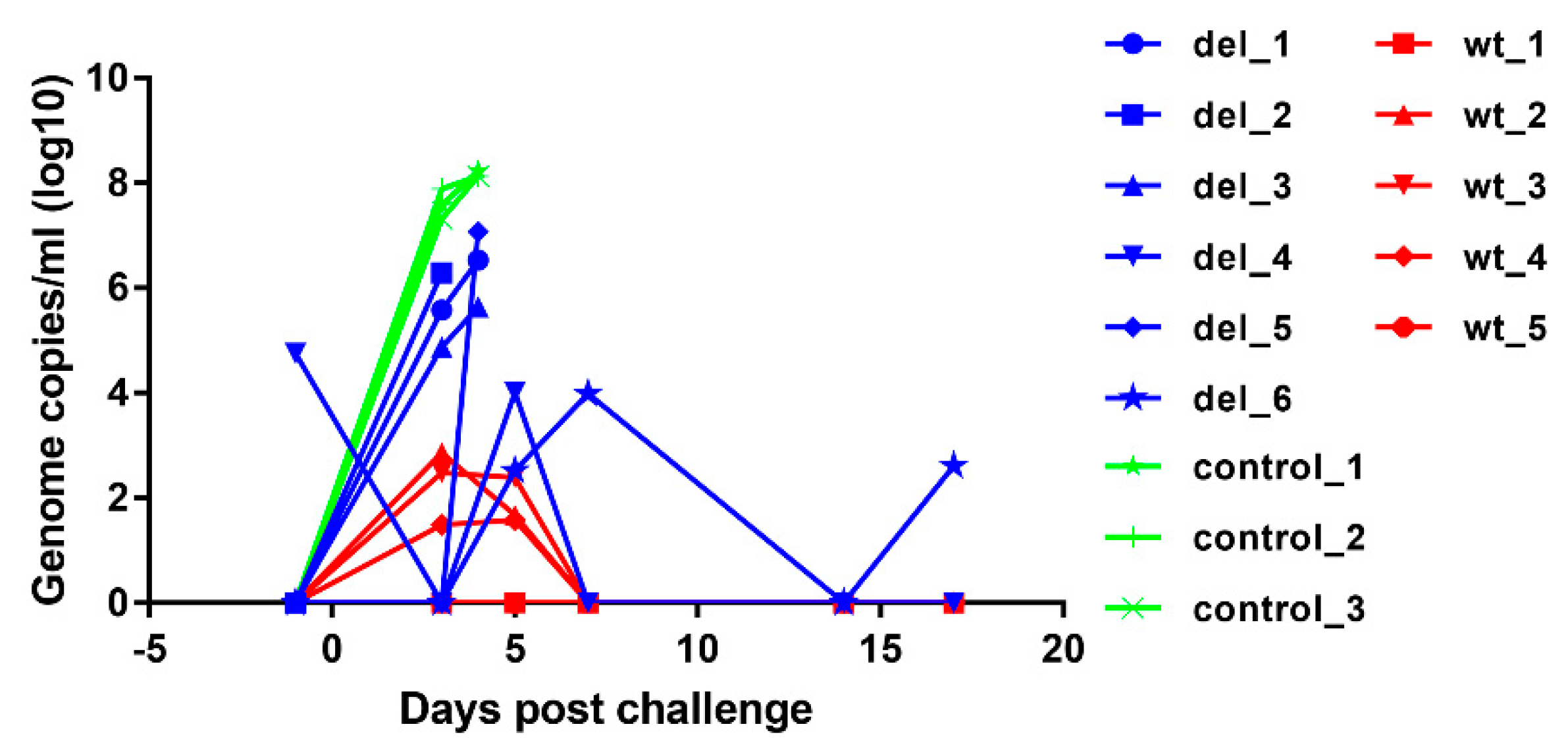
Virus genome copy numbers in whole blood were measured by quantitative PCR (qPCR) [
12,
14] and expressed as genome copies per ml of blood.
