Development of Universal Influenza Vaccines Targeting Conserved Viral Proteins
Abstract
1. Introduction
2. Commercial Influenza Vaccines
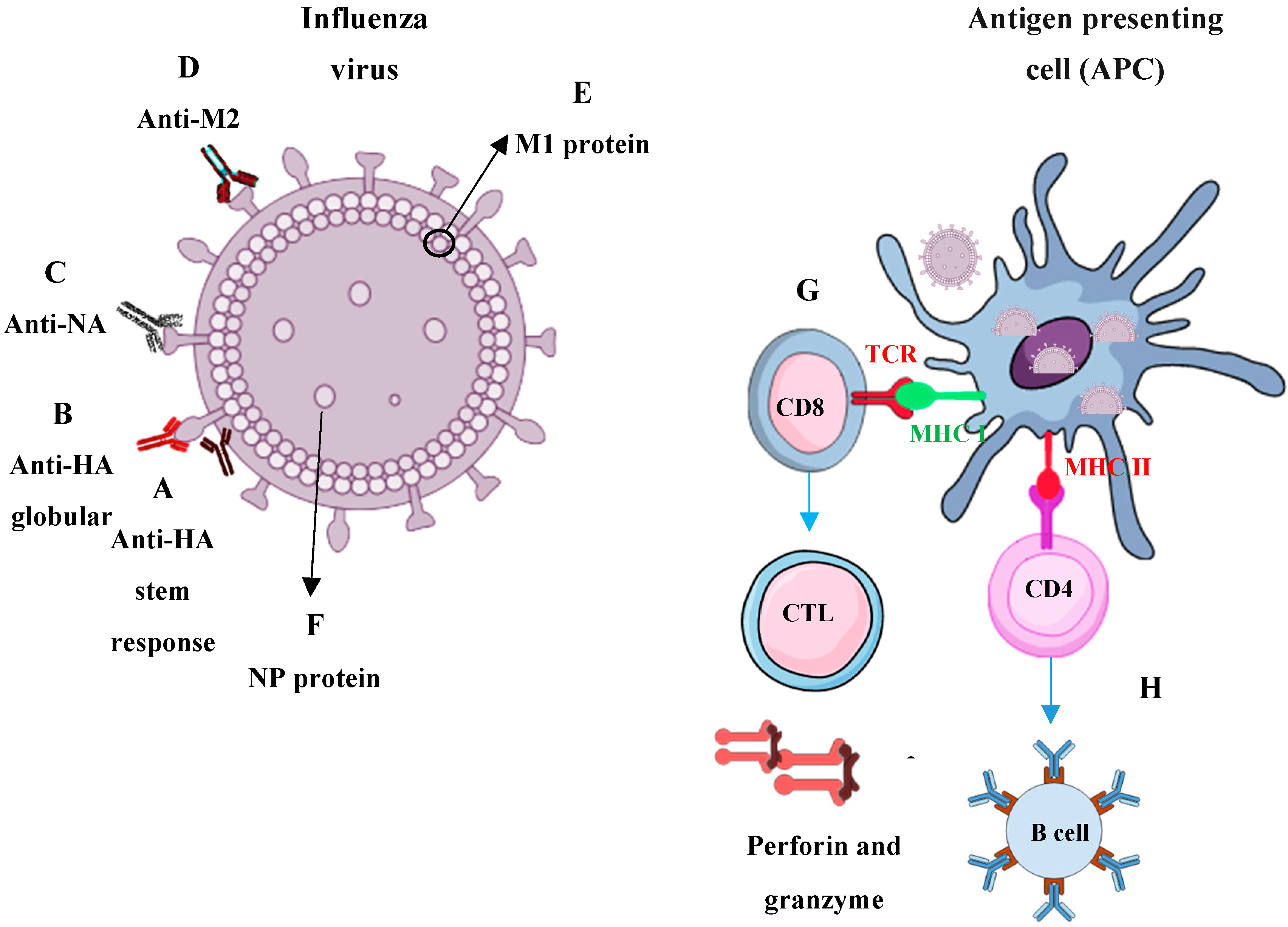
3. Immune Responses Required for Universal Influenza Protection
4. Potential Universal Vaccine Platforms
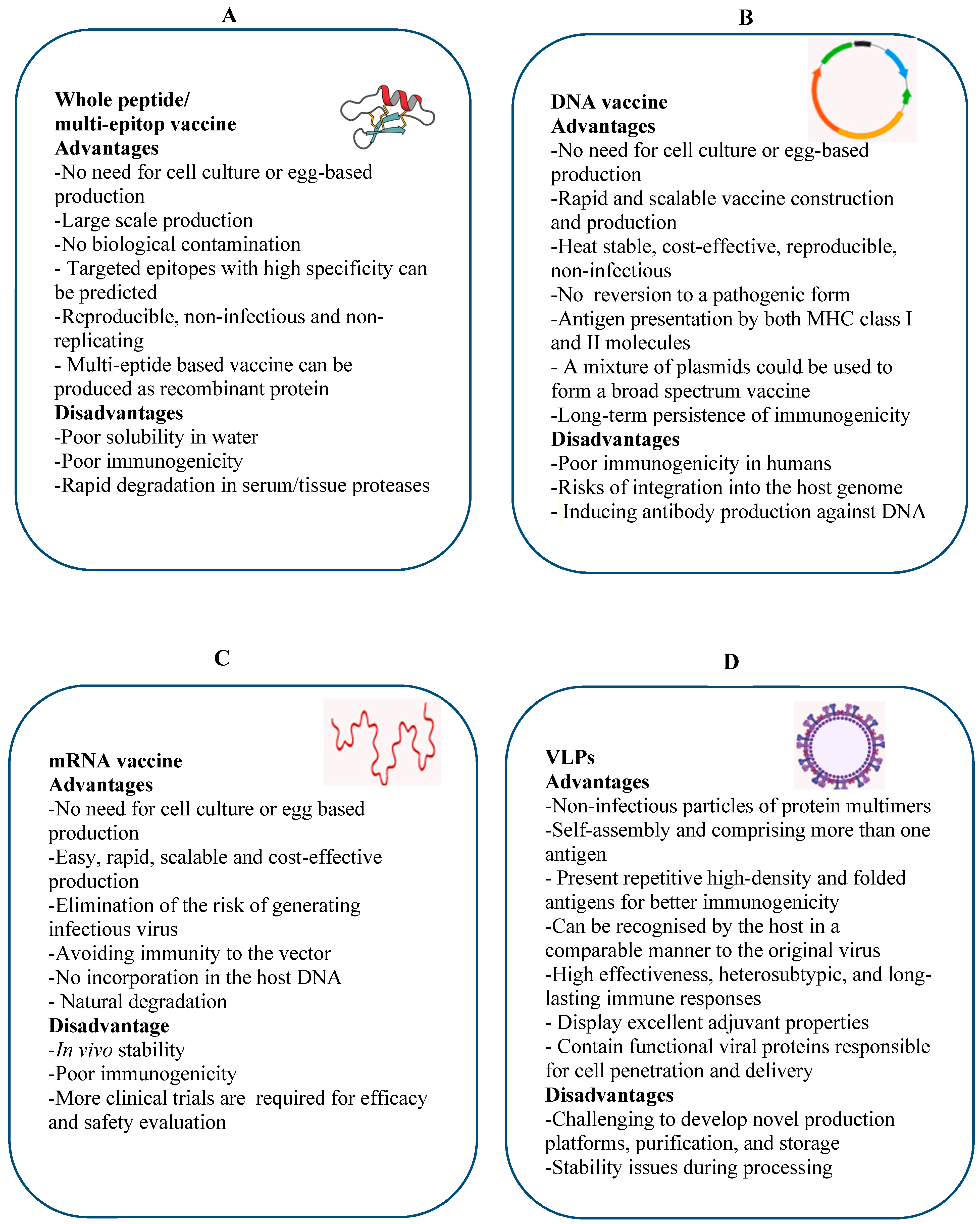
4.1. Multi Epitope-Based Vaccine
4.1.1. Recombinant Epitope-Based Vaccine
4.1.2. Recombinant Vectored Epitope-Based Vaccine
4.2. DNA-Based Vaccine
4.3. mRNA-Based Vaccines
4.4. Virus-Like Particles (VLPs)
5. Nanotechnology and Influenza Vaccine Development
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Palese, P. Influenza: Old and new threats. Nat. Med. 2004, 10, S82–S87. [Google Scholar] [CrossRef] [PubMed]
- Knipe, D.M.; Howley, P.M.; Griffin, D.E.; Lamb, R.A.; Martin, M.A.; Roizman, B. “Orthomyxoviruses” in Fields Virology, 5th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 2, pp. 1691–1740. [Google Scholar]
- Paules, C.; Subbarao, K. Influenza. Lancet 2017, 390, 697–708. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Disease Burden of Influenza. Available online: https://www.cdc.gov/flu/about/burden/index.html (accessed on 24 April 2019).
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal Influenza-Associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- WHO. Influenza (Seasonal) Fact Sheet. Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 10 May 2016).
- Centers for Disease Control and Prevention. Seasonal Influenza Vaccine Effectiveness. 2017–2018. Available online: https://www.cdc.gov/flu/about/season/flu-season-2017-2018.htm (accessed on 15 March 2019).
