Potential Role of Rainbow Trout Erythrocytes as Mediators in the Immune Response Induced by a DNA Vaccine in Fish
Abstract
1. Introduction
2. Material and Methods
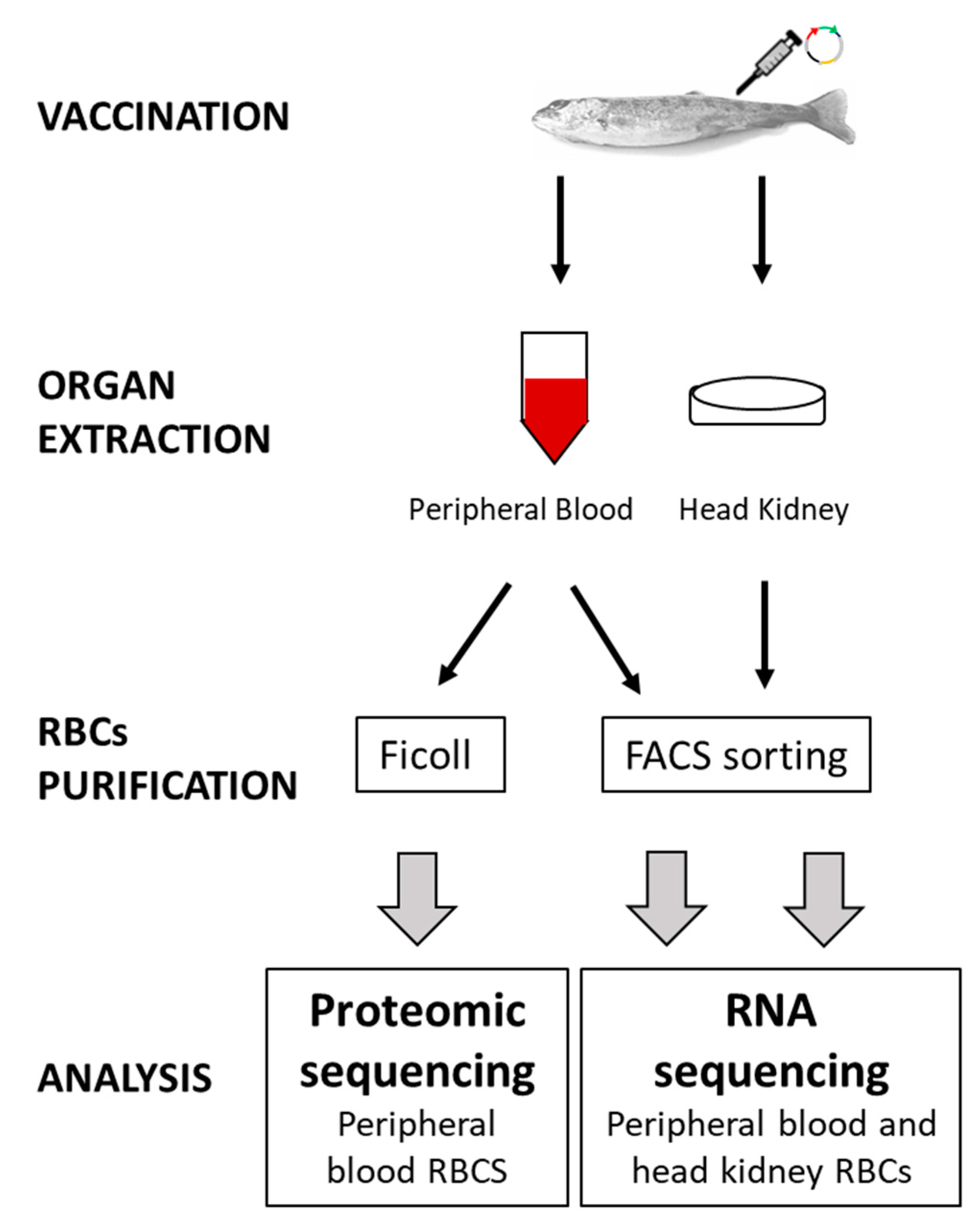
2.1. Animals
2.2. Plasmids
2.3. DNA Immunization
2.4. Purification of Head Kidney and Peripheral Blood RBCs for Transcriptome Analysis, by Means of FACS Single-Cell Sorting
2.5. Transcriptome Analysis of RBCs
2.6. Proteome Analysis of RBCs
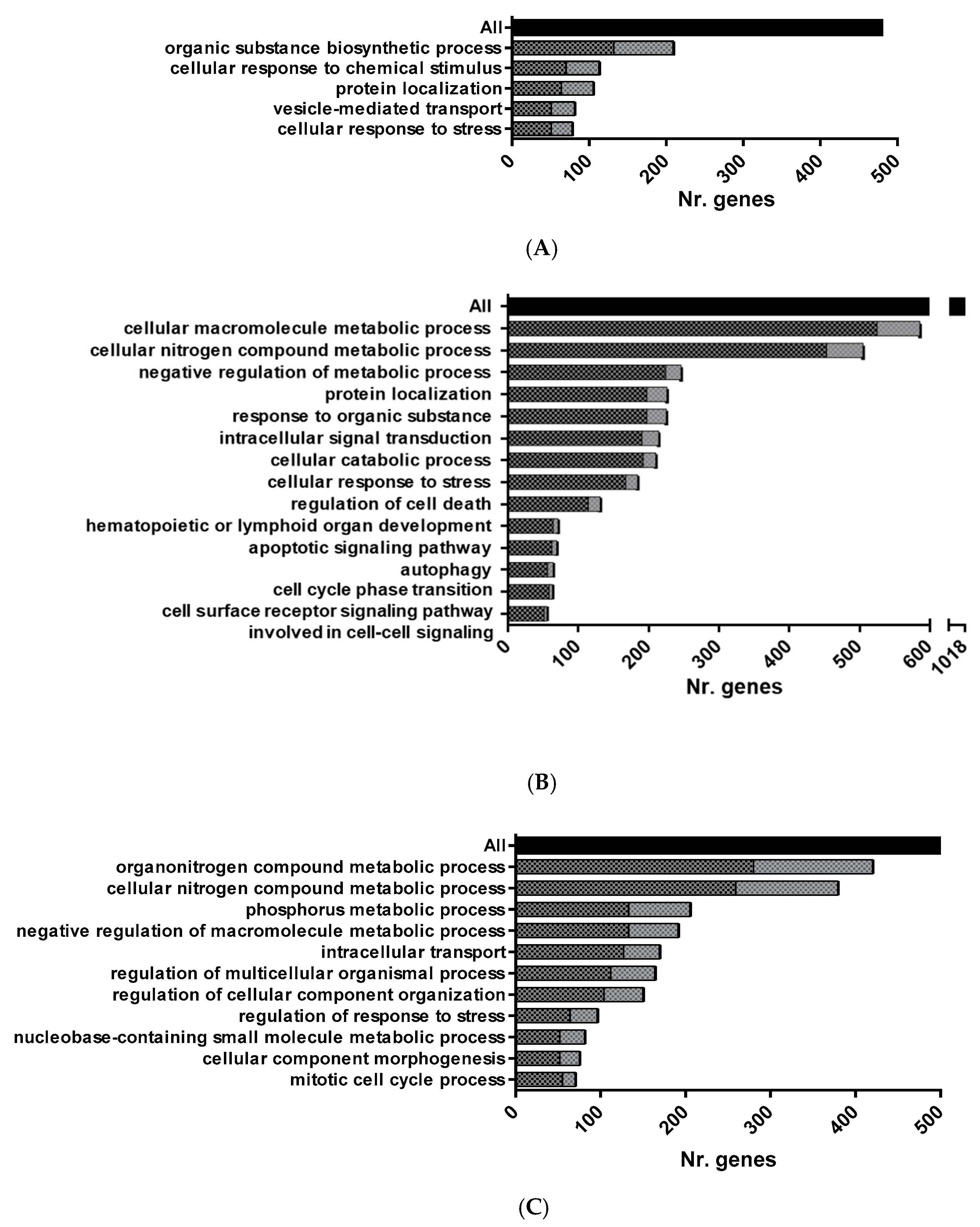
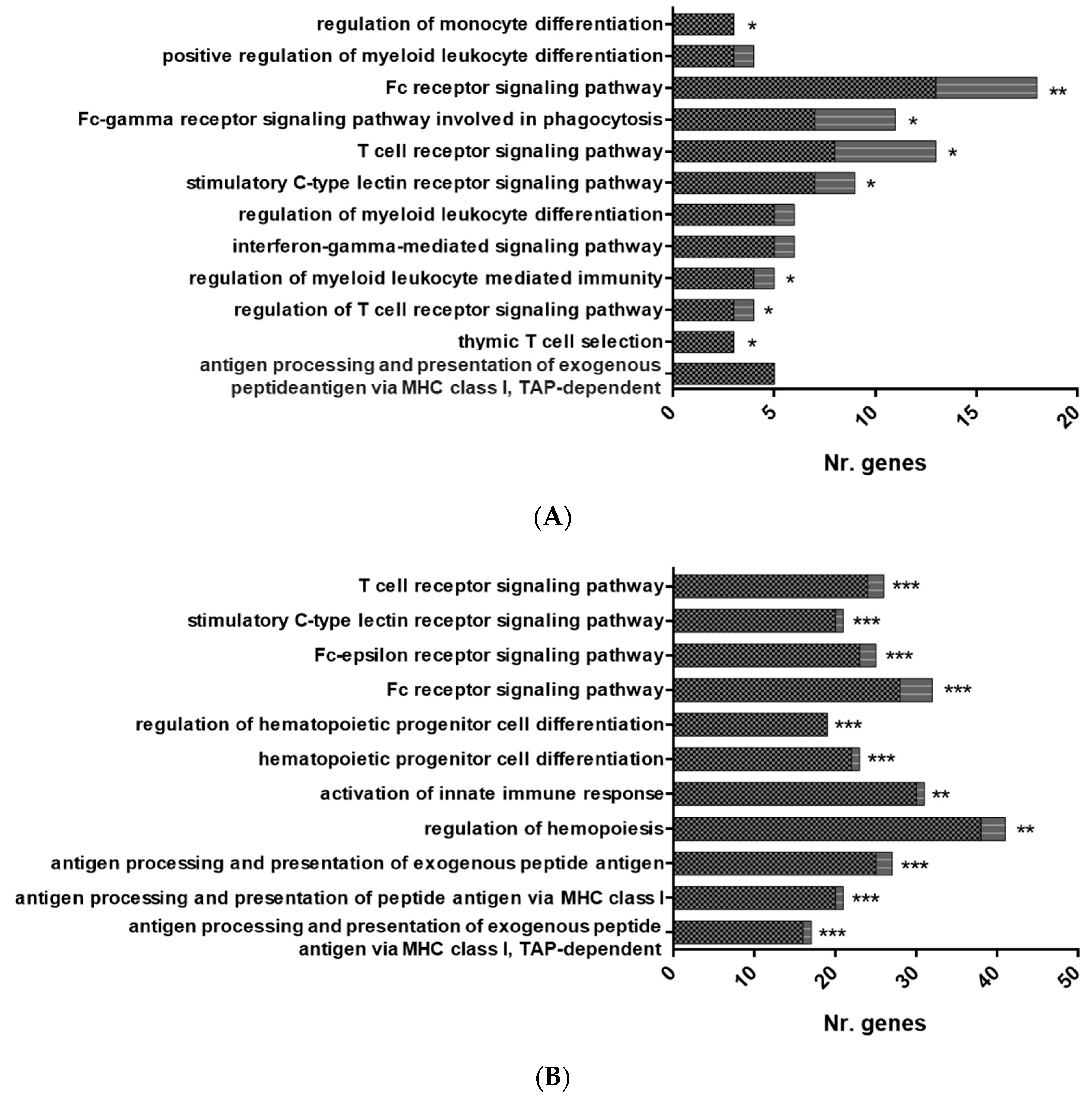
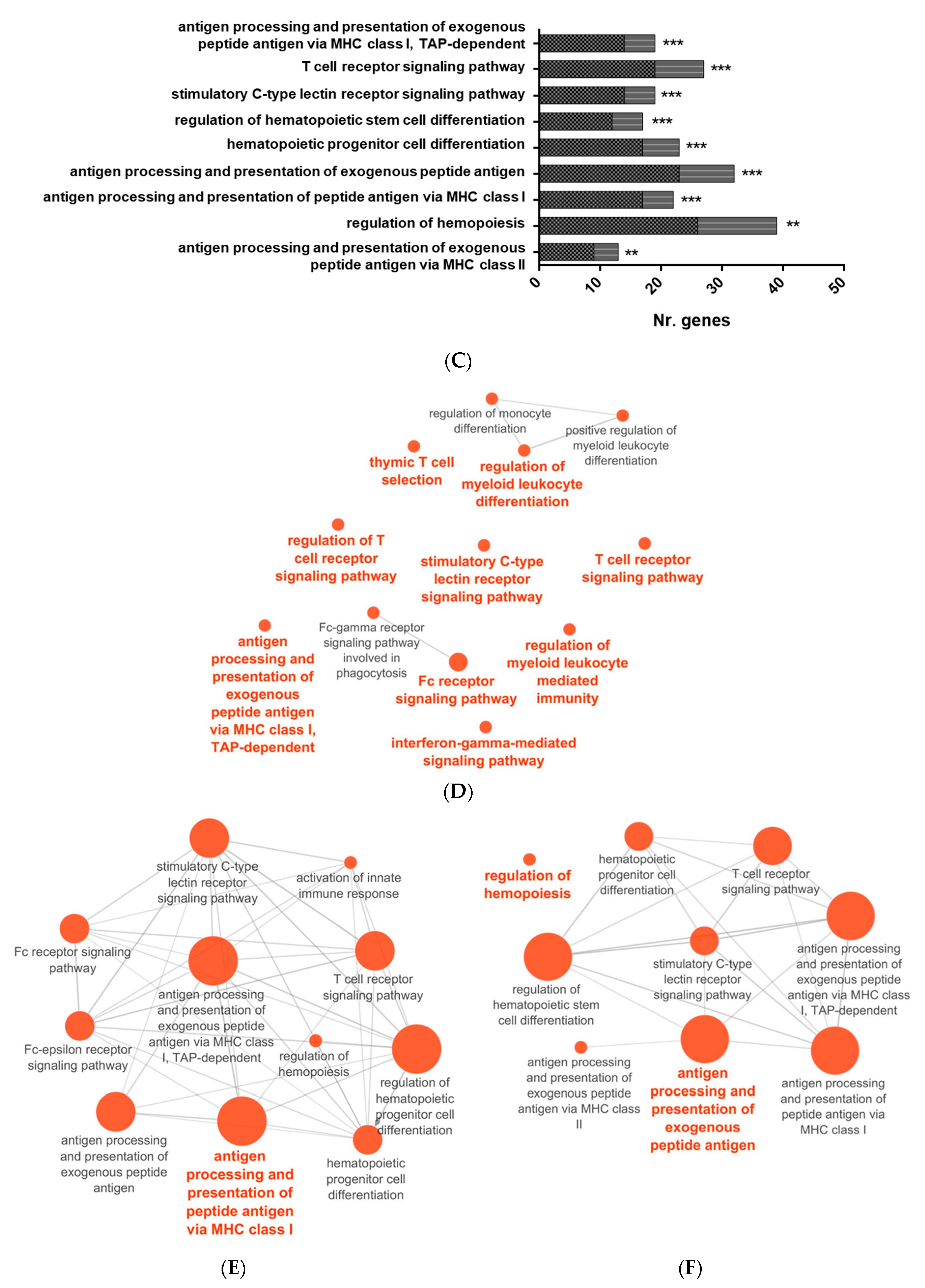
2.7. Pathway Enrichment Analysis
2.8. RNA Extraction, cDNA Synthesis, and RT-qPCR Gene Expression
2.9. Coculture of Transfected RBCs with White Blood Cells
2.10. Enzyme-Linked Immunosorbent Assay (ELISA)
2.11. Statistical Analysis
2.12. Ethics Statement
3. Results
3.1. RNA Sequencing of HK-RBC from GVHSV Immunized Rainbow Trout
3.2. RNA Sequencing of PB-RBCs from GVHSV-Immunized Rainbow Trout
3.3. Proteome Sequencing of PB-RBC from GVHSV-Immunized Rainbow Trout
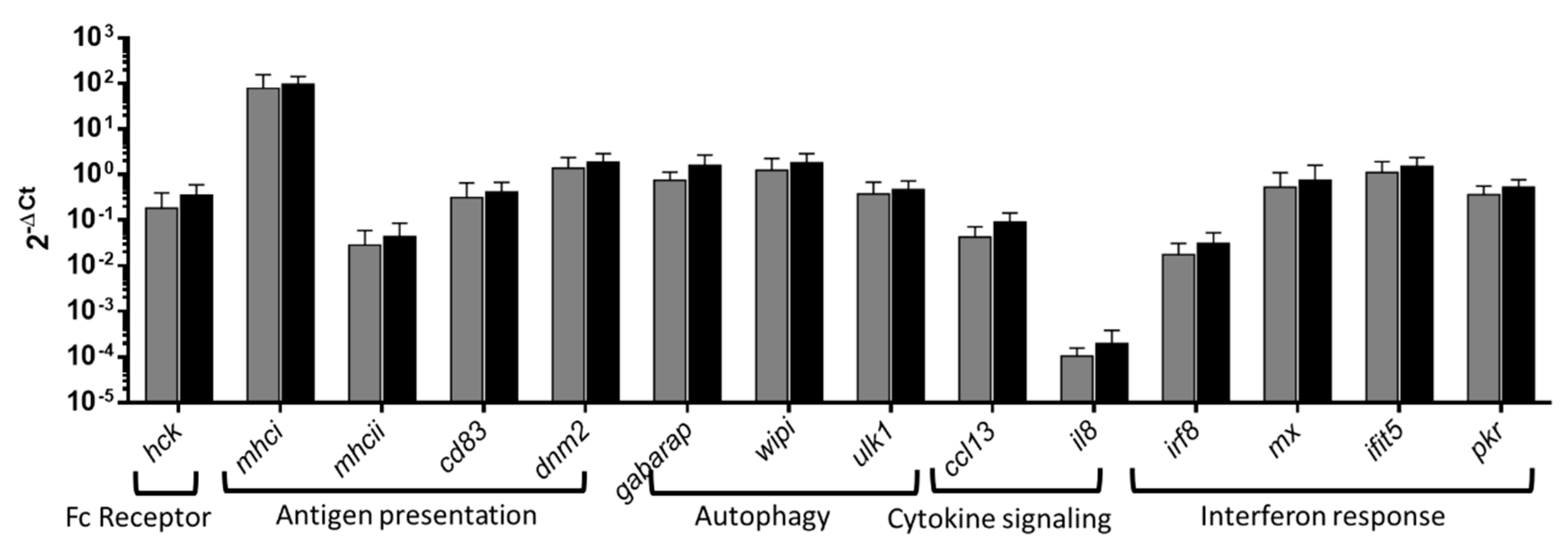
3.4. Overrepresented pathways RT-qPCR analysis
3.5. Leukocyte Proliferation
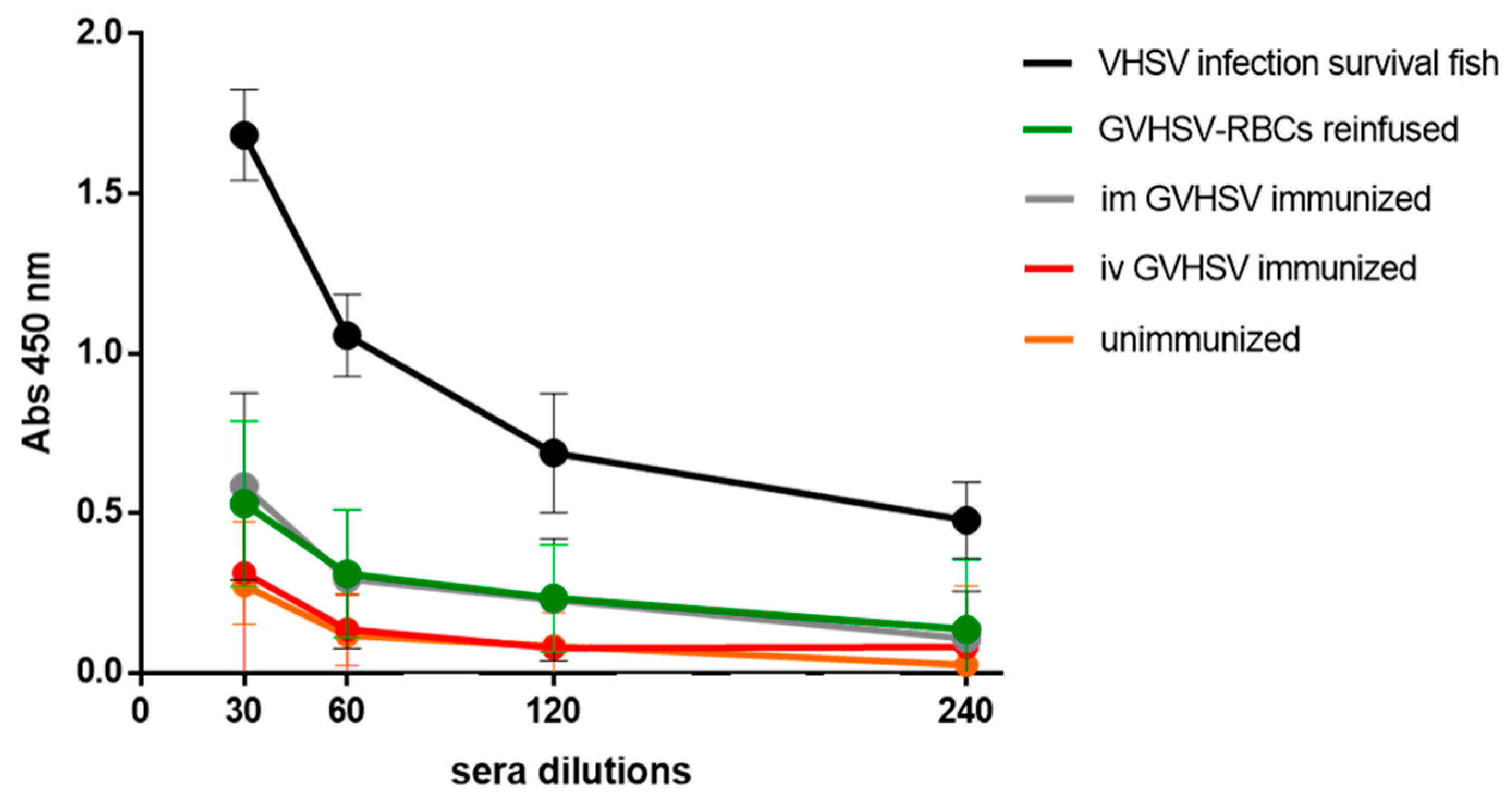
3.6. Antibody Detection in GVHSV-RBCs Reinfusion/Immunization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). The State Of World Fisheries and Aquaculture. Meeting the Sustainable Development Goals; FAO: Rome, Italy, 2018. [Google Scholar]
- Collins, C.; Lorenzen, N.; Collet, B. DNA vaccination for finfish aquaculture. Fish. Shellfish Immunol. 2019, 85, 106–125. [Google Scholar] [CrossRef]
- Tonheim, T.C.; Bogwald, J.; Dalmo, R.A. What happens to the DNA vaccine in fish? A review of current knowledge. Fish. Shellfish Immunol. 2008, 25, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kurath, G. Fish. Novirhabdoviruses; Caister Academic Press: Norfolk, UK, 2012. [Google Scholar]
- Sommerset, I.; Krossoy, B.; Biering, E.; Frost, P. Vaccines for fish in aquaculture. Expert Rev. Vaccines 2005, 4, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Biering, E.; Villoing, S.; Sommerset, I.; Christie, K.E. Update on viral vaccines for fish. Dev. Biol. 2005, 121, 97–113. [Google Scholar]
- Morera, D.; MacKenzie, S.A. Is there a direct role for erythrocytes in the immune response? Vet. Res. 2011, 42, 89. [Google Scholar] [CrossRef]
- Nombela, I.; Ortega-Villaizan, M.D.M. Nucleated red blood cells: Immune cell mediators of the antiviral response. PLoS Pathog. 2018, 14, e1006910. [Google Scholar] [CrossRef]
- Nombela, I.; Puente-Marin, S.; Chico, V.; Villena, A.J.; Carracedo, B.; Ciordia, S.; Mena, M.C.; Mercado, L.; Perez, L.; Coll, J.; et al. Identification of diverse defense mechanisms in rainbow trout red blood cells in response to halted replication of VHS virus. F1000Research 2017, 6, 1958. [Google Scholar] [CrossRef]
- Nombela, I.; Carrion, A.; Puente-Marin, S.; Chico, V.; Mercado, L.; Perez, L.; Coll, J.; Ortega-Villaizan, M.D.M. Infectious pancreatic necrosis virus triggers antiviral immune response in rainbow trout red blood cells, despite not being infective. F1000Research 2017, 6, 1968. [Google Scholar] [CrossRef]
