Deletion of Vaccinia Virus A40R Gene Improves the Immunogenicity of the HIV-1 Vaccine Candidate MVA-B
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cells
2.3. Viruses
2.4. Plasmids
2.4.1. Construction of Plasmid Transfer Vector pGem-RG-ΔA40R wm
2.4.2. Construction of Plasmid Transfer Vector pHA-A40R
2.5. Generation of Recombinant MVA-B Δ40R and MVA-B ΔA40R-rev Viruses
2.6. PCR Analysis
2.7. Expression of HIV-1BX08 gp120 and HIV-1IIIB GPN Proteins by Western Blot
2.8. Analysis of Virus Growth
2.9. RNA Analysis by Reverse Transcription Real-Time Quantitative PCR (RT-qPCR)
2.10. DNA Vectors
2.11. Peptides
2.12. Mouse Immunization Schedule
2.13. ICS Assay
2.14. Antibody Measurements by Enzyme-Linked Immunosorbent Assay (ELISA)
2.15. Statistical Procedures
3. Results
3.1. Generation and In Vitro Characterization of MVA-B ΔA40R
3.2. A40 Was Non-Essential in Cell Culture
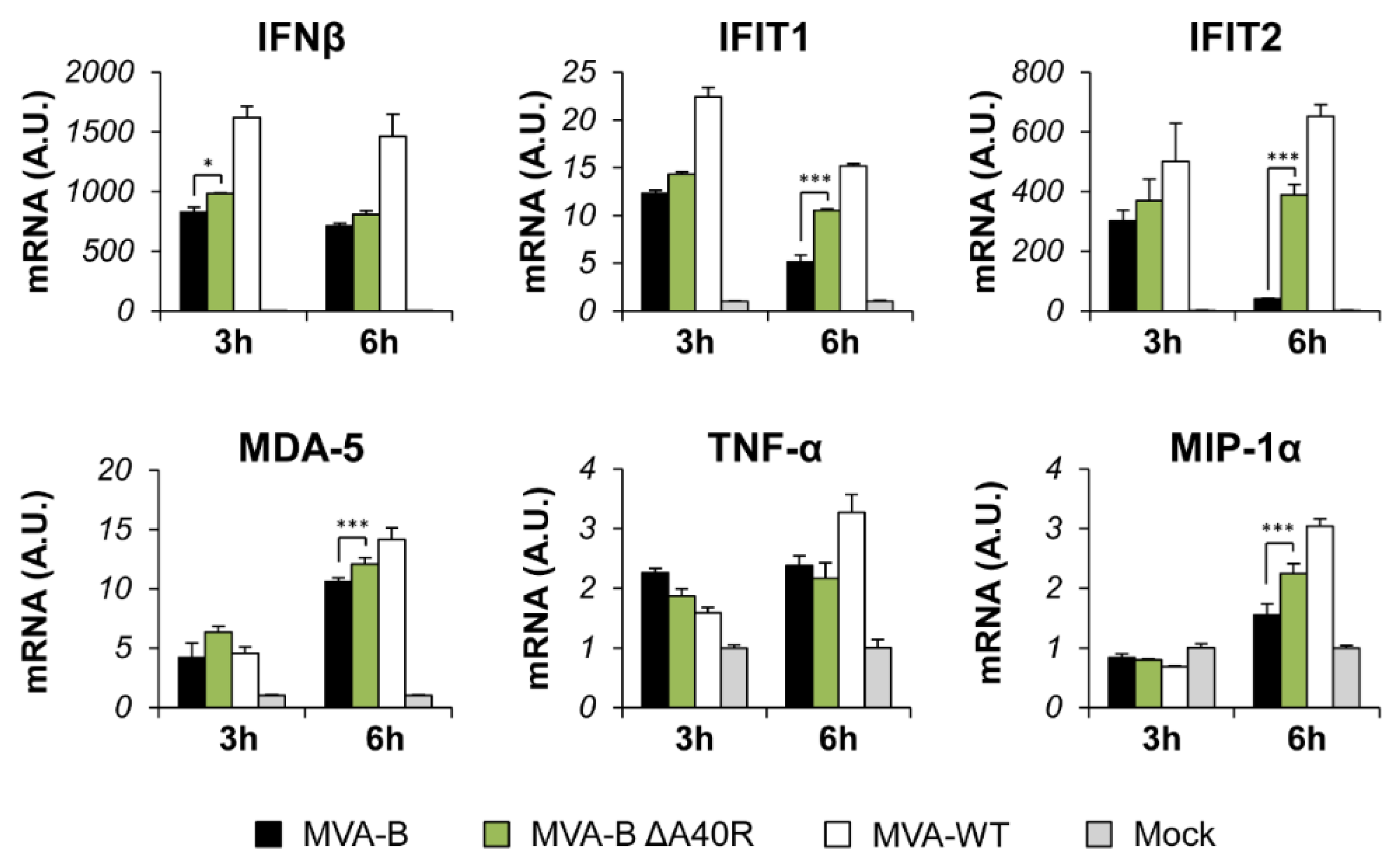
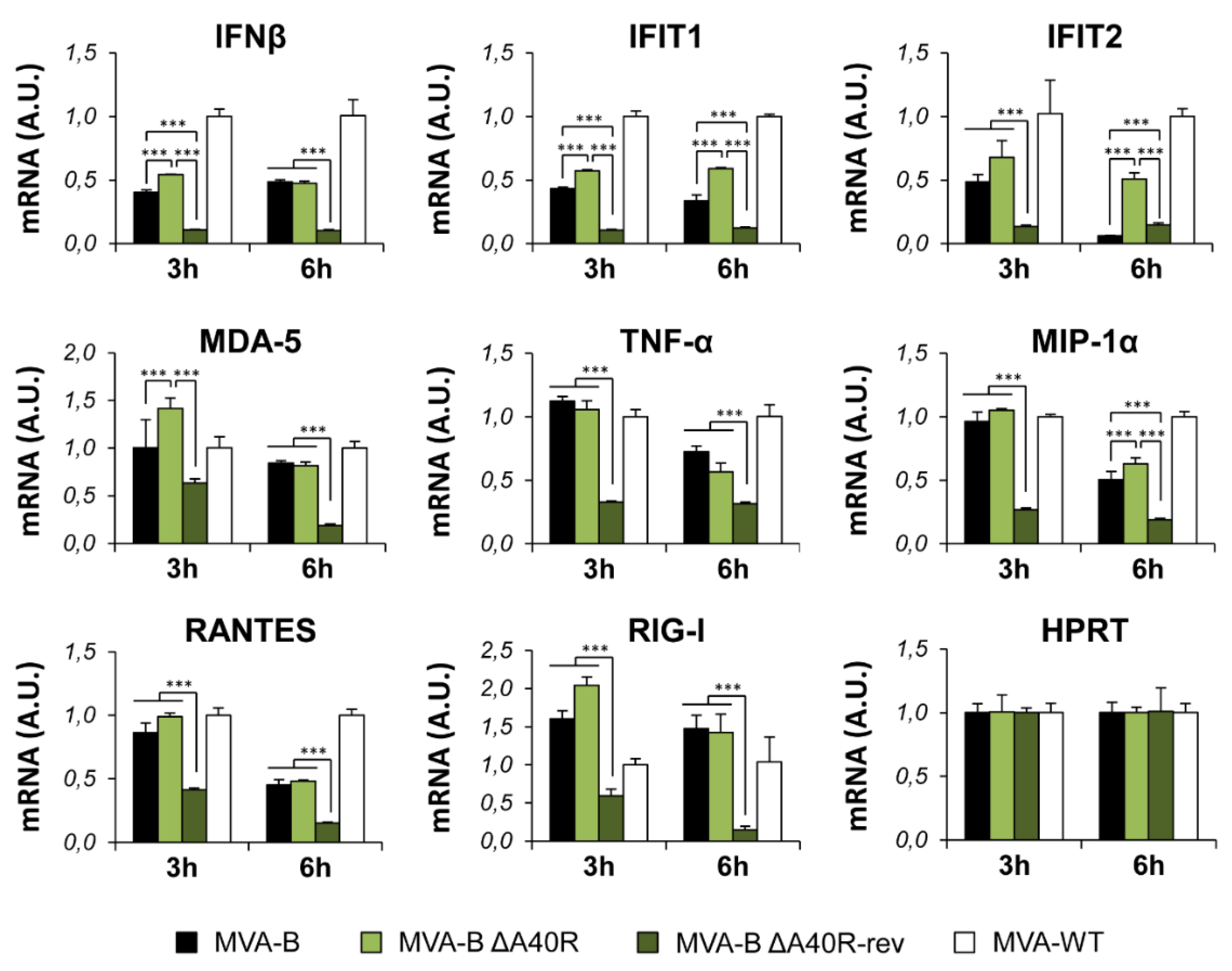
3.3. Deletion of MVA A40R Gene Enhanced the MVA-B Innate Immune Responses in Human Macrophages
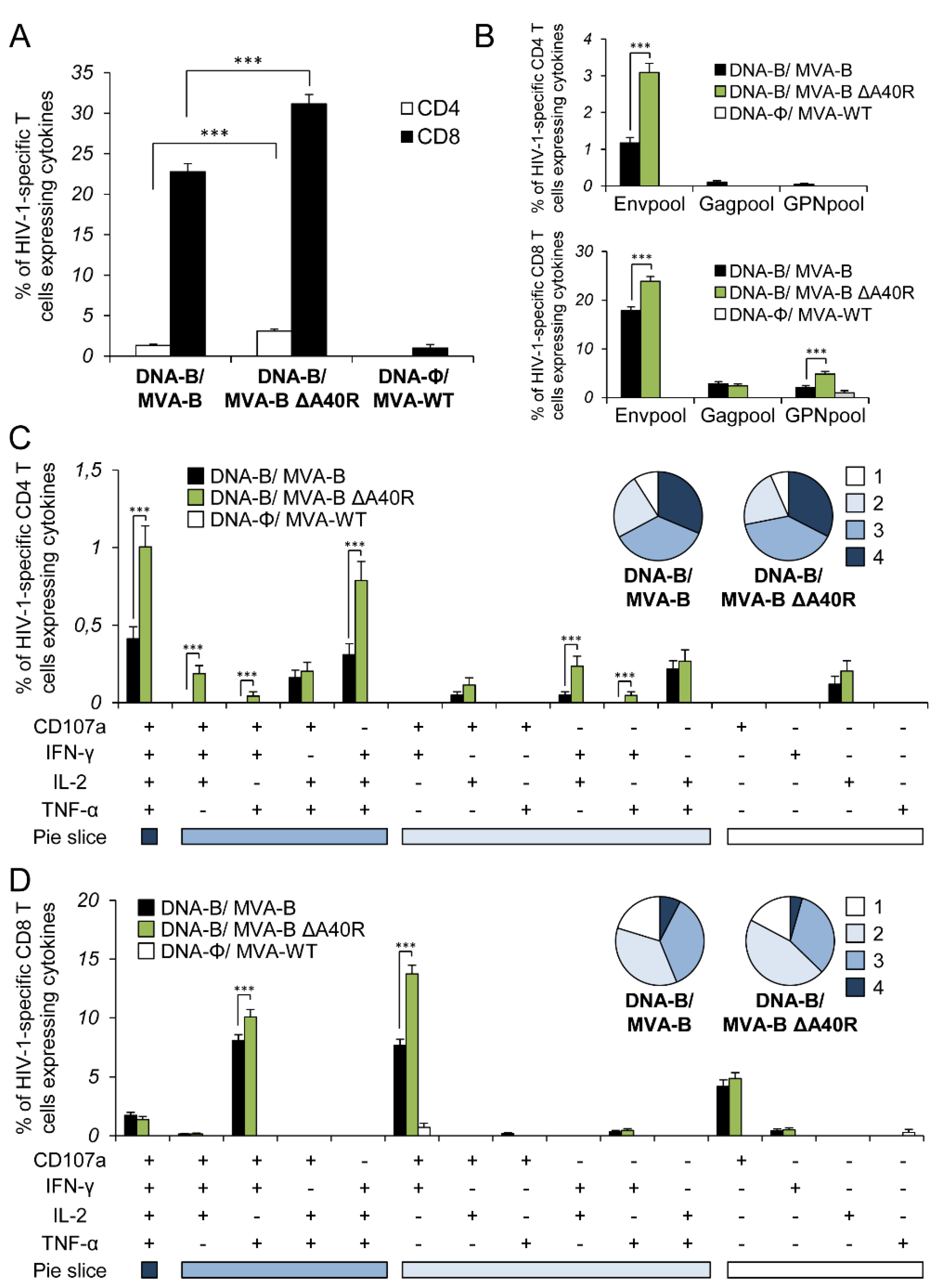
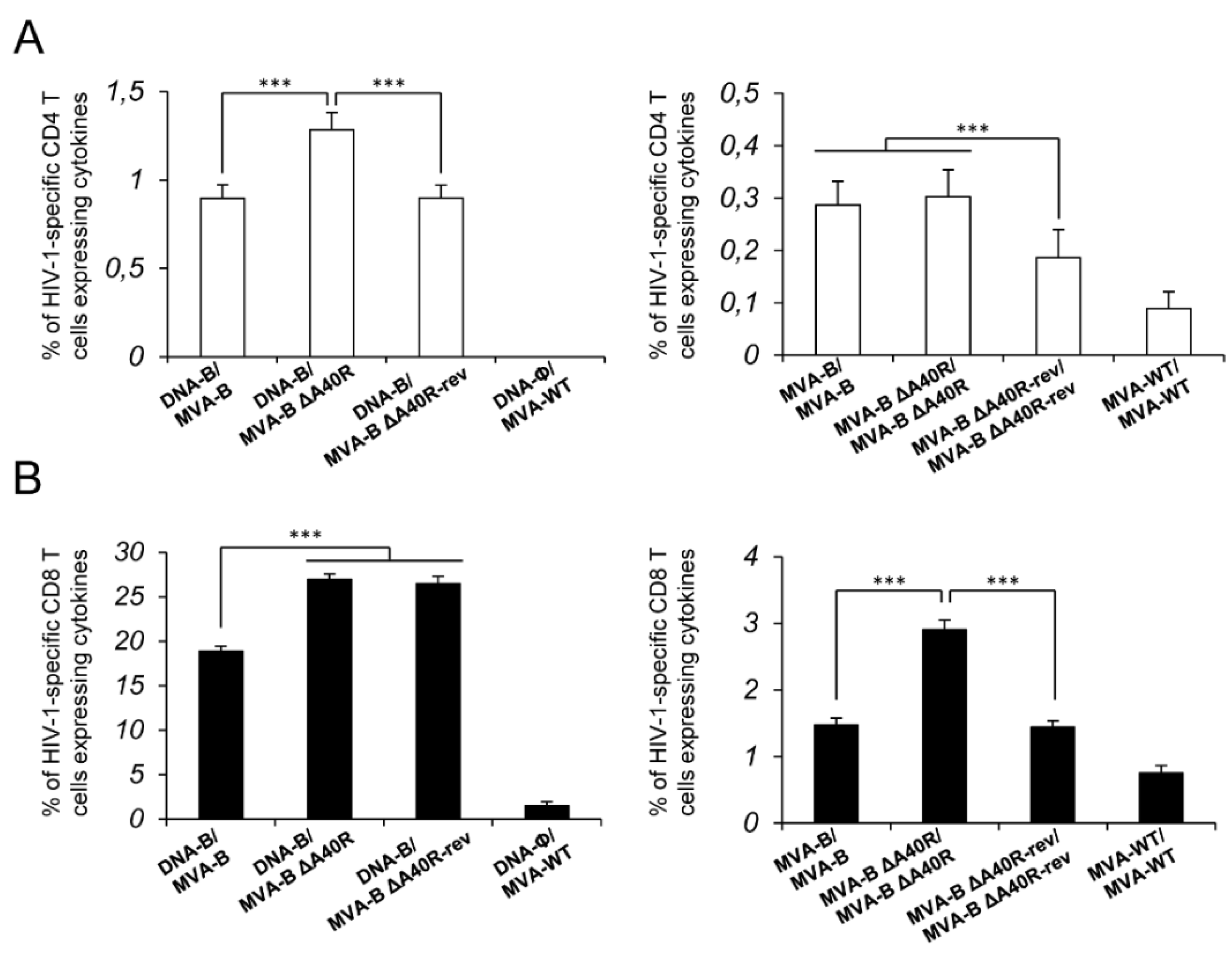
3.4. MVA-B ΔA40R Increased the Magnitude of HIV-1-Specific T-Cell Adaptive Immune Responses
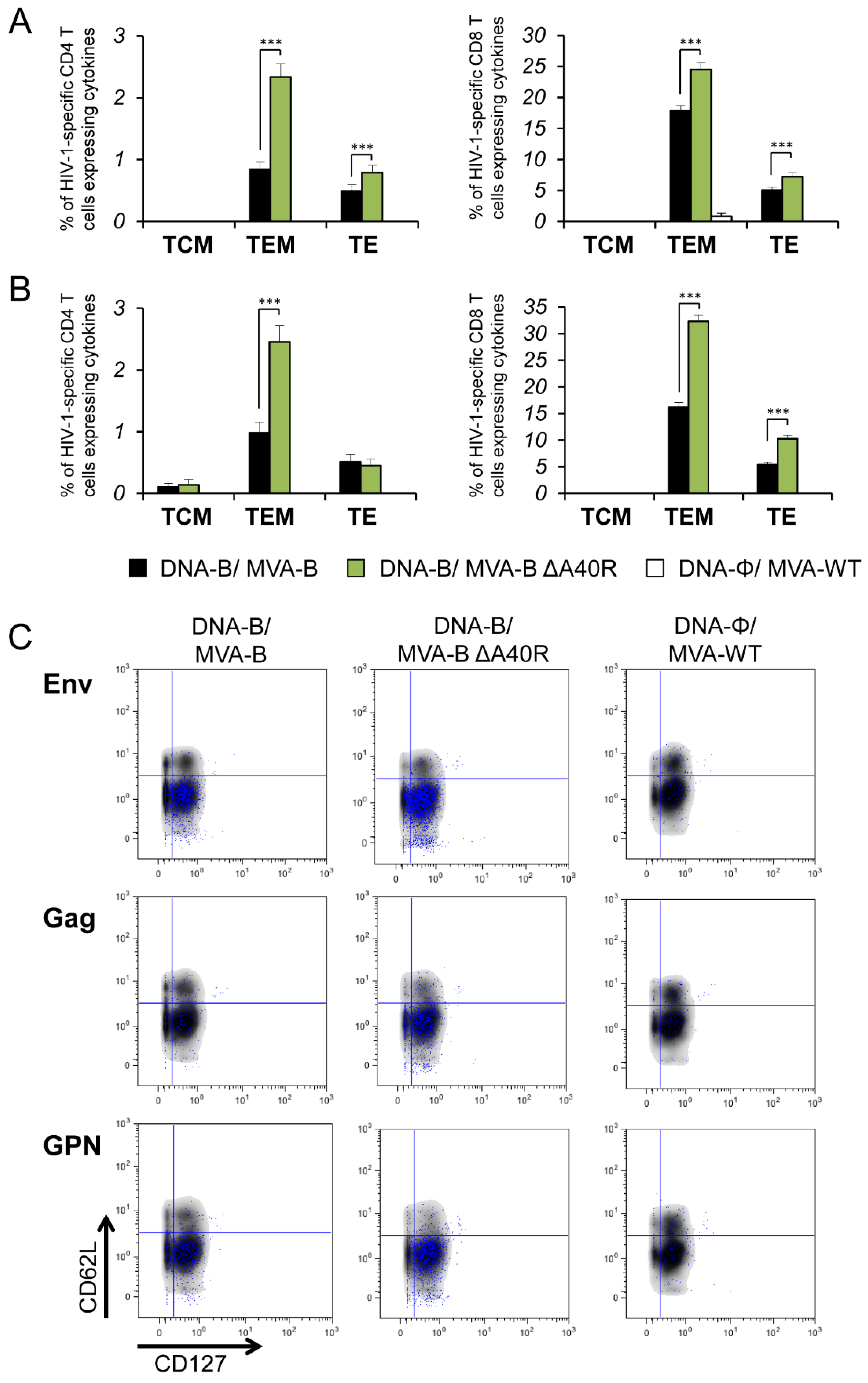
3.5. MVA-B ΔA40R Improved HIV-1-Specific T-Cell Memory Immune Responses
3.6. MVA-B ΔA40R Enhanced HIV-1-Specific T Cells with an Effector Memory Phenotype in the Adaptive and Memory Phases
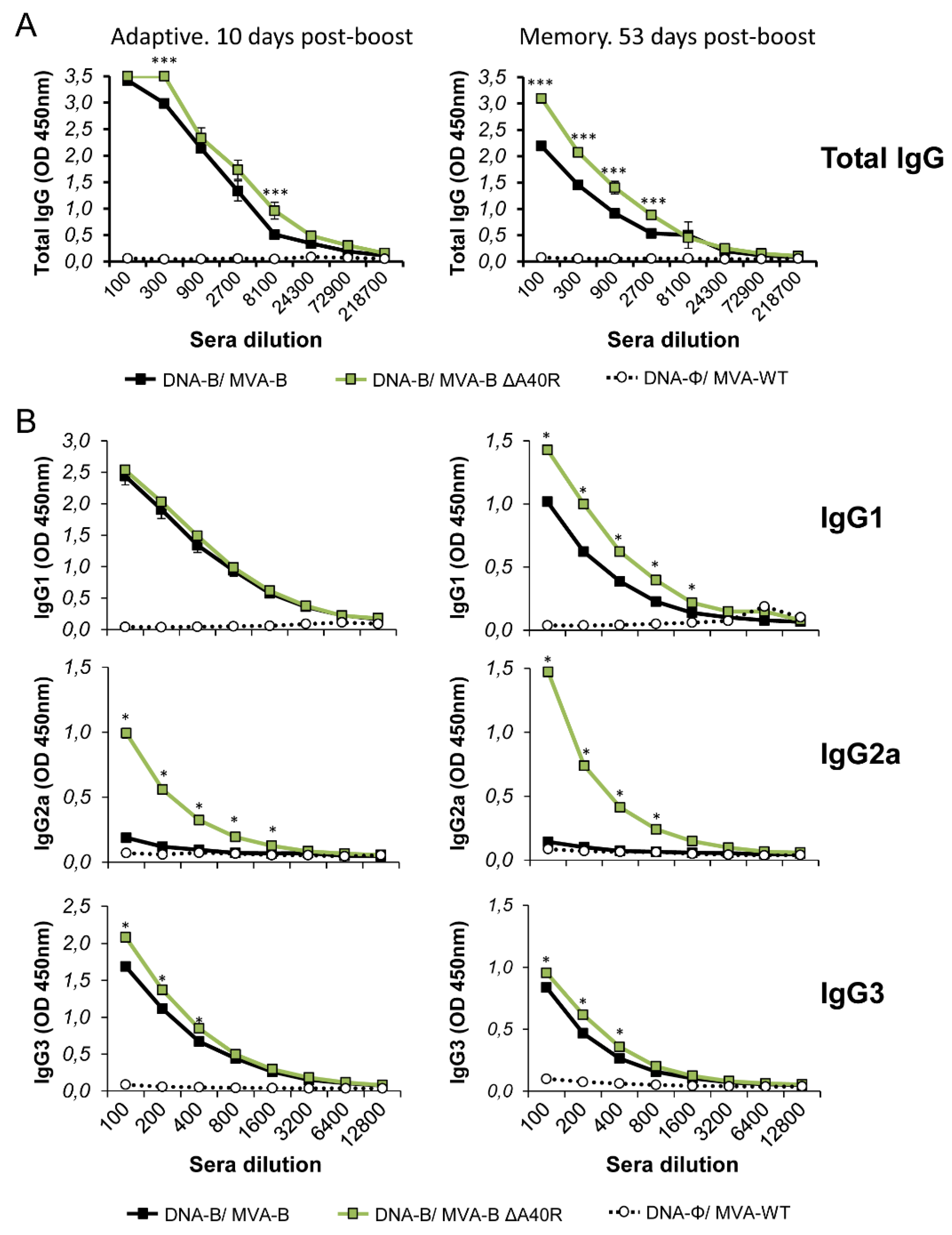
3.7. MVA-B ΔA40R Increased the Levels of Binding IgG Antibodies Against HIV-1 gp120
3.8. Generation of a Revertant MVA-B ΔA40R-rev Virus Expressing High Levels of MVA A40 Protein
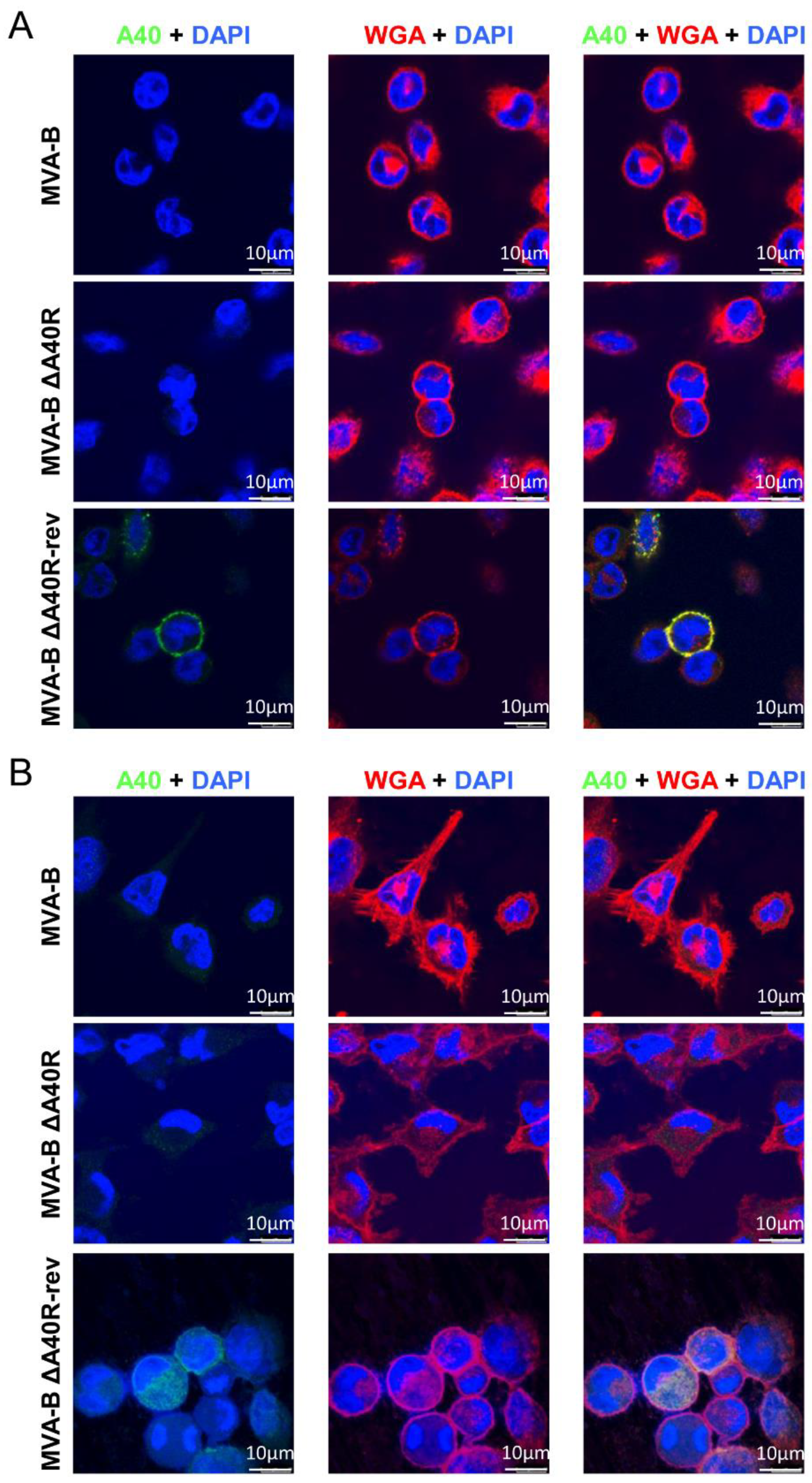
3.9. MVA A40 Protein Localized at the Cell Membrane
3.10. Reintroduction of MVA A40R Gene in MVA-B ΔA40R Inhibited Innate Immune Responses In Vitro
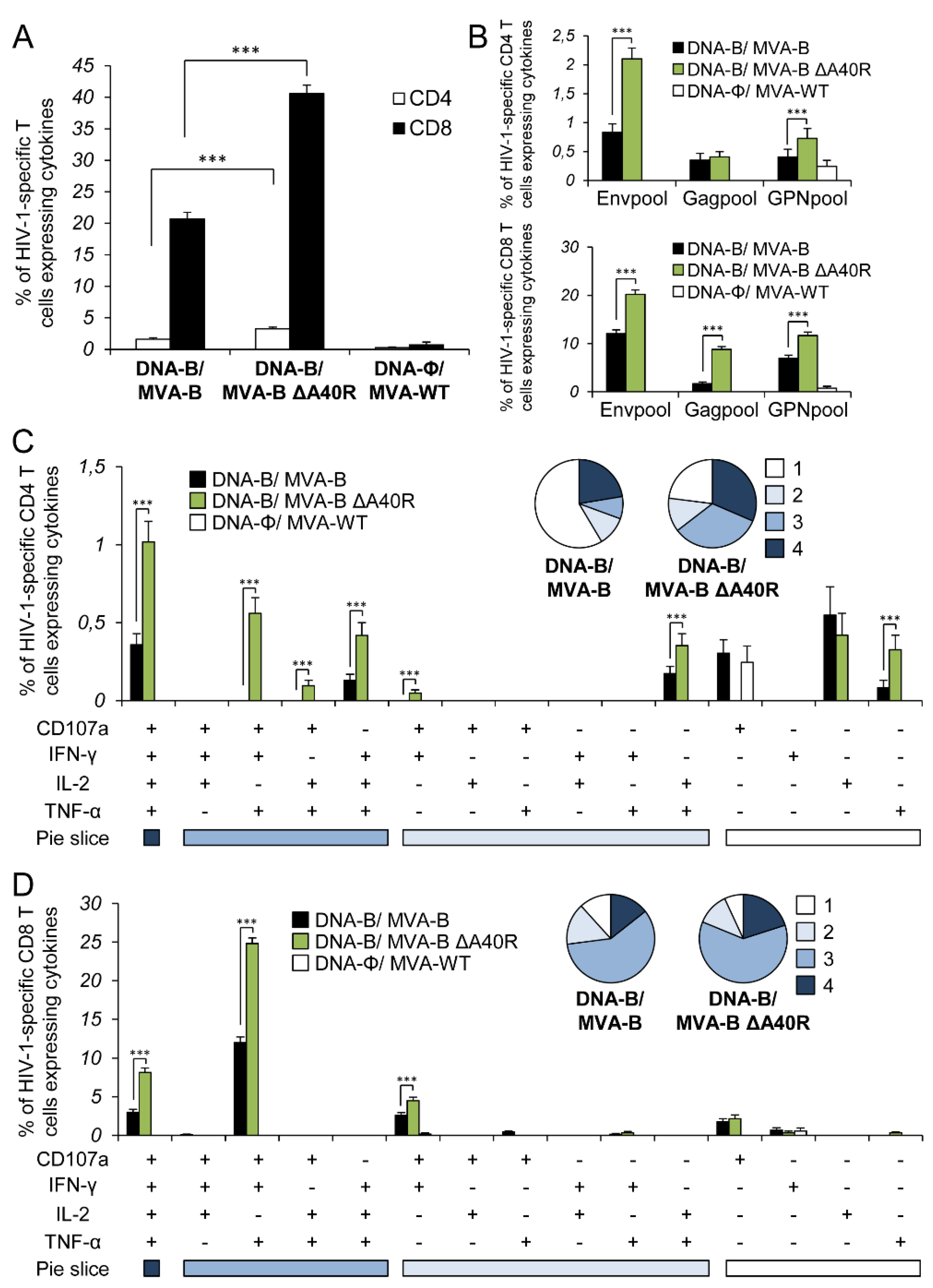
3.11. Reintroduction of MVA A40R Gene in MVA-B ΔA40R Impaired HIV-1-Specific T-Cell Immune Responses
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gao, Y.; McKay, P.F.; Mann, J.F.S. Advances in HIV-1 vaccine development. Viruses 2018, 10, 167. [Google Scholar] [CrossRef] [PubMed]
- Mcmichael, A.J.; Koff, W.C. Vaccines that stimulate T cell immunity to HIV-1: The next step. Nat. Immunol. 2014, 15, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Barouch, D.H.; Santra, S.; Schmitz, J.E.; Kuroda, M.J.; Fu, T.M.; Wagner, W.; Bilska, M.; Craiu, A.; Zheng, X.X.; Krivulka, G.R.; et al. Control of viremia prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 2000, 290, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Mooij, P.; Balla-Jhagjhoorsingh, S.S.; Koopman, G.; Beenhakker, N.; van Haaften, P.; Baak, I.; Nieuwenhuis, I.G.; Kondova, I.; Wagner, R.; Wolf, H.; et al. Differential CD4+ versus CD8+ T-cell responses elicited by different poxvirus-based human immunodeficiency virus Type 1 vaccine candidates provide comparable efficacies in Primates. J. Virol. 2008, 82, 2975–2988. [Google Scholar] [CrossRef]
- Amara, R.R.; Ibegbu, C.; Villinger, F.; Montefiori, D.C.; Sharma, S.; Nigam, P.; Xu, Y.; McClure, H.M.; Robinson, H.L. Studies using a viral challenge and CD8 T cell depletions on the roles of cellular and humoral immunity in the control of an SHIV-89.6P challenge in DNA/MVA-vaccinated macaques. Virology 2005, 343, 246–255. [Google Scholar] [CrossRef]