- Centers for Disease Control and Prevention. Pandemic Influenza—Past Pandemics. Available online: https://www.cdc.gov/flu/pandemic-resources/basics/past-pandemics.html (accessed on 28 July 2017).
- WHO. Influenza (Avian and Other Zoonotic). 2016. Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(avian-and-other-zoonotic) (accessed on 20 April 2019).
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A Universal influenza vaccine: The strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Soema, P.C.; Kompier, R.; Amorij, J.P.; Kersten, G.F. Current and next generation influenza vaccines: Formulation and production strategies. Eur. J. Pharm. Biopharm. 2015, 94, 251–263. [Google Scholar] [CrossRef]
- Xie, H.; Wan, X.F.; Ye, Z.; Plant, E.P.; Zhao, Y.; Xu, Y.; Li, X.; Finch, C.; Zhao, N.; Kawano, T.; et al. H3N2 Mismatch of 2014-15 Northern Hemisphere Influenza Vaccines and Head-to-head Comparison between Human and Ferret Antisera derived Antigenic Maps. Sci. Rep. 2015, 5, 15279. [Google Scholar] [CrossRef]
- Paules, C.I.; Sullivan, S.G.; Subbarao, K.; Fauci, A.S. Chasing seasonal influenza—The need for a universal influenza vaccine. N. Engl. J. Med. 2018, 378, 7–9. [Google Scholar] [CrossRef]
- Fiore, A.E.; Bridges, C.B.; Cox, N.J. Seasonal influenza vaccines. Curr. Top. Microbiol. Immunol. 2009, 333, 43–82. [Google Scholar]
- Centers for Disease Control and Prevention. Seasonal Influenza Vaccine Effectiveness. 2005–2017. Available online: https://www.cdc.gov/flu/professionals/vaccination/effectiveness-studies.htm (accessed on 20 May 2019).