- Workenhe, S.T.; Kibenge, M.J.; Wright, G.M.; Wadowska, D.W.; Groman, D.B.; Kibenge, F.S. Infectious salmon anaemia virus replication and induction of alpha interferon in Atlantic salmon erythrocytes. Virol. J. 2008, 5, 36. [Google Scholar] [CrossRef]
- Morera, D.; Roher, N.; Ribas, L.; Balasch, J.C.; Donate, C.; Callol, A.; Boltana, S.; Roberts, S.; Goetz, G.; Goetz, F.W.; et al. RNA-Seq reveals an integrated immune response in nucleated erythrocytes. PLoS ONE 2011, 6, e26998. [Google Scholar] [CrossRef] [PubMed]
- Dahle, M.K.; Wessel, O.; Timmerhaus, G.; Nyman, I.B.; Jorgensen, S.M.; Rimstad, E.; Krasnov, A. Transcriptome analyses of Atlantic salmon (Salmo salar L.) erythrocytes infected with piscine orthoreovirus (PRV). Fish. Shellfish Immunol. 2015, 45, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Puente-Marin, S.; Nombela, I.; Chico, V.; Ciordia, S.; Mena, M.C.; Coll, J.; Mercado, L.; Ortega-Villaizan, M.D.M. Rainbow trout erythrocytes ex vivo transfection with a DNA vaccine encoding VHSV glycoprotein G induces an antiviral immune response. Front. Immunol. 2018, 9, 2477. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.M.; Pearson, I.F.; Fairbanks, L.D.; Chalmers, R.A.; Bain, M.D.; Bax, B.E. The mouse immune response to carrier erythrocyte entrapped antigens. Vaccine 2006, 24, 6129–6139. [Google Scholar] [CrossRef] [PubMed]
- Cremel, M.; Guerin, N.; Horand, F.; Banz, A.; Godfrin, Y. Red blood cells as innovative antigen carrier to induce specific immune tolerance. Int. J. Pharm. 2013, 443, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, M.; Zarei, N.; Zarrin, A.; Mohammadi-Samani, S. Preparation and validation of carrier human erythrocytes loaded by bovine serum albumin as a model antigen/protein. Drug Deliv. 2007, 14, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, M.; Zarei, N.; Zarrin, A.H.; Mohammadi-Samani, S. Preparation and in vitro characterization of carrier erythrocytes for vaccine delivery. Int. J. Pharm. 2007, 338, 70–78. [Google Scholar] [CrossRef]
- Garu, A.; Moku, G.; Gulla, S.K.; Chaudhuri, A. Genetic immunization with in vivo dendritic cell-targeting liposomal DNA vaccine carrier induces long-lasting antitumor immune response. Mol. Ther. 2016, 24, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Zaneti, A.B.; Yamamoto, M.M.; Sulczewski, F.B.; Almeida, B.D.S.; Souza, H.F.S.; Ferreira, N.S.; Maeda, D.; Sales, N.S.; Rosa, D.S.; Ferreira, L.C.S.; et al. Dendritic cell targeting using a DNA vaccine induces specific antibodies and CD4+ T cells to the dengue virus envelope protein domain III. Front. Immunol. 2019, 10, 59. [Google Scholar] [CrossRef]
- Andersen, T.K.; Zhou, F.; Cox, R.; Bogen, B.; Grodeland, G. A DNA vaccine that targets hemagglutinin to antigen-presenting cells protects mice against H7 influenza. J. Virol. 2017, 91, e01340-17. [Google Scholar] [CrossRef]