2.8. IFN-γ ELiSpot Assays
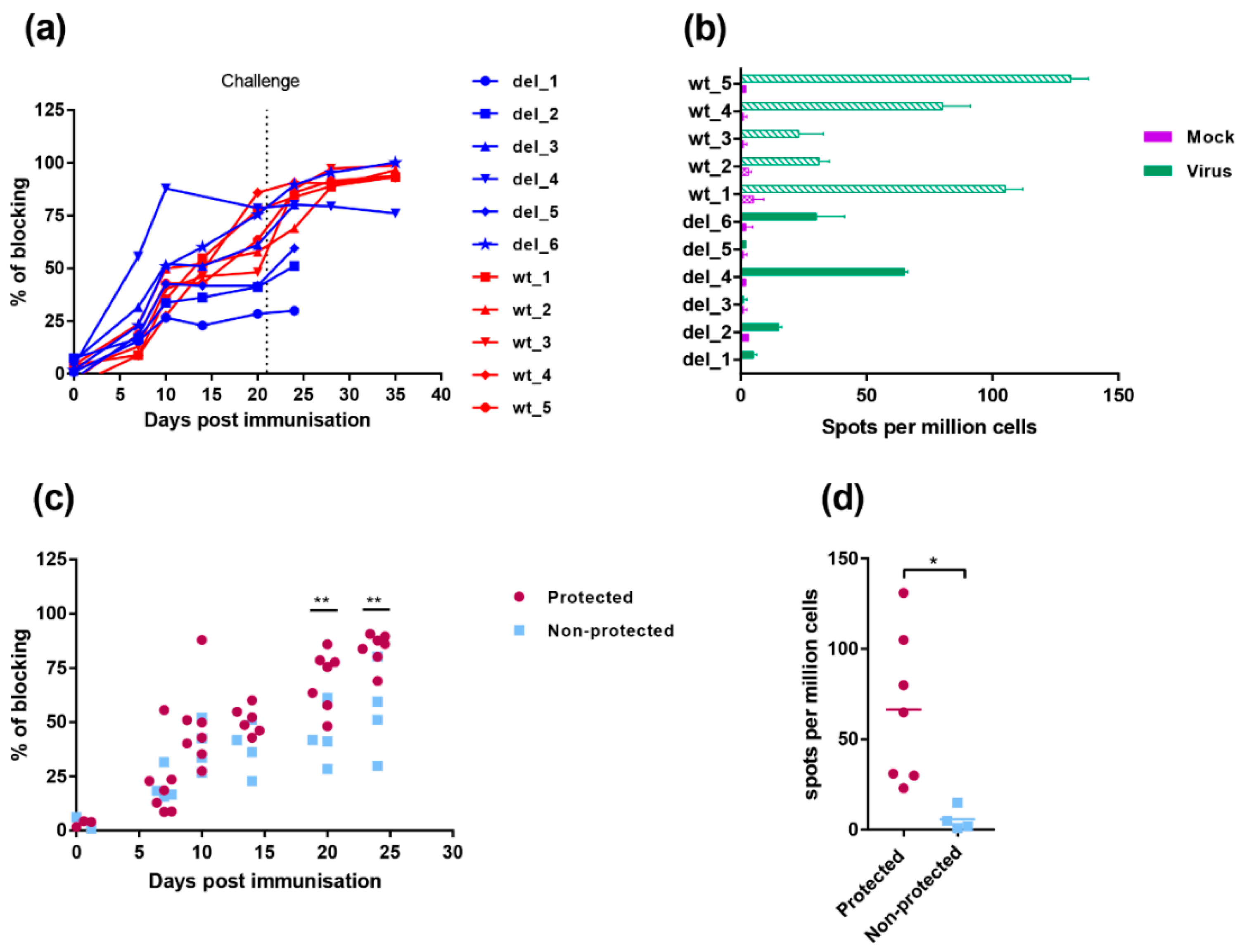
Peripheral blood mononuclear cells (PBMC) were purified from EDTA blood using gradient centrifugation. ELIspot plates (MAIPS4510, Millipore, Burlington, MA, USA) were coated overnight at 4 °C with 4 µg/mL anti-porcine IFN-γ mAb P2F6 in 0.5 M carbonate-bicarbonate coating buffer and then washed with PBS. Cells were plated in duplicate at two different dilutions, typically 8 × 105 and 4 × 105 per well in Roswell Park Memorial Institute (RMPI) supplemented with 10% foetal calf serum, 1 mM sodium pyruvate, 50 µM 2-mercaptoethanol, 100 IU/mL penicillin and 100 µg/mL streptomycin. Cells were then incubated overnight in a final volume of 200 µL with 105 haemadsorption (HAD) units of OURT88/1 or an equivalent volume of mock inoculum, or 2.5 µg/mL phytohaemagglutinin as a positive control. Cells were lysed by incubating for 5 min in water and then washed with PBS. Biotinylated anti-porcine IFN-γ mAb P2C11, followed by streptavidin conjugated to alkaline phosphatase was used to visualise spots that were then counted using an ELIspot Reader System (AID). The number of spots per well was converted into the number of spots per million cells and the mean of duplicate wells plotted.