- Sullivan, S.G.; Chilver, M.B.; Carville, K.S.; Deng, Y.M.; Grant, K.A.; Higgins, G.; Komadina, N.; Leung, V.K.; Minney-Smith, C.A.; Teng, D.; et al. Low interim influenza vaccine effectiveness, Australia, 1 May to 24 September 2017. Eurosurveillance 2017, 22. [Google Scholar] [CrossRef]
- Flannery, B.; Chung, J.R.; Belongia, E.A.; McLean, H.Q.; Gaglani, M.; Murthy, K.; Zimmerman, R.K.; Nowalk, M.P.; Jackson, M.L.; Jackson, L.A.; et al. Interim estimates of 2017-18 seasonal influenza vaccine effectiveness-United States, February 2018. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 180–185. [Google Scholar] [CrossRef]
- Yamazaki, T.; Chiba, J.; Akashi-Takamura, S. Neutralizing Anti-Hemagglutinin monoclonal antibodies induced by Gene-Based transfer have prophylactic and therapeutic effects on influenza virus infection. Vaccines 2018, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Marcelin, G.; Sandbulte, M.R.; Webby, R.J. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev. Med. Virol. 2012, 22, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Ryder, A.B.; Buonocore, L.; Vogel, L.; Nachbagauer, R.; Krammer, F.; Rose, J.K. A viable recombinant rhabdovirus lacking its glycoprotein gene and expressing influenza virus hemagglutinin and neuraminidase is a potent influenza vaccine. J. Virol. 2015, 89, 2820–2830. [Google Scholar] [CrossRef] [PubMed]
- Grant, E.J.; Josephs, T.M.; Loh, L.; Clemens, E.B.; Sant, S.; Bharadwaj, M.; Chen, W.; Rossjohn, J.; Gras, S.; Kedzierska, K. Broad CD8(+) T cell cross-recognition of distinct influenza A strains in humans. Nat. Commun. 2018, 9, 5427. [Google Scholar] [CrossRef] [PubMed]
- Rajao, D.S.; Perez, D.R. Universal vaccines and vaccine platforms to protect against influenza viruses in humans and agriculture. Front. Microbiol. 2018, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Alsharifi, M.; Furuya, Y.; Bowden, T.R.; Lobigs, M.; Koskinen, A.; Regner, M.; Trinidad, L.; Boyle, D.B.; Mullbacher, A. Intranasal flu vaccine protective against seasonal and H5N1 avian influenza infections. PLoS ONE 2009, 4, e5336. [Google Scholar] [CrossRef]
- Furuya, Y.; Chan, J.; Regner, M.; Lobigs, M.; Koskinen, A.; Kok, T.; Manavis, J.; Li, P.; Mullbacher, A.; Alsharifi, M. Cytotoxic T cells are the predominant players providing Cross-Protective immunity induced by γ-irradiated influenza A viruses. J. Virol. 2010, 84, 4212–4221. [Google Scholar] [CrossRef]
- Kon, T.C.; Onu, A.; Berbecila, L.; Lupulescu, E.; Ghiorgisor, A.; Kersten, G.F.; Cui, Y.Q.; Amorij, J.P.; Van der Pol, L. Influenza vaccine manufacturing: Effect of inactivation, splitting and site of manufacturing. comparison of influenza vaccine production processes. PLoS ONE 2016, 11, e0150700. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. 2018. Available online: https://www.cdc.gov/flu/protect/vaccine/how-fluvaccine-made.htm#recombinant (accessed on 20 March 2019).
- Cox, M.M.; Patriarca, P.A.; Treanor, J. FluBlok, a recombinant hemagglutinin influenza vaccine. Influenza Other Respir. Viruses 2008, 2, 211–219. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/flu/prevent/qa_flublok-vaccine.htm (accessed on 10 June 2019).
- Zheng, D.; Yi, Y.; Chen, Z. Development of live-attenuated influenza vaccines against outbreaks of H5N1 influenza. Viruses 2012, 4, 3589–3605. [Google Scholar] [CrossRef]
- Grohskopf, L.A.; Sokolow, L.Z.; Fry, A.M.; Walter, E.B.; Jernigan, D.B. Update: ACIP Recommendations for the Use of Quadrivalent Live Attenuated Influenza Vaccine (LAIV4)-United States, 2018–2019 Influenza Season. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 643–645. [Google Scholar] [CrossRef] [PubMed]
- Shcherbik, S.; Pearce, N.; Carney, P.; Bazhenova, E.; Larionova, N.; Kiseleva, I.; Rudenko, L.; Kumar, A.; Goldsmith, C.S.; Dugan, V.; et al. Evaluation of A(H1N1)pdm09 LAIV vaccine candidates stability and replication efficiency in primary human nasal epithelial cells. Vaccine X 2019, 2, 100031. [Google Scholar] [CrossRef] [PubMed]
- Skowronski, D.M.; Janjua, N.Z.; De Serres, G.; Sabaiduc, S.; Eshaghi, A.; Dickinson, J.A.; Fonseca, K.; Winter, A.L.; Gubbay, J.B.; Krajden, M.; et al. Low 2012-13 influenza vaccine effectiveness associated with mutation in the Egg-Adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS ONE 2014, 9, e92153. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Transcript for CDC Update on Flu Activity. Available online: https://www.cdc.gov/media/releases/2018/t0202-flu-update-activity.html (accessed on 2 February 2018).