- Nombela, I.; Requena-Platek, R.; Morales-Lange, B.; Chico, V.; Puente-Marin, S.; Ciordia, S.; Mena, M.C.; Coll, J.; Perez, L.; Mercado, L.; et al. Rainbow trout red blood cells exposed to viral hemorrhagic septicemia virus up-regulate antigen-processing mechanisms and MHC I&II, CD86, and CD83 antigen-presenting cell markers. Cells 2019, 8, 386. [Google Scholar] [CrossRef]
- Ai, H.W.; Henderson, J.N.; Remington, S.J.; Campbell, R.E. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: Structural characterization and applications in fluorescence imaging. Biochem. J. 2006, 400, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Valtanen, P.; Ortega-Villaizan Mdel, M.; Martinez-Lopez, A.; Medina-Gali, R.; Perez, L.; Mackenzie, S.; Figueras, A.; Coll, J.M.; Estepa, A. Autophagy-inducing peptides from mammalian VSV and fish VHSV rhabdoviral G glycoproteins (G) as models for the development of new therapeutic molecules. Autophagy 2014, 10, 1666–1680. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, A.; Garcia-Valtanen, P.; Ortega-Villaizan Mdel, M.; Chico, V.; Medina-Gali, R.M.; Perez, L.; Coll, J.; Estepa, A. Increasing versatility of the DNA vaccines through modification of the subcellular location of plasmid-encoded antigen expression in the in vivo transfected cells. PLoS ONE 2013, 8, e77426. [Google Scholar] [CrossRef] [PubMed]
- Chico, V.; Ortega-Villaizan, M.; Falco, A.; Tafalla, C.; Perez, L.; Coll, J.M.; Estepa, A. The immunogenicity of viral haemorragic septicaemia rhabdovirus (VHSV) DNA vaccines can depend on plasmid regulatory sequences. Vaccine 2009, 27, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, A.; Encinas, P.; Garcia-Valtanen, P.; Gomez-Casado, E.; Coll, J.M.; Estepa, A. Improving the safety of viral DNA vaccines: Development of vectors containing both 5′ and 3′ homologous regulatory sequences from non-viral origin. Appl. Microbiol. Biotechnol. 2013, 97, 3007–3016. [Google Scholar] [CrossRef] [PubMed]
- Garver, K.A.; Conway, C.M.; Elliott, D.G.; Kurath, G. Analysis of DNA-vaccinated fish reveals viral antigen in muscle, kidney and thymus, and transient histopathologic changes. Mar. Biotechnol. 2005, 7, 540–553. [Google Scholar] [CrossRef]
- Puente-Marin, S.; Nombela, I.; Ciordia, S.; Mena, M.C.; Chico, V.; Coll, J.; Ortega-Villaizan, M.D.M. In silico functional networks identified in fish nucleated red blood cells by means of transcriptomic and proteomic profiling. Genes 2018, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: protein—Pprotein association networks with increased coverage, supporting functional discovery in genome—wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Villaizan, M.; Martinez-Lopez, A.; Garcia-Valtanen, P.; Chico, V.; Perez, L.; Coll, J.M.; Estepa, A. Ex vivo transfection of trout pronephros leukocytes, a model for cell culture screening of fish DNA vaccine candidates. Vaccine 2012, 30, 5983–5990. [Google Scholar] [CrossRef] [PubMed]