2.9. ELISA Assays
The presence in serum of antibodies against ASFV protein VP72/B646L were measured using a commercial competitive ELISA (INGEZIM PPA Compac, Ingenasa, Madrid, Spain). IL-10 was detected in serum using a commercial kit (R and D systems). IFN-
α in serum was quantified as described in
Section 2.4.
2.10. Statistical Analysis
Statistical analysis was performed using GraphPad Prism7 software (GraphPad Software Inc., San Diego, CA, USA). Differences between groups were determined using unpaired t-test or two-way ANOVA followed by Tukey’s multiple comparison test. The log-rank test (Mantel–Cox) was used to compare survival after challenge.
4. Discussion
ASFV codes for many inhibitors of innate immunity, including inhibitors of type I interferon induction. In general, little is known about the mechanisms by which these proteins act. Most, for example, the MGF360, MGF505 and DP96R genes have no similarities with other known proteins, although some defined motifs are present. One exception is the type I transmembrane I329L protein, which shares similarity with, and is proposed to act as an antagonist of, TLR3, thus inhibiting the downstream pathway of IFN induction. The deletion of multiple copies of MGF360 and 505 or of DP96R genes can result in attenuation of virulent ASFV, with some variation in the level of attenuation between isolates. In most cases, pigs immunised with attenuated viruses, from which genes coding for inhibitors of type I IFN have been deleted, are protected against challenge with parental virulent virus. This is presumed to result from the increased induction of interferon stimulated genes which induce innate and adaptive immune responses.
In our current study, we investigated the effect of deleting the I329L gene from an already attenuated isolate, OURT88/3, and from a highly virulent isolate. Our results confirm that the gene could be deleted without affecting the levels of virus replication in porcine macrophage cultures. Importantly, the deletion mutant OURT88/3ΔI329L induced higher levels of IFN-
β transcripts, in infected porcine primary macrophages, than the wild-type virus. Furthermore, we observed a small but significant increase in IFN-
α levels in supernatants from OURT88/3ΔI329L macrophages compared to mock-infected cells. Remarkably, Razzuoli et al. showed that the infection of porcine macrophages with NHP/68 (an attenuated genotype I isolate, similar to OURT88/3) significantly increased the number of several IFN-
α subtype transcripts [
15]. However, in agreement with our results from the OURT88/3 wild type, they could not detect any significant increase in IFN-
α in supernatants from infected cells. It is therefore possible that ASFV also interferes with post-translation modifications/secretion of IFN. It is also worth mentioning that type I IFN secretion initiates a positive feedback loop, priming neighbouring cells to produce more IFN. Hence, even low levels of IFN secreted by the first infected cells may have a significant impact at the virus replication sites. Taken together, our results suggest that I329L has evolved to modulate the host innate immune response and supports previous data from transiently transfected cells [
7,
8].