- Wong, S.S.; Webby, R.J. Traditional and new influenza vaccines. Clin. Microbiol. Rev. 2013, 26, 476–492. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M.; Izikson, R.; Post, P.; Dunkle, L. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther. Adv. Vaccines 2015, 3, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Mahmood, K.; Chen, Z.; Yang, C.F.; Spaete, J.; Greenberg, H.B.; Herlocher, M.L.; Jin, H.; Kemble, G. Multiple gene segments control the temperature sensitivity and attenuation phenotypes of ca B/Ann Arbor/1/66. J. Virol. 2005, 79, 11014–11021. [Google Scholar] [CrossRef]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef]
- Krammer, F.; Garcia-Sastre, A.; Palese, P. Is it possible to develop a “universal” influenza virus vaccine? Potential target antigens and critical aspects for a universal influenza vaccine. Cold Spring Harb. Perspect. Biol. 2018, 10, a028845. [Google Scholar] [CrossRef]
- Andrews, S.F.; McDermott, A.B. Shaping a universally broad antibody response to influenza amidst a variable immunoglobulin landscape. Curr. Opin. Immunol. 2018, 53, 96–101. [Google Scholar] [CrossRef]
- Valkenburg, S.A.; Mallajosyula, V.V.A.; Li, O.T.W.; Chin, A.W.H.; Carnell, G.; Temperton, N.; Varadarajan, R.; Poon, L.L.M. Stalking influenza by vaccination with Pre-Fusion headless HA Mini-Stem. Sci. Rep. 2016, 6, 22666. [Google Scholar] [CrossRef]
- Rathore, U.; Kesavardhana, S.; Mallajosyula, V.V.; Varadarajan, R. Immunogen design for HIV-1 and influenza. Biochim. Biophys. Acta. 2014, 1844, 1891–1906. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.Y.; Ha do, L.A.; Simmons, C.; de Jong, M.D.; Chau, N.V.; Schumacher, R.; Peng, Y.C.; McMichael, A.J.; Farrar, J.J.; Smith, G.L.; et al. Memory T cells established by seasonal human influenza A infection Cross-React with avian influenza A (H5N1) in healthy individuals. J. Clin. Investig. 2008, 118, 3478–3490. [Google Scholar] [CrossRef] [PubMed]
- Terajima, M.; Babon, J.A.; Co, M.D.; Ennis, F.A. Cross-Reactive human B cell and T cell epitopes between influenza A and B viruses. Virol. J. 2013, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, L.A.; Kotlyarov, R.Y.; Kovaleva, A.A.; Potapchuk, M.V.; Korotkov, A.V.; Sergeeva, M.V.; Kasianenko, M.A.; Kuprianov, V.V.; Ravin, N.V.; Tsybalova, L.M.; et al. Protection against multiple influenza A virus strains induced by candidate recombinant vaccine based on heterologous M2e peptides linked to flagellin. PLoS ONE 2015, 10, e0119520. [Google Scholar] [CrossRef] [PubMed]
- Price, G.E.; Lo, C.Y.; Misplon, J.A.; Epstein, S.L. Mucosal immunization with a candidate universal influenza vaccine reduces virus transmission in a mouse model. J. Virol. 2014, 88, 6019–6030. [Google Scholar] [CrossRef] [PubMed]
- Quinones-Parra, S.; Grant, E.; Loh, L.; Nguyen, T.H.; Campbell, K.A.; Tong, S.Y.; Miller, A.; Doherty, P.C.; Vijaykrishna, D.; Rossjohn, J.; et al. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc. Natl. Acad. Sci. USA 2014, 111, 1049–1054. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Bui, H.H.; Sidney, J.; Zhang, Q.; Glenn, J.; Oseroff, C.; Mbawuike, I.N.; Alexander, J.; Newman, M.J.; Grey, H.; et al. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 2008, 82, 12241–12251. [Google Scholar] [CrossRef]
- Wang, Z.; Wan, Y.; Qiu, C.; Quinones-Parra, S.; Zhu, Z.; Loh, L.; Tian, D.; Ren, Y.; Hu, Y.; Zhang, X.; et al. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8(+) T cells. Nat. Commun. 2015, 6, 6833. [Google Scholar] [CrossRef]
- Hayward, A.C.; Wang, L.; Goonetilleke, N.; Fragaszy, E.B.; Bermingham, A.; Copas, A.; Dukes, O.; Millett, E.R.; Nazareth, I.; Nguyen-Van-Tam, J.S.; et al. Natural T Cell-mediated protection against seasonal and pandemic influenza. results of the Flu watch cohort study. Am. J. Respir. Crit. Care Med. 2015, 191, 1422–1431. [Google Scholar] [CrossRef]
- Grant, E.J.; Quinones-Parra, S.M.; Clemens, E.B.; Kedzierska, K. Human influenza viruses and CD8(+) T cell responses. Curr. Opin. Virol. 2016, 16, 132–142. [Google Scholar] [CrossRef]