- Raida, M.K.; Buchmann, K. Temperature-dependent expression of immune-relevant genes in rainbow trout following Yersinia ruckeri vaccination. Dis. Aquat. Org. 2007, 77, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Chico, V.; Gomez, N.; Estepa, A.; Perez, L. Rapid detection and quantitation of viral hemorrhagic septicemia virus in experimentally challenged rainbow trout by real-time RT-PCR. J. Virol. Methods 2006, 132, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Barreda, D.R.; Zhang, Y.A.; Boshra, H.; Gelman, A.E.; Lapatra, S.; Tort, L.; Sunyer, J.O. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat. Immunol. 2006, 7, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Bird, S.; Koussounadis, A.; Holland, J.W.; Carrington, A.; Zou, J.; Secombes, C.J. Identification of a novel IL-1 cytokine family member in teleost fish. J. Immunol. 2009, 183, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Chico, V.; Puente-Marin, S.; Nombela, I.; Ciordia, S.; Mena, M.C.; Carracedo, B.; Villena, A.; Mercado, L.; Coll, J.; Ortega-Villaizan, M.D.M. Shape-shifted red blood cells: A novel red blood cell stage? Cells 2018, 7, 31. [Google Scholar] [CrossRef]
- Holland, J.W.; Karim, A.; Wang, T.; Alnabulsi, A.; Scott, J.; Collet, B.; Mughal, M.S.; Secombes, C.J.; Bird, S. Molecular cloning and characterization of interferon regulatory factors 4 and 8 (IRF-4 and IRF-8) in rainbow trout, Oncorhynchus mykiss. Fish. Shellfish Immunol. 2010, 29, 157–166. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Montero, J.; Cuesta, A.; Tafalla, C. Viral hemorrhagic septicemia and infectious pancreatic necrosis viruses replicate differently in rainbow trout gonad and induce different chemokine transcription profiles. Dev. Comp. Immunol. 2010, 34, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, T.R.; Raida, M.K.; Kania, P.W.; Buchmann, K. Response of rainbow trout (Oncorhynchus mykiss) in skin and fin tissue during infection with a variant of Gyrodactylus salaris (Monogenea: Gyrodactylidae). Folia Parasitol. 2009, 56, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Villaizan, M.; Chico, V.; Martinez-Lopez, A.; Falco, A.; Perez, L.; Coll, J.M.; Estepa, A. In vitro analysis of the factors contributing to the antiviral state induced by a plasmid encoding the viral haemorrhagic septicaemia virus glycoprotein G in transfected trout cells. Vaccine 2011, 29, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Zwollo, P.; Haines, A.; Rosato, P.; Gumulak-Smith, J. Molecular and cellular analysis of B-cell populations in the rainbow trout using Pax5 and immunoglobulin markers. Dev. Comp. Immunol. 2008, 32, 1482–1496. [Google Scholar] [CrossRef] [PubMed]
- Barabas, S.; Spindler, T.; Kiener, R.; Tonar, C.; Lugner, T.; Batzilla, J.; Bendfeldt, H.; Rascle, A.; Asbach, B.; Wagner, R.; et al. An optimized IFN-gamma ELISpot assay for the sensitive and standardized monitoring of CMV protein-reactive effector cells of cell-mediated immunity. BMC Immunol. 2017, 18, 14. [Google Scholar] [CrossRef] [PubMed]