- Streeck, H.; D’souza, M.P.; Littman, D.R.; Crotty, S. Harnessing CD4+ T cell responses in HIV vaccine development. Nat. Med. 2013, 19, 143–149. [Google Scholar] [CrossRef]
- Buckheit, R.W.; Siliciano, R.F.; Blankson, J.N. Primary CD8+ T cells from elite suppressors effectively eliminate non-productively HIV-1 infected resting and activated CD4+ T cells. Retrovirology 2013, 10, 68. [Google Scholar] [CrossRef]
- Autran, B.; Carcelain, G.; Li, T.S.; Blanc, C.; Mathez, D.; Tubiana, R.; Katlama, C.; Debré, P.; Leibowitch, J. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science 1997, 277, 112–116. [Google Scholar] [CrossRef]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; de Souza, M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef]
- Esteban, M. Attenuated poxvirus vectors MVA and NYVAC as promising vaccine candidates against HIV/AIDS. Hum. Vaccin. 2009, 5, 867–871. [Google Scholar] [CrossRef]
- Gómez, C.E.; Nájera, J.L.; Krupa, M.; Perdiguero, B.; Esteban, M. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr. Gene Ther. 2011, 11, 189–217. [Google Scholar] [CrossRef] [PubMed]
- Gómez, C.E.; Perdiguero, B.; García-Arriaza, J.; Esteban, M. Clinical applications of attenuated MVA poxvirus strain. Expert Rev. Vaccines 2013, 12, 1395–1416. [Google Scholar] [CrossRef] [PubMed]
- Volz, A.; Sutter, G. Protective efficacy of modified Vaccinia virus Ankara in preclinical studies. Vaccine 2013, 31, 4235–4240. [Google Scholar] [CrossRef] [PubMed]
- Volz, A.; Sutter, G. Modified Vaccinia virus Ankara: History, value in basic research, and current perspectives for vaccine development. Adv. Virus. Res. 2017, 97, 187–243. [Google Scholar] [PubMed]
- Gilbert, S.C. Clinical development of modified Vaccinia virus Ankara vaccines. Vaccine 2013, 31, 4241–4246. [Google Scholar] [CrossRef]
- Gómez, C.E.; Nájera, J.L.; Jiménez, E.P.; Jiménez, V.; Wagner, R.; Graf, M.; Frachette, M.-J.J.; Liljeström, P.; Pantaleo, G.; Esteban, M. Head-to-head comparison on the immunogenicity of two HIV/AIDS vaccine candidates based on the attenuated poxvirus strains MVA and NYVAC co-expressing in a single locus the HIV-1BX08 gp120 and HIV-1IIIB Gag-Pol-Nef proteins of clade B. Vaccine 2007, 25, 2863–2885. [Google Scholar] [CrossRef]
- Vijayan, A.; García-Arriaza, J.; Raman, S.C.; Conesa, J.J.; Chichón, F.J.; Santiago, C.; Sorzano, C.Ó.S.; Carrascosa, J.L.; Esteban, M. A chimeric HIV-1 gp120 fused with Vaccinia virus 14K (A27) protein as an HIV immunogen. PLoS ONE 2015, 10, e0133595. [Google Scholar] [CrossRef]
- García-Arriaza, J.; Nájera, J.L.; Gómez, C.E.; Tewabe, N.; Sorzano, C.O.S.; Calandra, T.; Roger, T.; Esteban, M. A candidate HIV/AIDS vaccine (MVA-B) lacking Vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS ONE 2011, 6, e24244. [Google Scholar] [CrossRef]
- Climent, N.; Guerra, S.; García, F.; Rovira, C.; Miralles, L.; Gómez, C.E.; Piqué, N.; Gil, C.; Gatell, J.M.; Esteban, M.; et al. Dendritic cells exposed to MVA-based HIV-1 vaccine induce highly functional HIV-1-specific CD8 (+) T cell responses in HIV-1-infected individuals. PLoS ONE 2011, 6, e19644. [Google Scholar] [CrossRef]
- García-Arriaza, J.; Arnáez, P.; Gómez, C.E.; Sorzano, C.Ó.S.; Esteban, M. Improving adaptive and memory immune responses of an HIV/AIDS vaccine candidate MVA-B by deletion of Vaccinia virus genes (C6L and K7R) blocking interferon signaling pathways. PLoS ONE 2013, 8, e66894. [Google Scholar] [CrossRef]