- Wilkinson, T.M.; Li, C.K.; Chui, C.S.; Huang, A.K.; Perkins, M.; Liebner, J.C.; Lambkin-Williams, R.; Gilbert, A.; Oxford, J.; Nicholas, B.; et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012, 18, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Valkenburg, S.A.; Li, O.T.W.; Li, A.; Bull, M.; Waldmann, T.A.; Perera, L.P.; Peiris, M.; Poon, L.L.M. Protection by universal influenza vaccine is mediated by memory CD4 T cells. Vaccine 2018, 36, 4198–4206. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.L.; Leung, Y.H.; Nicholls, J.M.; Perera, P.Y.; Lichy, J.H.; Yamamoto, M.; Waldmann, T.A.; Peiris, J.S.; Perera, L.P. Vaccinia Virus-Based multivalent H5N1 avian influenza vaccines adjuvanted with IL-15 confer sterile cross-clade protection in mice. J. Immunol. 2009, 182, 3063–3071. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, Q.M.; Gatherer, D.; Reche, P.A.; Flower, D.R. Towards the knowledge-based design of universal influenza epitope ensemble vaccines. Bioinformatics 2016, 32, 3233–3239. [Google Scholar] [CrossRef] [PubMed]
- Huarte, E.; Sarobe, P.; Lu, J.; Casares, N.; Lasarte, J.J.; Dotor, J.; Ruiz, M.; Prieto, J.; Celis, E.; Borras-Cuesta, F. Enhancing immunogenicity of a CTL epitope from carcinoembryonic antigen by selective amino acid replacements. Clin. Cancer Res. 2002, 8, 2336–2344. [Google Scholar] [PubMed]
- Hemmer, B.; Kondo, T.; Gran, B.; Pinilla, C.; Cortese, I.; Pascal, J.; Tzou, A.; McFarland, H.F.; Houghten, R.; Martin, R. Minimal peptide length requirements for CD4(+) T cell clones—Implications for molecular mimicry and T cell survival. Int. Immunol. 2000, 12, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Ansari, H.R.; Raghava, G.P. Improved method for linear B-cell epitope prediction using antigen’s primary sequence. PLoS ONE 2013, 8, e62216. [Google Scholar] [CrossRef]
- Herrera-Rodriguez, J.; Meijerhof, T.; Niesters, H.G.; Stjernholm, G.; Hovden, A.O.; Sorensen, B.; Okvist, M.; Sommerfelt, M.A.; Huckriede, A. A novel Peptide-Based vaccine candidate with protective efficacy against influenza A in a mouse model. Virology 2018, 515, 21–28. [Google Scholar] [CrossRef]
- Lu, I.N.; Farinelle, S.; Sausy, A.; Muller, C.P. Identification of a CD4 T-cell epitope in the hemagglutinin stalk domain of pandemic H1N1 influenza virus and its antigen-driven TCR usage signature in BALB/c mice. Cell. Mol. Immunol. 2017, 14, 511–520. [Google Scholar] [CrossRef]
- Ben-Yedidia, T.; Marcus, H.; Reisner, Y.; Arnon, R. Intranasal administration of peptide vaccine protects human/mouse radiation chimera from influenza infection. Int. Immunol. 1999, 11, 1043–1051. [Google Scholar] [CrossRef]
- Atsmon, J.; Kate-Ilovitz, E.; Shaikevich, D.; Singer, Y.; Volokhov, I.; Haim, K.Y.; Ben-Yedidia, T. Safety and immunogenicity of multimeric-001--a novel universal influenza vaccine. J. Clin. Immunol. 2012, 32, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Atsmon, J.; Caraco, Y.; Ziv-Sefer, S.; Shaikevich, D.; Abramov, E.; Volokhov, I.; Bruzil, S.; Haima, K.Y.; Gottlieb, T.; Ben-Yedidia, T. Priming by a novel universal influenza vaccine (Multimeric-001)-a gateway for improving immune response in the elderly population. Vaccine 2014, 32, 5816–5823. [Google Scholar] [CrossRef] [PubMed]
- Clinical Trials.Gov. Available online: https://clinicaltrials.gov/ct2/show/study/NCT03058692 (accessed on 1 March 2019).
- Tsybalova, L.M.; Stepanova, L.A.; Shuklina, M.A.; Mardanova, E.S.; Kotlyarov, R.Y.; Potapchuk, M.V.; Petrov, S.A.; Blokhina, E.A.; Ravin, N.V. Combination of M2e peptide with stalk HA epitopes of influenza A virus enhances protective properties of recombinant vaccine. PLoS ONE 2018, 13, e0201429. [Google Scholar] [CrossRef] [PubMed]
- Saelens, X. The Role of Matrix Protein 2 Ectodomain in the Development of Universal Influenza Vaccines. J. Infect. Dis. 2019, 219, S68–S74. [Google Scholar] [CrossRef]
- Safety Study of Recombinant M2e Influenza—A Vaccine in Healthy Adults (FLU-A). Available online: https://clinicaltrials.gov/ct2/show/NCT00819013?term=Acam-flu-A&rank=1 (accessed on 10 October 2008).