- Ceuppens, J.L.; Baroja, M.L.; Lorre, K.; Van Damme, J.; Billiau, A. Human T cell activation with phytohemagglutinin. The function of IL-6 as an accessory signal. J. Immunol. 1988, 141, 3868–3874. [Google Scholar] [PubMed]
- Wykes, M.; Renia, L. ELISPOT assay to measure peptide-specific IFN-γ production. Bio-Protoc. 2017, 7. [Google Scholar] [CrossRef]
- Morten, B.C.; Scott, R.J.; Avery-Kiejda, K.A. Comparison of three different methods for determining cell proliferation in breast cancer cell lines. J. Vis. Exp. 2016. [Google Scholar] [CrossRef]
- Sánchez, C.; Coll, J.; Domínguez, J. One-step purification of the major rainbow trout immunoglobulin. Vet. Immunol. Immunopathol. 1991, 27, 383–391. [Google Scholar] [CrossRef]
- Jung, M.H.; Chico, V.; Ciordia, S.; Mena, M.C.; Jung, S.J.; Ortega-Villaizan, M.D.M. The Megalocytivirus RBIV Induces Apoptosis and MHC Class I Presentation in Rock Bream (Oplegnathus fasciatus) Red Blood Cells. Front. Immunol. 2019, 10, 160. [Google Scholar] [CrossRef]
- Press, C.M.; Evensen, Ø. The morphology of the immune system in teleost fishes. Fish. Shellfish Immunol. 1999, 9, 309–318. [Google Scholar] [CrossRef]
- Tort, L.; Balasch, J.C.; Mackenzie, S. Fish immune system. A crossroads between innate and adaptive responses. Inmunología 2003, 22, 277–286. [Google Scholar]
- Mendez-Enriquez, E.; Garcia-Zepeda, E.A. The multiple faces of CCL13 in immunity and inflammation. Inflammopharmacology 2013, 21, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Bing, Y.; Wang, X.; Yu, Q.; Wang, Y.; Xu, S.; Song, L.; Wang, X.; Xia, B.; Zhu, Y.; et al. CCL25/CCR9 interactions regulate the function of iNKT cells in oxazolone-induced colitis in mice. PLoS ONE 2014, 9, e100167. [Google Scholar] [CrossRef] [PubMed]
- Kucia, M.; Jankowski, K.; Reca, R.; Wysoczynski, M.; Bandura, L.; Allendorf, D.J.; Zhang, J.; Ratajczak, J.; Ratajczak, M.Z. CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J. Mol. Histol. 2004, 35, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Uehara, S.; Song, K.; Farber, J.M.; Love, P.E. Characterization of CCR9 expression and CCL25/thymus-expressed chemokine responsiveness during T cell development: CD3highCD69+ thymocytes and gammadeltaTCR+ thymocytes preferentially respond to CCL25. J. Immunol. 2002, 168, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Villegas, J.; Mulero, I.; Garcia-Alcazar, A.; Munoz, I.; Penalver-Mellado, M.; Streitenberger, S.; Scapigliati, G.; Meseguer, J.; Mulero, V. Recombinant TNFalpha as oral vaccine adjuvant protects European sea bass against vibriosis: Insights into the role of the CCL25/CCR9 axis. Fish. Shellfish Immunol. 2013, 35, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhou, L.; Wang, H.Q.; Luo, X.C.; Dan, X.M.; Li, Y.W. Molecular cloning and expression analysis of CCL25 and its receptor CCR9s from Epinephelus coioides post Cryptocaryon irritans infection. Fish. Shellfish Immunol. 2017, 67, 402–410. [Google Scholar] [CrossRef]