- García-Arriaza, J.; Nájera, J.L.; Gómez, C.E.; Sorzano, C.O.S.; Esteban, M. Immunogenic profiling in mice of a HIV/AIDS vaccine candidate (MVA-B) expressing four HIV-1 antigens and potentiation by specific gene deletions. PLoS ONE 2010, 5, e12395. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, B.; Raman, S.C.; Sánchez-Corzo, C.; Sorzano, C.O.S.; Valverde, J.R.; Esteban, M.; Gómez, C.E. Potent HIV-1-specific CD8 T cell responses induced in mice after priming with a multiepitopic DNA-TMEP and boosting with the HIV vaccine MVA-B. Viruses 2018, 10, 424. [Google Scholar] [CrossRef] [PubMed]
- García-Arriaza, J.; Gomez, C.E.; Sorzano, C.O.S.; Esteban, M. Deletion of the Vaccinia virus N2L gene encoding an inhibitor of IRF3 improves the immunogenicity of modified Vaccinia virus Ankara expressing HIV-1 antigens. J. Virol. 2014, 88, 3392–3410. [Google Scholar] [CrossRef]
- Guerra, S.; González, J.M.; Climent, N.; Reyburn, H.; López-Fernández, L.A.; Nájera, J.L.; Gómez, C.E.; García, F.; Gatell, J.M.; Gallart, T.; et al. Selective induction of host genes by MVA-B, a candidate vaccine against HIV/AIDS. J. Virol. 2010, 84, 8141–8152. [Google Scholar] [CrossRef]
- Gómez, C.E.; Nájera, J.L.; Sánchez, R.; Jiménez, V.; Esteban, M. Multimeric soluble CD40 ligand (sCD40L) efficiently enhances HIV specific cellular immune responses during DNA prime and boost with attenuated poxvirus vectors MVA and NYVAC expressing HIV antigens. Vaccine 2009, 27, 3165–3174. [Google Scholar] [CrossRef] [PubMed]
- García, F.; Bernaldo de Quirós, J.C.L.; Gómez, C.E.; Perdiguero, B.; Nájera, J.L.; Jiménez, V.; García-Arriaza, J.; Guardo, A.C.; Pérez, I.; Díaz-Brito, V.; et al. Safety and immunogenicity of a modified pox vector-based HIV/AIDS vaccine candidate expressing Env, Gag, Pol and Nef proteins of HIV-1 subtype B (MVA-B) in healthy HIV-1-uninfected volunteers: A phase I clinical trial (RISVAC02). Vaccine 2011, 29, 8309–8316. [Google Scholar] [CrossRef] [PubMed]
- Gómez, C.E.; Nájera, J.L.; Perdiguero, B.; García-Arriaza, J.; Sorzano, C.O.S.; Jiménez, V.; González-Sanz, R.; Jiménez, J.L.; Muñoz-Fernández, M.A.; López Bernaldo de Quirós, J.C.; et al. The HIV/AIDS vaccine candidate MVA-B administered as a single immunogen in humans triggers robust, polyfunctional, and selective effector memory T cell responses to HIV-1 antigens. J. Virol. 2011, 85, 11468–11478. [Google Scholar] [CrossRef] [PubMed]
- Guardo, A.C.; Gómez, C.E.; Díaz-Brito, V.; Pich, J.; Arnaiz, J.A.; Perdiguero, B.; García-Arriaza, J.; González, N.; Sorzano, C.O.S.; Jiménez, L.; et al. Safety and vaccine-induced HIV-1 immune responses in healthy volunteers following a late MVA-B boost 4 years after the last immunization. PLoS ONE 2017, 12, e0186602. [Google Scholar] [CrossRef]
- Gómez, C.E.; Perdiguero, B.; García-Arriaza, J.; Cepeda, V.; Sorzano, C.Ó.S.; Mothe, B.; Jiménez, J.L.; Muñoz-Fernández, M.Á.; Gatell, J.M.; López Bernaldo de Quirós, J.C.; et al. A phase I randomized therapeutic MVA-B vaccination improves the magnitude and quality of the T cell immune responses in HIV-1-infected subjects on HAART. PLoS ONE 2015, 10, e0141456. [Google Scholar] [CrossRef]