- Deng, L.; Cho, K.J.; Fiers, W.; Saelens, X. M2e-Based Universal Influenza A Vaccines. Vaccines 2015, 3, 105–136. [Google Scholar] [CrossRef]
- Rosendahl Huber, S.K.; Camps, M.G.; Jacobi, R.H.; Mouthaan, J.; van Dijken, H.; van Beek, J.; Ossendorp, F.; de Jonge, J. Synthetic long peptide influenza vaccine containing conserved T and B cell epitopes reduces viral load in lungs of mice and ferrets. PLoS ONE 2015, 10, e0127969. [Google Scholar] [CrossRef]
- Tutykhina, I.; Esmagambetov, I.; Bagaev, A.; Pichugin, A.; Lysenko, A.; Shcherbinin, D.; Sedova, E.; Logunov, D.; Shmarov, M.; Ataullakhanov, R.; et al. Vaccination potential of B and T epitope-enriched NP and M2 against Influenza A viruses from different clades and hosts. PLoS ONE 2018, 13, e0191574. [Google Scholar] [CrossRef]
- Zerbe, K.; Moehle, K.; Robinson, J.A. Protein Epitope Mimetics: From New Antibiotics to Supramolecular Synthetic Vaccines. Acc. Chem. Res. 2017, 50, 1323–1331. [Google Scholar] [CrossRef]
- Guo, C.; Zhang, H.; Xie, X.; Liu, Y.; Sun, L.; Li, H.; Yu, P.; Hu, H.; Sun, J.; Li, Y.; et al. H1N1 influenza virus epitopes classified by monoclonal antibodies. Exp. Ther. Med. 2018, 16, 2001–2007. [Google Scholar] [CrossRef]
- Andersen, T.K.; Zhou, F.; Cox, R.; Bogen, B.; Grodeland, G. A DNA vaccine that targets hemagglutinin to antigen-presenting cells protects mice against H7 influenza. J. Virol. 2017, 91, e01340-17. [Google Scholar] [CrossRef]
- Lee, L.Y.Y.; Izzard, L.; Hurt, A.C. A Review of DNA Vaccines against Influenza. Front. Immunol. 2018, 9, 1568. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Bilsel, P.; del Guercio, M.F.; Stewart, S.; Marinkovic-Petrovic, A.; Southwood, S.; Crimi, C.; Vang, L.; Walker, L.; Ishioka, G.; et al. Universal influenza DNA vaccine encoding conserved CD4+ T cell epitopes protects against lethal viral challenge in HLA-DR transgenic mice. Vaccine 2010, 28, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hackett, A.; Jia, N.; Zhang, C.; Zhang, L.; Parker, C.; Zhou, A.; Li, J.; Cao, W.C.; Huang, Z.; et al. Polyvalent DNA vaccines expressing HA antigens of H5N1 influenza viruses with an optimized leader sequence elicit Cross-Protective antibody responses. PLoS ONE 2011, 6, e28757. [Google Scholar] [CrossRef] [PubMed]
- Koday, M.T.; Leonard, J.A.; Munson, P.; Forero, A.; Koday, M.; Bratt, D.L.; Fuller, J.T.; Murnane, R.; Qin, S.; Reinhart, T.A.; et al. Multigenic DNA vaccine induces protective Cross-Reactive T cell responses against heterologous influenza virus in nonhuman primates. PLoS ONE 2017, 12, e0189780. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, I.; Borggren, M.; Rosenstierne, M.W.; Trebbien, R.; Williams, J.A.; Vidal, E.; Vergara-Alert, J.; Foz, D.S.; Darji, A.; Sistere-Oro, M.; et al. Protective effect of a polyvalent influenza DNA vaccine in pigs. Vet. Immunol. Immunopathol. 2018, 195, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Houser, K.V.; Yamshchikov, G.V.; Bellamy, A.R.; May, J.; Enama, M.E.; Sarwar, U.; Larkin, B.; Bailer, R.T.; Koup, R.; Paskel, M.; et al. DNA vaccine priming for seasonal influenza vaccine in children and adolescents 6 to 17 years of age: A phase 1 randomized clinical trial. PLoS ONE 2018, 13, e0206837. [Google Scholar] [CrossRef]
- AgriLabs: First DNA Vaccine Licensed for Chickens: Cision. 2017. Available online: https://www.prnewswire.com/news-releases/first-dna-vaccine-licensed-for-chickens-300554855.html (accessed on 22 March 2019).
- Rodriguez-Gascon, A.; del Pozo-Rodriguez, A.; Solinis, M.A. Development of nucleic acid vaccines: Use of self-amplifying RNA in lipid nanoparticles. Int. J. Nanomed. 2014, 9, 1833–1843. [Google Scholar] [CrossRef]
- Deering, R.P.; Kommareddy, S.; Ulmer, J.B.; Brito, L.A.; Geall, A.J. Nucleic acid vaccines: Prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 2014, 11, 885–899. [Google Scholar] [CrossRef]
- Zhang, C.; Maruggi, G.; Shan, H.; Li, J. Advances in mRNA Vaccines for Infectious Diseases. Front. Immunol. 2019, 10, 594. [Google Scholar] [CrossRef]
- Magini, D.; Giovani, C.; Mangiavacchi, S.; Maccari, S.; Cecchi, R.; Ulmer, J.B.; De Gregorio, E.; Geall, A.J.; Brazzoli, M.; Bertholet, S. Self-Amplifying mRNA Vaccines Expressing Multiple Conserved Influenza Antigens Confer Protection against Homologous and Heterosubtypic Viral Challenge. PLoS ONE 2016, 11, e0161193. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015, 14, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Hekele, A.; Bertholet, S.; Archer, J.; Gibson, D.G.; Palladino, G.; Brito, L.A.; Otten, G.R.; Brazzoli, M.; Buccato, S.; Bonci, A.; et al. Rapidly produced SAM((R)) vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013, 2, e52. [Google Scholar] [CrossRef] [PubMed]