We anticipated that the deletion of I329L from the OURT88/3 isolate would further increase the expression of type I IFN and enhance the host immune response, thus preventing the occurrence of adverse clinical signs following immunisation. Our hypothesis was not confirmed, and, instead, we observed a decrease in protection in pigs immunised with OURT88/3ΔI329L. The reduced protection observed following immunisation with a deletion mutant from an already attenuated virus is not without precedent [
16]. Gallardo et al. showed that the deletion of another IFN inhibitor (A276R) from the NH/P68 isolate results in the complete loss of protection against challenge with Arm07, in contrast to the 100% protection afforded by vaccination with the wild-type virus. In our study, the reduced protection in pigs immunised with OURT88/3ΔI329L was associated with an impairment of both antibody and cellular immune responses, as measured by VP72 ELISA and IFN-
γ ELIspot, respectively. The reasons for this are unknown and may involve reduced virus replication in the non-protected pigs, altered immune responses or both. Interestingly, type I IFN has been implicated in the dysregulation of the immune responses during persistent viral infections [
17,
18]. Similarly, during ASFV infection, a delicate control of the IFN response may be necessary to avoid viral persistence and to promote the induction of protective responses. Hence, enough IFN needs to be produced to control early virus replication and to stimulate adaptive immune responses but excessive/prolonged IFN exposure may result in immunosuppression. Whilst we did not observe an increase in IFN-
α levels in the serum of non-protected animals following vaccination, we cannot dismiss the notion that high levels of IFN were present at the viral replication sites. Comparably to Golding et al., [
19] we observed a sharp increase in type I IFN levels following the challenge of control pigs with OURT88/1, which is probably associated with high levels of viral replication and does not correlate with the ability of the virus to control the IFN response in vitro.
Levels of IL-10 were significantly higher at day 3 post-challenge in the group immunised with OURT88/3ΔI329L, and this was mainly driven by those animals that did not survive challenge. IL-10 prevents excessive inflammatory responses and is a broadly expressed cytokine, known to be produced by cells of the innate immune system (macrophages, dendritic cells and natural killer cells among others) and cells of the adaptive immune system including regulatory T (Tregs) cells but also T helper (Th) cells, cytotoxic T lymphocytes (CTLs) and B cells [
20]. In the current study, infected macrophages are unlikely to be the source of the high levels of IL-10 observed in the vaccinated/non-protected pigs, since IL-10 values were much lower in control pigs, which presented the highest viremias at this day post-challenge. Furthermore, a recent study [
21] showed that IL-10 was expressed at significantly lower levels in cells infected with the highly virulent Georgia/2007 isolate than in non-infected cells. Taken together, these results indicate that, in the group immunised with OURT88/3Δ329L, IL-10 is probably produced by cells of the adaptive immune system. It is therefore tempting to speculate that immunisation with the deletion mutant virus resulted in an increase in the number of local and/or circulating Tregs. Two main subsets of Tregs have been described: naturally occurring Tregs (nTregs) and inducible Tregs (iTregs). iTregs may be classified as T
R1 (IL-10 producing) or T
H3 (TGF-
β producing) [
22] and they can be induced by prolonged exposure to circulating antigen, chronic inflammation or weak co-stimulation in the periphery [
23]. The deletion of the I329L gene, an inhibitor of TLR signalling [
7] might have indeed increased the inflammatory response at the viral replication sites, thus promoting Treg induction. Interestingly, a recent study found a correlation between the lack of protection following the immunisation of pigs with OURT88/3 and increased levels of IL-10 and Tregs [
24]. It is also interesting to note that type I IFN signalling has been implicated in the promotion of T
R1 responses during chronic virus infection [
25], and, therefore, the relationship between IFN responses, the activation of regulatory T cells and IL-10 production in the context of ASFV infection merits further studies.
The single deletion of the I329L gene did not attenuate the virulent Georgia/2007 isolate. This is probably a result of its functional redundancy, since several other IFN inhibitors are encoded by ASFV. As discussed above, the attenuation of this isolate required the deletion of multiple members of the MGF 360 and 505 [
3]. Additionally, I329L was shown to inhibit the IFN induction pathway when activated via TLR signalling but not through the engagement of intracellular receptors [
7]. Our results indicate that attenuation may require the deletion of gene(s) acting downstream in the signalling cascade (e.g., at the level of transcription factors) that allow the virus to control IFN expression induced by different receptors, including the cytoplasmic DNA sensor cyclic guanosine monophosphate–adenosine monophosphate (GMP-AMP) synthase (cGAS). It was also shown [
26] that the Armenia/07 isolate, a virus closely related to Georgia/2007, is able to control IFN induction through the cGAS-STING pathway. Therefore, the identification of ASFV genes involved in the modulation of host DNA sensing may provide additional targets for the development of rationally attenuated ASFV vaccines.