- Mothe, B.; Climent, N.; Plana, M.; Rosas, M.; Jimenez, J.L.; Munoz-Fernandez, M.A.; Puertas, M.C.; Carrillo, J.; Gonzalez, N.; Leon, A.; et al. Safety and immunogenicity of a modified Vaccinia Ankara-based HIV-1 vaccine (MVA-B) in HIV-1-infected patients alone or in combination with a drug to reactivate latent HIV-1. J. Antimicrob. Chemother. 2015, 70, 1833–1842. [Google Scholar] [CrossRef]
- Rosás-Umbert, M.; Mothe, B.; Noguera-Julian, M.; Bellido, R.; Puertas, M.C.; Carrillo, J.; Rodriguez, C.; Perez-Alvarez, N.; Cobarsí, P.; Gomez, C.E.; et al. Virological and immunological outcome of treatment interruption in HIV-1-infected subjects vaccinated with MVA-B. PLoS ONE 2017, 12, e0184929. [Google Scholar] [CrossRef] [PubMed]
- García-Arriaza, J.; Arnáez, P.; Jiménez, J.L.; Gómez, C.E.; Muñoz-Fernández, M.Á.; Esteban, M. Vector replication and expression of HIV-1 antigens by the HIV/AIDS vaccine candidate MVA-B is not affected by HIV-1 protease inhibitors. Virus Res. 2012, 167, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Rallón, N.; Mothe, B.; Lopez Bernaldo de Quiros, J.C.; Plana, M.; Ligos, J.M.; Montoya, M.M.; Muñoz-Fernández, M.A.; Esteban, M.; Garcia, F.; Brander, C.; et al. Balance between activation and regulation of HIV-specific CD8+ T-cell response after modified Vaccinia Ankara B therapeutic vaccination. AIDS 2016, 30, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Vitallé, J.; Zenarruzabeitia, O.; Terrén, I.; Plana, M.; Guardo, A.C.; Leal, L.; Peña, J.; García, F.; Borrego, F. Monocytes phenotype and cytokine production in human immunodeficiency virus-1 infected patients receiving a modified Vaccinia ankara-based HIV-1 vaccine: Relationship to CD300 molecules expression. Front. Immunol. 2017, 8, 836. [Google Scholar] [CrossRef]
- García-Arriaza, J.; Esteban, M. Enhancing poxvirus vectors vaccine immunogenicity. Hum. Vaccin. Immunother. 2014, 10, 2235–2244. [Google Scholar] [CrossRef]
- Blanchard, T.J.; Alcamí, A.; Andrea, P.; Smith, G.L. Modified Vaccinia virus Ankara undergoes limited replication in human cells and lacks several immunomodulatory proteins: Implications for use as a human vaccine. J. Gen. Virol. 1998, 79, 1159–1167. [Google Scholar] [CrossRef]
- Antoine, G.; Scheiflinger, F.; Dorner, F.; Falkner, F.G. The complete genomic sequence of the modified Vaccinia Ankara strain: Comparison with other orthopoxviruses. Virology 1998, 244, 365–396. [Google Scholar] [CrossRef]
- Mayr, A.; Stickl, H.; Müller, H.K.; Danner, K.; Singer, H. The smallpox vaccination strain MVA: Marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism. Zentralbl. Bakteriol. B. 1978, 167, 375–390. [Google Scholar]
- Smith, G.L.; Benfield, C.T.O.; Maluquer de Motes, C.; Mazzon, M.; Ember, S.W.J.; Ferguson, B.J.; Sumner, R.P. Vaccinia virus immune evasion: Mechanisms, virulence and immunogenicity. J. Gen. Virol. 2013, 94, 2367–2392. [Google Scholar] [CrossRef]
- Albarnaz, J.D.; Torres, A.A.; Smith, G.L. Modulating Vaccinia virus immunomodulators to improve immunological memory. Viruses 2018, 10, 101. [Google Scholar] [CrossRef]
- García-Arriaza, J.; Cepeda, V.; Hallengärd, D.; Sorzano, C.Ó.S.; Kümmerer, B.M.; Liljeström, P.; Esteban, M. A novel Poxvirus-based vaccine, MVA-CHIKV, is highly immunogenic and protects mice against Chikungunya Infection. J. Virol. 2014, 88, 3527–3547. [Google Scholar] [CrossRef] [PubMed]
- Lázaro-Frías, A.; Gómez-Medina, S.; Sánchez-Sampedro, L.; Ljungberg, K.; Ustav, M.; Liljeström, P.; Muñoz-Fontela, C.; Esteban, M.; García-Arriaza, J. Distinct immunogenicity and efficacy of Poxvirus-based vaccine candidates against Ebola virus expressing GP and VP40 proteins. J. Virol. 2018, 92, e00363-18. [Google Scholar] [CrossRef] [PubMed]