- Monaco, J.J. A molecular model of MHC class-I-restricted antigen processing. Immunol. Today 1992, 13, 173–179. [Google Scholar] [CrossRef]
- Hewitt, E.W. The MHC class I antigen presentation pathway: Strategies for viral immune evasion. Immunology 2003, 110, 163–169. [Google Scholar] [CrossRef]
- Storni, T.; Bachmann, M.F. Loading of MHC class I and II presentation pathways by exogenous antigens: A quantitative in vivo comparison. J. Immunol. 2004, 172, 6129–6135. [Google Scholar] [CrossRef]
- Huang, A.Y.; Bruce, A.T.; Pardoll, D.M.; Levitsky, H.I. In vivo cross-priming of MHC class I-restricted antigens requires the TAP transporter. Immunity 1996, 4, 349–355. [Google Scholar] [CrossRef]
- Sever, L.; Vo, N.T.K.; Bols, N.C.; Dixon, B. Tapasin’s protein interactions in the rainbow trout peptide-loading complex. Dev. Comp. Immunol. 2018, 81, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Joffre, O.P.; Segura, E.; Savina, A.; Amigorena, S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012, 12, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Landis, E.D.; Palti, Y.; Dekoning, J.; Drew, R.; Phillips, R.B.; Hansen, J.D. Identification and regulatory analysis of rainbow trout tapasin and tapasin-related genes. Immunogenetics 2006, 58, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Ritz, U.; Seliger, B. The transporter associated with antigen processing (TAP): Structural integrity, expression, function, and its clinical relevance. Mol. Med. 2001, 7, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, A.L.; Kyritsis, C.; Tampe, R.; Cresswell, P. Early phagosomes in dendritic cells form a cellular compartment sufficient for cross presentation of exogenous antigens. Proc. Natl. Acad. Sci. USA 2003, 100, 12889–12894. [Google Scholar] [CrossRef]
- Voeten, J.T.; Rimmelzwaan, G.F.; Nieuwkoop, N.J.; Fouchier, R.A.; Osterhaus, A.D. Antigen processing for MHC class I restricted presentation of exogenous influenza A virus nucleoprotein by B-lymphoblastoid cells. Clin. Exp. Immunol. 2001, 125, 423–431. [Google Scholar] [CrossRef]
- Yewdell, J.W.; Norbury, C.C.; Bennink, J.R. Mechanisms of exogenous antigen presentation by MHC class I molecules in vitro and in vivo: Implications for generating CD8+ T cell responses to infectious agents, tumors, transplants, and vaccines. Adv. Immunol. 1999, 73, 1–77. [Google Scholar]
- St Paul, M.; Paolucci, S.; Barjesteh, N.; Wood, R.D.; Sharif, S. Chicken erythrocytes respond to Toll-like receptor ligands by up-regulating cytokine transcripts. Res. Vet. Sci. 2013, 95, 87–91. [Google Scholar] [CrossRef]
- Sanchez-Mejorada, G.; Rosales, C. Signal transduction by immunoglobulin Fc receptors. J. Leukoc. Biol. 1998, 63, 521–533. [Google Scholar] [CrossRef]