- Lutz, J.; Lazzaro, S.; Habbeddine, M.; Schmidt, K.E.; Baumhof, P.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Heidenreich, R.; et al. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. npj Vaccines 2017, 2, 29. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Parkhouse, K.; Kirkpatrick, E.; McMahon, M.; Zost, S.J.; Mui, B.L.; Tam, Y.K.; Kariko, K.; Barbosa, C.J.; Madden, T.D.; et al. Nucleoside-Modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018, 9, 3361. [Google Scholar] [CrossRef] [PubMed]
- Brazzoli, M.; Magini, D.; Bonci, A.; Buccato, S.; Giovani, C.; Kratzer, R.; Zurli, V.; Mangiavacchi, S.; Casini, D.; Brito, L.M.; et al. Induction of Broad-Based immunity and protective efficacy by Self-Amplifying mRNA vaccines encoding influenza virus hemagglutinin. J. Virol. 2016, 90, 332–344. [Google Scholar] [CrossRef]
- Petsch, B.; Schnee, M.; Vogel, A.B.; Lange, E.; Hoffmann, B.; Voss, D.; Schlake, T.; Thess, A.; Kallen, K.J.; Stitz, L.; et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012, 30, 1210–1216. [Google Scholar] [CrossRef]
- Kumar, A.; Meldgaard, T.S.; Bertholet, S. Novel Platforms for the Development of a Universal Influenza Vaccine. Front. Immunol. 2018, 9, 600. [Google Scholar] [CrossRef]
- Plummer, E.M.; Manchester, M. Viral nanoparticles and Virus-Like particles: Platforms for contemporary vaccine design. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2011, 3, 174–196. [Google Scholar] [CrossRef]
- Galarza, J.M.; Latham, T.; Cupo, A. Virus-Like particle (VLP) vaccine conferred complete protection against a lethal influenza virus challenge. Viral Immunol. 2005, 18, 244–251. [Google Scholar] [CrossRef]
- Gao, X.; Wang, W.; Li, Y.; Zhang, S.; Duan, Y.; Xing, L.; Zhao, Z.; Zhang, P.; Li, Z.; Li, R.; et al. Enhanced Influenza VLP vaccines comprising matrix-2 ectodomain and nucleoprotein epitopes protects mice from lethal challenge. Antivir. Res. 2013, 98, 4–11. [Google Scholar] [CrossRef]
- Schwartzman, L.M.; Cathcart, A.L.; Pujanauski, L.M.; Qi, L.; Kash, J.C.; Taubenberger, J.K. An intranasal Virus-Like particle vaccine broadly protects mice from multiple subtypes of influenza A virus. mBio 2015, 6, e01044. [Google Scholar] [CrossRef] [PubMed]
- Valero-Pacheco, N.; Perez-Toledo, M.; Villasis-Keever, M.A.; Nunez-Valencia, A.; Bosco-Garate, I.; Lozano-Dubernard, B.; Lara-Puente, H.; Espitia, C.; Alpuche-Aranda, C.; Bonifaz, L.C.; et al. Antibody persistence in adults two years after vaccination with an H1N1 2009 pandemic influenza Virus-Like particle vaccine. PLoS ONE 2016, 11, e0150146. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Mohan, T.; Zhu, W.; Wang, C.; Deng, L.; Wang, B.Z. Sequential Immunizations with heterosubtypic Virus-Like particles elicit cross protection against divergent influenza A viruses in mice. Sci. Rep. 2018, 8, 4577. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Song, J.M.; O, E.; Kwon, Y.M.; Lee, Y.J.; Compans, R.W.; Kang, S.M. Virus-Like particles containing multiple M2 extracellular domains confer improved cross-protection against various subtypes of influenza virus. Mol. Ther. 2013, 21, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Lee, Y.N.; Hwang, H.S.; Lee, Y.T.; Ko, E.J.; Jung, Y.J.; Cho, M.K.; Kim, Y.J.; Lee, J.S.; Ha, S.H.; et al. Influenza M2 Virus-Like particles confer a broader range of cross protection to the Strain-Specific pre-existing immunity. Vaccine 2014, 32, 5824–5831. [Google Scholar] [CrossRef][Green Version]
- Hu, C.J.; Chien, C.Y.; Liu, M.T.; Fang, Z.S.; Chang, S.Y.; Juang, R.H.; Chang, S.C.; Chen, H.W. Multi-Antigen avian influenza a (H7N9) Virus-Like particles: Particulate characterizations and immunogenicity evaluation in murine and avian models. BMC Biotechnol. 2017, 17, 2. [Google Scholar] [CrossRef]
- Pushko, P.; Tretyakova, I.; Hidajat, R.; Zsak, A.; Chrzastek, K.; Tumpey, T.M.; Kapczynski, D.R. Virus-Like particles displaying H5, H7, H9 hemagglutinins and N1 neuraminidase elicit protective immunity to heterologous avian influenza viruses in chickens. Virology 2017, 501, 176–182. [Google Scholar] [CrossRef]
- Pyo, H.M.; Masic, A.; Woldeab, N.; Embury-Hyatt, C.; Lin, L.; Shin, Y.K.; Song, J.Y.; Babiuk, S.; Zhou, Y. Pandemic H1N1 influenza Virus-Like particles are immunogenic and provide protective immunity to pigs. Vaccine 2012, 30, 1297–1304. [Google Scholar] [CrossRef]
- Pillet, S.; Aubin, E.; Trepanier, S.; Bussiere, D.; Dargis, M.; Poulin, J.F.; Yassine-Diab, B.; Ward, B.J.; Landry, N. A Plant-Derived quadrivalent virus like particle influenza vaccine induces cross-reactive antibody and T cell response in healthy adults. Clin. Immunol. 2016, 168, 72–87. [Google Scholar] [CrossRef]
- Scotti, N.; Rybicki, E.P. Virus-Like particles produced in plants as potential vaccines. Expert Rev. Vaccines 2013, 12, 211–224. [Google Scholar] [CrossRef]
- Clinical Trials.Gov. Available online: https://clinicaltrials.gov/ct2/show/study/NCT03739112 (accessed on 15 November 2018).