- Pérez, P.; Marín, M.Q.; Lázaro-Frías, A.; Jiménez de Oya, N.; Blázquez, A.B.; Escribano-Romero, E.; Sorzano, C.Ó.S.; Ortego, J.; Saiz, J.C.; Esteban, M.; et al. A vaccine based on a modified Vaccinia virus Ankara vector expressing Zika virus structural proteins controls Zika virus replication in mice. Sci. Rep. 2018, 8, 17385. [Google Scholar] [PubMed]
- Wilcock, D.; Duncan, S.A.; Traktman, P.; Zhang, W.-H.H.; Smith, G.L. The Vaccinia virus A4OR gene product is a nonstructural, type II membrane glycoprotein that is expressed at the cell surface. J. Gen. Virol. 1999, 80, 2137–2148. [Google Scholar] [CrossRef]
- Mayer, S.; Raulf, M.-K.; Lepenies, B. C-type lectins: Their network and roles in pathogen recognition and immunity. Histochem. Cell Biol. 2017, 147, 223–237. [Google Scholar] [CrossRef]
- Tscharke, D.C.; Reading, P.C.; Smith, G.L. Dermal infection with Vaccinia virus reveals roles for virus proteins not seen using other inoculation routes. J. Gen. Virol. 2002, 83, 1977–1986. [Google Scholar] [CrossRef]
- Schramm, B.; Locker, J.K. Cytoplasmic organization of POXvirus DNA replication. Traffic 2005, 6, 839–846. [Google Scholar] [CrossRef]
- Palacios, S.; Perez, L.H.; Welsch, S.; Schleich, S.; Chmielarska, K.; Melchior, F.; Locker, J.K. Quantitative SUMO-1 modification of a Vaccinia virus protein is required for its specific localization and prevents its self-association. Mol. Biol. Cell 2005, 16, 2822–2835. [Google Scholar] [CrossRef]
- Ramírez, J.C.; Gherardi, M.M.; Esteban, M. Biology of attenuated modified Vaccinia virus Ankara recombinant vector in mice: Virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 2000, 74, 923–933. [Google Scholar] [CrossRef]
- Marín, M.Q.; Pérez, P.; Gómez, C.E.; Sorzano, C.Ó.S.; Esteban, M.; García-Arriaza, J. Removal of the C6 Vaccinia virus interferon-β inhibitor in the Hepatitis C vaccine candidate MVA-HCV elicited in mice high immunogenicity in spite of reduced host gene expression. Viruses 2018, 10, 414. [Google Scholar] [CrossRef]
- Delaloye, J.; Roger, T.; Steiner-Tardivel, Q.G.; Le Roy, D.; Reymond, M.K.; Akira, S.; Petrilli, V.; Gomez, C.E.; Perdiguero, B.; Tschopp, J.; et al. Innate immune sensing of modified Vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 2009, 5, e1000480. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nájera, J.L.; Gómez, C.E.; García-Arriaza, J.; Sorzano, C.O.S.; Esteban, M. Insertion of Vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS ONE 2010, 5, e11406. [Google Scholar] [CrossRef]
- Najera, J.L.; Gomez, C.E.; Domingo-Gil, E.; Gherardi, M.M.; Esteban, M. Cellular and biochemical differences between two attenuated poxvirus vaccine candidates (MVA and NYVAC) and role of the C7L gene. J. Virol. 2006, 80, 6033–6047. [Google Scholar] [CrossRef] [PubMed]
- Platt, A.; Wetzler, L. Innate immunity and vaccines. Curr. Top. Med. Chem. 2013, 13, 2597–2608. [Google Scholar] [CrossRef] [PubMed]
- Price, P.J.R.R.; Torres-Domínguez, L.E.; Brandmüller, C.; Lehmann, M.H.; Sutter, G.; Lehmann, M.H. Modified Vaccinia virus ankara: Innate immune activation and induction of cellular signalling. Vaccine 2013, 31, 4231–4234. [Google Scholar] [CrossRef] [PubMed]