- Lowell, C.A. Src-family kinases: Rheostats of immune cell signaling. Mol. Immunol. 2004, 41, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.V.; Scholl, P.R.; Geha, R.S. Physical and functional association of the high affinity immunoglobulin G receptor (FcγRI) with the kinases Hck and Lyn. J. Exp. Med. 1994, 180, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Baruzzi, A.; Iacobucci, I.; Soverini, S.; Lowell, C.A.; Martinelli, G.; Berton, G. c-Abl and Src-family kinases cross-talk in regulation of myeloid cell migration. FEBS Lett. 2010, 584, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Zarbock, A.; Ley, K. Protein tyrosine kinases in neutrophil activation and recruitment. Arch. Biochem. Biophys. 2011, 510, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Buttari, B.; Profumo, E.; Rigano, R. Crosstalk between red blood cells and the immune system and its impact on atherosclerosis. Biomed. Res. Int. 2015, 2015, 616834. [Google Scholar] [CrossRef]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, B.; Wu, S.; Lin, L.; Liu, G.; Zhou, Y.; Wang, W.; Asim, M.; Yuan, J.; Li, L.; et al. Spring viraemia of carp virus induces autophagy for necessary viral replication. Cell Microbiol. 2015, 17, 595–605. [Google Scholar] [CrossRef]
- Li, C.; Fu, X.; Lin, Q.; Liu, L.; Liang, H.; Huang, Z.; Li, N. Autophagy promoted infectious kidney and spleen necrosis virus replication and decreased infectious virus yields in CPB cell line. Fish. Shellfish Immunol. 2017, 60, 25–32. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, N.; Hegazy, A.M.; Liu, X.; Wu, Z.; Liu, X.; Zhao, L.; Qin, Q.; Lan, J.; Lin, L. Autophagy induced by snakehead fish vesiculovirus inhibited its replication in SSN-1 cell line. Fish. Shellfish Immunol. 2016, 55, 415–422. [Google Scholar] [CrossRef]
- Pereiro, P.; Romero, A.; Diaz-Rosales, P.; Estepa, A.; Figueras, A.; Novoa, B. Nucleated teleost erythrocytes play an Nk-Lysin- and autophagy-dependent role in antiviral immunity. Front. Immunol. 2017, 8, 1458. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.W.; Shi, Y.F. Apoptosis: The quiet death silences the immune system. Pharm. Ther. 2001, 92, 135–145. [Google Scholar] [CrossRef]
- Foller, M.; Huber, S.M.; Lang, F. Erythrocyte programmed cell death. IUBMB Life 2008, 60, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhang, Q.; Xu, L.; Li, S.; Wang, D.; Zhao, J.; Liu, H.; Feng, J.; Lu, T. Effects of different cytokines on immune responses of rainbow trout in a virus DNA vaccination model. Oncotarget 2017, 8, 112222–112235. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Sun, B.; Robertsen, B. Adjuvant activity of fish type I interferon shown in a virus DNA vaccination model. Vaccine 2015, 33, 2442–2448. [Google Scholar] [CrossRef] [PubMed]
- Kambayashi, T.; Laufer, T.M. Atypical MHC class II-expressing antigen-presenting cells: Can anything replace a dendritic cell? Nat. Rev. Immunol. 2014, 14, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Cassatella, M.A. Human mature neutrophils as atypical APC. Blood 2017, 129, 1895–1896. [Google Scholar] [CrossRef]


| Gene | Forward Primer | Reverse Primer | Probe | Reference or Accession Number |
|---|---|---|---|---|
| ccl13 | CCTCTTCAACAAGTGGTTTCTCTCA | AGAAGGGTCAACACAAAATGTCTTC | - | NM_001160689.1 |