- Hodgins, B.; Pillet, S.; Landry, N.; Ward, B.J. A Plant-Derived VLP influenza vaccine elicits a balanced immune response even in very old mice with Co-Morbidities. PLoS ONE 2019, 14, e0210009. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Wang, B.Z. A Perspective on Nanoparticle Universal Influenza Vaccines. ACS Infect. Dis. 2018, 4, 1656–1665. [Google Scholar] [CrossRef] [PubMed]
- Sahdev, P.; Ochyl, L.J.; Moon, J.J. Biomaterials for nanoparticle vaccine delivery systems. Pharm. Res. 2014, 31, 2563–2582. [Google Scholar] [CrossRef] [PubMed]
- Tregoning, J.S.; Russell, R.F.; Kinnear, E. Adjuvanted influenza vaccines. Hum. Vaccines Immunother. 2018, 14, 550–564. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.G.; Bertholet, S.; Coler, R.N.; Friede, M. New horizons in adjuvants for vaccine development. Trends Immunol. 2009, 30, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deng, L.; Kang, S.M.; Wang, B.Z. Universal influenza vaccines: From viruses to nanoparticles. Expert Rev. Vaccines 2018, 17, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Mohan, T.; Chang, T.Z.; Gonzalez, G.X.; Wang, Y.; Kwon, Y.M.; Kang, S.M.; Compans, R.W.; Champion, J.A.; Wang, B.Z. Double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nat. Commun. 2018, 9, 359. [Google Scholar] [CrossRef]
- Hiremath, J.; Kang, K.I.; Xia, M.; Elaish, M.; Binjawadagi, B.; Ouyang, K.; Dhakal, S.; Arcos, J.; Torrelles, J.B.; Jiang, X.; et al. Entrapment of H1N1 Influenza Virus Derived Conserved Peptides in PLGA Nanoparticles Enhances T Cell Response and Vaccine Efficacy in Pigs. PLoS ONE 2016, 11, e0151922. [Google Scholar] [CrossRef]
- Dhakal, S.; Cheng, X.; Salcido, J.; Renu, S.; Bondra, K.; Lakshmanappa, Y.S.; Misch, C.; Ghimire, S.; Feliciano-Ruiz, N.; Hogshead, B.; et al. Liposomal Nanoparticle-Based conserved peptide influenza vaccine and monosodium urate crystal adjuvant elicit protective immune response in pigs. Int. J. Nanomed. 2018, 13, 6699–6715. [Google Scholar] [CrossRef]
- Bahl, K.; Senn, J.J.; Yuzhakov, O.; Bulychev, A.; Brito, L.A.; Hassett, K.J.; Laska, M.E.; Smith, M.; Almarsson, Ö.; Thompson, J.; et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017, 25, 1316–1327. [Google Scholar] [CrossRef]
- Tao, W.; Hurst, B.L.; Shakya, A.K.; Uddin, M.J.; Ingrole, R.S.; Hernandez-Sanabria, M.; Arya, R.P.; Bimler, L.; Paust, S.; Tarbet, E.B.; et al. Consensus M2e peptide conjugated to gold nanoparticles confers protection against H1N1, H3N2 and H5N1 influenza A viruses. Antivir. Res. 2017, 141, 62–72. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/show/study/NCT03814720 (accessed on 19 August 2019).
| Type of Vaccine | Virus Strain | Trade Name | Production | Age | Immunological Outcomes | Route | Ref. |
|---|---|---|---|---|---|---|---|
| IIV | Influenza A: H1N1 and H3N2 virus/Influenza B: Victoria and/or Yamagata | Fluzone | Eggs | 6–35 m | Ab immune response | IM | [34] |
| Fluarix | Eggs | >6 m | |||||
| Flucelvax | MDCK cells | >4 y | |||||
| RIV | Contains the HA ectodomain amino acid sequence of cell-cultured vaccine prototype viruses suggested by WHO | Flublok | Recombinant-expression in insect cell line | >18 y | Ab immune response | IM | [35,36] |
| LAIV | Subtypes of H1N1 and H3N2 (influenza A) and one Influenza B | FluMist | Eggs | 2–49 y | Mucosal (nasal) IgA Ab and strong cell-mediated immunity | IN | [37,38,39] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jazayeri, S.D.; Poh, C.L. Development of Universal Influenza Vaccines Targeting Conserved Viral Proteins. Vaccines 2019, 7, 169. https://doi.org/10.3390/vaccines7040169
Jazayeri SD, Poh CL. Development of Universal Influenza Vaccines Targeting Conserved Viral Proteins. Vaccines. 2019; 7(4):169. https://doi.org/10.3390/vaccines7040169
Chicago/Turabian StyleJazayeri, Seyed Davoud, and Chit Laa Poh. 2019. "Development of Universal Influenza Vaccines Targeting Conserved Viral Proteins" Vaccines 7, no. 4: 169. https://doi.org/10.3390/vaccines7040169
APA StyleJazayeri, S. D., & Poh, C. L. (2019). Development of Universal Influenza Vaccines Targeting Conserved Viral Proteins. Vaccines, 7(4), 169. https://doi.org/10.3390/vaccines7040169