- Seder, R.A.; Darrah, P.A.; Roederer, M. T-cell quality in memory and protection: Implications for vaccine design. Nat. Rev. Immunol. 2008, 8, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Sallusto, F.; Lenig, D.; Förster, R.; Lipp, M.; Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999, 401, 708–712. [Google Scholar] [CrossRef]
- Sallusto, F.; Geginat, J.; Lanzavecchia, A. Central memory and effector memory T cell subsets: Function, generation, and maintenance. Annu. Rev. Immunol. 2004, 22, 745–763. [Google Scholar] [CrossRef]
- Champagne, P.; Ogg, G.S.; King, A.S.; Knabenhans, C.; Ellefsen, K.; Nobile, M.; Appay, V.; Rizzardi, G.P.; Fleury, S.; Lipp, M.; et al. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 2001, 410, 106–111. [Google Scholar] [CrossRef]
- Ghiglione, Y.; Falivene, J.; Ruiz, M.J.; Laufer, N.; Socías, M.E.; Cahn, P.; Giavedoni, L.; Sued, O.; Gherardi, M.M.; Salomón, H.; et al. Early skewed distribution of total and HIV-specific CD8+ T-cell memory phenotypes during primary HIV infection is related to reduced antiviral activity and faster disease progression. PLoS ONE 2014, 9, e104235. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.A.; Piriou, E.; Bronke, C.; Vingerhoed, J.; Kostense, S.; Van Baarle, D.; Miedema, F. Characterization of virus-specific CD8+ effector T cells in the course of HIV-1 infection: Longitudinal analyses in slow and rapid progressors. Clin. Immunol. 2004, 113, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Addo, M.M.; Draenert, R.; Rathod, A.; Verrill, C.L.; Davis, B.T.; Gandhi, R.T.; Robbins, G.K.; Basgoz, N.O.; Stone, D.R.; Cohen, D.E.; et al. Fully differentiated HIV-1 specific CD8+ T effector cells are more frequently detectable in controlled than in progressive HIV-1 infection. PLoS ONE 2007, 2, e321. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Wolint, P.; Schwarz, K.; Oxenius, A. Recall proliferation potential of memory CD8+ T cells and antiviral protection. J. Immunol. 2005, 175, 4677–4685. [Google Scholar] [CrossRef] [PubMed]
- McElrath, M.J.; Haynes, B.F. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity 2010, 33, 542–554. [Google Scholar] [CrossRef]
- Asbach, B.; Kibler, K.V.; Köstler, J.; Perdiguero, B.; Yates, N.L.; Stanfield-Oakley, S.; Tomaras, G.D.; Kao, S.-F.; Foulds, K.E.; Roederer, M.; et al. Priming with a potent HIV-1 DNA vaccine frames the quality of immune responses prior to a Poxvirus and protein boost. J. Virol. 2019, 93, e01529-18. [Google Scholar] [CrossRef]
- Gómez, C.E.; Perdiguero, B.; García-Arriaza, J.; Esteban, M. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum. Vaccines Immunother. 2012, 8, 1192–1207. [Google Scholar] [CrossRef]
- Pantaleo, G.; Esteban, M.; Jacobs, B.; Tartaglia, J. Poxvirus vector-based HIV vaccines. Curr. Opin. HIV AIDS 2010, 5, 391–396. [Google Scholar] [CrossRef]
- Falivene, J.; Del Mé Zajac, M.P.; Pascutti, M.F.; Rodríguez, A.M.; Maeto, C.; Perdiguero, B.; Gómez, C.E.; Esteban, M.; Calamante, G.; Gherardi, M.M. Improving the MVA vaccine potential by deleting the viral gene coding for the IL-18 binding protein. PLoS ONE 2012, 7, e32220. [Google Scholar] [CrossRef]
- Perdiguero, B.; Gómez, C.E.; Nájera, J.L.; Sorzano, C.O.S.; Delaloye, J.; González-Sanz, R.; Jiménez, V.; Roger, T.; Calandra, T.; Pantaleo, G.; et al. Deletion of the viral anti-apoptotic gene F1L in the HIV/AIDS vaccine candidate MVA-C enhances immune responses against HIV-1 antigens. PLoS ONE 2012, 7, e48524. [Google Scholar] [CrossRef]