| cd8 | GAC TGC TGG CTG TGG CTT CC | CCC CGG AGC TGC CAT TCT | - | [25] |
| cd83 | TTGGCTGATGAT TCTTTCGATATC | TGCTGCCAGGAG ACACTTGT | TCCTGCCCAATG TAACGGCTGTTG | [35] |
| dnm2 | GTCAACAAGTCCATCAGGGATCT | CAACTCAGAATGGATGAAGTCTTTAGC | - | [14] |
| ef1α | ACCCTCCTCTTGGTCGTTTC | TGATGACACCAACAGCAACA | GCTGTGCGTGACATGAGGCA | [36] |
| gabarap | CCTCATCCATCCATTT TTACCTCTT | ATTCAACCGAAATCCCC ATCT | TCTGAATTTTATTTG CCTCCGGGTCTCC | [22] |
| gvhsv | GGGCCTTCCTTCTACTGGTACTC | CGGAATCCCGTAATTTGGAAT | CTGTTGCTGCAAGGCGTCCCCT | [37] |
| hck | CCATCTCCACTGGCCCTACA | TACCCTCATAGTCATACAGTGCGATAG | - | XM_021567092.1 |
| ifit5 | CCCTGCCCTCATCTTTCTTCT | CCCTCAATGACTCTGACAAGCA | CCAGCTTCGGCCTGTTTCTGTTCCA | [14] |
| igm | AAAGCCTACAAGAGGGAGACCGAT | AGAGTTATGAGGAAGAGTATGATGAAGGTG | CTCGTGTTGACTGACTGTCCATGCAGCAAC | [38] |
| il8 | AGAGACACTGA GATCATTGCCAC | CCCTCTTCATTTG TTGTTGGC | TCCTGGCCCTCC TGACCATTACTG AG | [39,40] |
| irf8 | CCGAGGAGGAGCAGAAGAGTAAAAG | GCGGCATTGAAAGAACCCAT | - | [41] |
| mhcI | GACAGTCCGTCCCTCAGTGT | CTGGAAGGTTCCATCATCGT | - | [42] |
| mhcII | TGCCATGCTGATGTGCAG | GTCCCTCAGCCAGGTCACT | CGCCTATGACTTCTACCCCAAACAAAT | [43] |
| mx1-3 | TGAAGCCCAGGATGAAATGG | TGGCAGGTCGATGAGTGTGA | ACCTCATCAGCCTAGAGATTGGCTCCCC | [44] |
| pax5 | ACGGAGATCGGATGTTCCTCTG | GATGCCGCGCTGTAGTAGTAC | - | [45] |
| pkr | ACACCGCGTACCGATGTG | GGACGAACTGCTGCCTGAAT | CACCACCTCTGAGAGCGACACCACTTC | [9] |
| tcr | AGCACCCAGACTGCCAAGCT | GAGGAGCCCTGGAACTCCA | TCT TCA TCG CTA AGA GTA CCT TCT ATG GCC TGG T | [25] |
| ulk1 | CTTCTGCTGCTGGGTCTTCTG | GGTGACGGAAGAACTCCTCAAA | CGAAACCACAAGGACCGCATGGA | [22] |
| wipi1 | CAAAGACATGAAGCTG CTGAAGA | GGTTCACAGAGAGGGC ACAGA | CTCAACACGCCCCACAA CCCCT | XM_021581280.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puente-Marin, S.; Nombela, I.; Chico, V.; Ciordia, S.; Mena, M.C.; Perez, L.; Coll, J.; Ortega-Villaizan, M.d.M. Potential Role of Rainbow Trout Erythrocytes as Mediators in the Immune Response Induced by a DNA Vaccine in Fish. Vaccines 2019, 7, 60. https://doi.org/10.3390/vaccines7030060
Puente-Marin S, Nombela I, Chico V, Ciordia S, Mena MC, Perez L, Coll J, Ortega-Villaizan MdM. Potential Role of Rainbow Trout Erythrocytes as Mediators in the Immune Response Induced by a DNA Vaccine in Fish. Vaccines. 2019; 7(3):60. https://doi.org/10.3390/vaccines7030060
Chicago/Turabian StylePuente-Marin, Sara, Ivan Nombela, Veronica Chico, Sergio Ciordia, Maria Carmen Mena, Luis Perez, Julio Coll, and Maria del Mar Ortega-Villaizan. 2019. "Potential Role of Rainbow Trout Erythrocytes as Mediators in the Immune Response Induced by a DNA Vaccine in Fish" Vaccines 7, no. 3: 60. https://doi.org/10.3390/vaccines7030060
APA StylePuente-Marin, S., Nombela, I., Chico, V., Ciordia, S., Mena, M. C., Perez, L., Coll, J., & Ortega-Villaizan, M. d. M. (2019). Potential Role of Rainbow Trout Erythrocytes as Mediators in the Immune Response Induced by a DNA Vaccine in Fish. Vaccines, 7(3), 60. https://doi.org/10.3390/vaccines7030060