- Garber, D.A.; O’Mara, L.A.; Gangadhara, S.; McQuoid, M.; Zhang, X.; Zheng, R.; Gill, K.; Verma, M.; Yu, T.; Johnson, B.; et al. Deletion of specific immune-modulatory genes from modified Vaccinia virus Ankara-based HIV vaccines engenders improved immunogenicity in Rhesus Macaques. J. Virol. 2012, 86, 12605–12615. [Google Scholar] [CrossRef] [PubMed]
- Holgado, M.P.; Falivene, J.; Maeto, C.; Amigo, M.; Pascutti, M.F.; Vecchione, M.B.; Bruttomesso, A.; Calamante, G.; Del Médico-Zajac, M.P.; Gherardi, M.M. Deletion of A44L, A46R and C12L Vaccinia virus genes from the MVA genome improved the vector immunogenicity by modifying the innate immune response generating enhanced and optimized specific T-cell responses. Viruses 2016, 8, 139. [Google Scholar] [CrossRef] [PubMed]
- Altfeld, M.; Gale, M., Jr. Innate immunity against HIV-1 infection. Nat. Immunol. 2015, 16, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Gómez, C.E.; Perdiguero, B.; Sánchez-Corzo, C.; Sorzano, C.O.S.; Esteban, M. Immune modulation of NYVAC-based HIV vaccines by combined deletion of viral genes that act on several signalling pathways. Viruses 2018, 10, 7. [Google Scholar] [CrossRef]
- Perdiguero, B.; Gómez, C.E.; Di Pilato, M.; Sorzano, C.O.S.; Delaloye, J.; Roger, T.; Calandra, T.; Pantaleo, G.; Esteban, M. Deletion of the Vaccinia virus gene A46R, encoding for an inhibitor of TLR signalling, is an effective approach to enhance the immunogenicity in mice of the HIV/AIDS vaccine candidate NYVAC-C. PLoS ONE 2013, 8, e74831. [Google Scholar] [CrossRef]
- Gomez, C.E.; Perdiguero, B.; Najera, J.L.; Sorzano, C.O.S.; Jimenez, V.; Gonzalez-Sanz, R.; Esteban, M. Removal of Vaccinia virus genes that block interferon type I and II pathways improves adaptive and memory responses of the HIV/AIDS vaccine candidate NYVAC-C in mice. J. Virol. 2012, 86, 5026–5038. [Google Scholar] [CrossRef]
- Di Pilato, M.; Mejías-Pérez, E.; Zonca, M.; Perdiguero, B.; Gómez, C.E.; Trakala, M.; Nieto, J.; Nájera, J.L.; Sorzano, C.O.S.; Combadière, C.; et al. NFκB activation by modified Vaccinia virus as a novel strategy to enhance neutrophil migration and HIV-specific T-cell responses. Proc. Natl. Acad. Sci. USA 2015, 112, E1333–E1342. [Google Scholar] [CrossRef]
- Di Pilato, M.; Mejías-Pérez, E.; Sorzano, C.O.S.; Esteban, M. Distinct roles of Vaccinia virus NF-κB inhibitor proteins A52, B15, and K7 in the immune response. J. Virol. 2017, 91, e00575-17. [Google Scholar] [CrossRef]
- Hansen, S.G.; Ford, J.C.; Lewis, M.S.; Ventura, A.B.; Hughes, C.M.; Coyne-Johnson, L.; Whizin, N.; Oswald, K.; Shoemaker, R.; Swanson, T.; et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 2011, 473, 523–527. [Google Scholar] [CrossRef]
- Hansen, S.G.; Vieville, C.; Whizin, N.; Coyne-Johnson, L.; Siess, D.C.; Drummond, D.D.; Legasse, A.W.; Axthelm, M.K.; Oswald, K.; Trubey, C.M.; et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 2009, 15, 293–299. [Google Scholar] [CrossRef]
- Haynes, B.F.; Gilbert, P.B.; Mcelrath, M.J.; Zolla-Pazner, S.; Tomaras, G.D.; Alam, S.M.; Evans, D.T.; Montefiori, D.C.; Karnasuta, C.; Sutthent, R.; et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012, 366, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Vicenzi, E.; Panina-Bodignon, P.; Vallanti, G.; Di Lucia, P.; Poli, G. Restricted replication of primary HIV-1 isolates using both CCR5 and CXCR4 in Th2 but not in Th1 CD4 (+) T cells. J. Leukoc. Biol. 2002, 72, 913–920. [Google Scholar] [PubMed]
- Ofori, H.; Jagodziński, P.P. Increased in vitro replication of CC chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates in Th2 lymphocytes may correlate with AIDS progression. Scand. J. Infect. Dis. 2004, 36, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Moonis, M.; Lee, B.; Bailer, R.T.; Luo, Q.; Montaner, L.J. CCR5 and CXCR4 expression correlated with X4 and R5 HIV-1 infection yet not sustained replication in Th1 and Th2 cells. AIDS 2001, 15, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- Maggi, E.; Mazzetti, M.; Ravina, A.; Annunziato, F.; De Carli, M.; Piccinni, M.P.; Manetti, R.; Carbonari, M.; Pesce, A.M.; Del Prete, G.; et al. Ability of HIV to promote a TH1 to TH0 shift and to replicate preferentially in TH2 and TH0 cells. Science 1994, 265, 244–248. [Google Scholar] [CrossRef]
- Clerici, M.; Shearer, G.M. A TH1→TH2 switch is a critical step in the etiology of HIV infection. Immunol. Today 1993, 14, 107–111. [Google Scholar] [CrossRef]
- Klein, S.A.; Dobmeyer, J.M.; Dobmeyer, T.S.; Pape, M.; Ottmann, O.G.; Helm, E.B.; Hoelzer, D.; Rossol, R. Demonstration of the Th1 to Th2 cytokine shift during the course of HIV-1 infection using cytoplasmic cytokine detection on single cell level by flow cytometry. AIDS 1997, 11, 1111–1118. [Google Scholar] [CrossRef]
- Auclair, S.; Liu, F.; Niu, Q.; Hou, W.; Churchyard, G.; Morgan, C.; Frahm, N.; Nitayaphan, S.; Pitisuthithum, P.; Rerks-Ngarm, S.; et al. Distinct susceptibility of HIV vaccine vector-induced CD4 T cells to HIV infection. PLoS Pathog. 2018, 14, e1006888. [Google Scholar] [CrossRef]
- Antoine, G.; Scheiflinger, F.; Holzer, G.; Langmann, T.; Falkner, F.G.; Dorner, F. Characterization of the Vaccinia MVA hemagglutinin gene locus and its evaluation as an insertion site for foreign genes. Gene 1996, 177, 43–46. [Google Scholar] [CrossRef]
- Pérez-Jiménez, E.; Kochan, G.; Gherardi, M.M.; Esteban, M. MVA-LACK as a safe and efficient vector for vaccination against leishmaniasis. Microbes Infect. 2006, 8, 810–822. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez, P.; Marín, M.Q.; Lázaro-Frías, A.; Sorzano, C.Ó.S.; Gómez, C.E.; Esteban, M.; García-Arriaza, J. Deletion of Vaccinia Virus A40R Gene Improves the Immunogenicity of the HIV-1 Vaccine Candidate MVA-B. Vaccines 2020, 8, 70. https://doi.org/10.3390/vaccines8010070
Pérez P, Marín MQ, Lázaro-Frías A, Sorzano CÓS, Gómez CE, Esteban M, García-Arriaza J. Deletion of Vaccinia Virus A40R Gene Improves the Immunogenicity of the HIV-1 Vaccine Candidate MVA-B. Vaccines. 2020; 8(1):70. https://doi.org/10.3390/vaccines8010070
Chicago/Turabian StylePérez, Patricia, María Q. Marín, Adrián Lázaro-Frías, Carlos Óscar S. Sorzano, Carmen E. Gómez, Mariano Esteban, and Juan García-Arriaza. 2020. "Deletion of Vaccinia Virus A40R Gene Improves the Immunogenicity of the HIV-1 Vaccine Candidate MVA-B" Vaccines 8, no. 1: 70. https://doi.org/10.3390/vaccines8010070
APA StylePérez, P., Marín, M. Q., Lázaro-Frías, A., Sorzano, C. Ó. S., Gómez, C. E., Esteban, M., & García-Arriaza, J. (2020). Deletion of Vaccinia Virus A40R Gene Improves the Immunogenicity of the HIV-1 Vaccine Candidate MVA-B. Vaccines, 8(1), 70. https://doi.org/10.3390/vaccines8010070





