Harnessing the Loop: The Perspective of Circular RNA in Modern Therapeutics
Abstract
1. Introduction
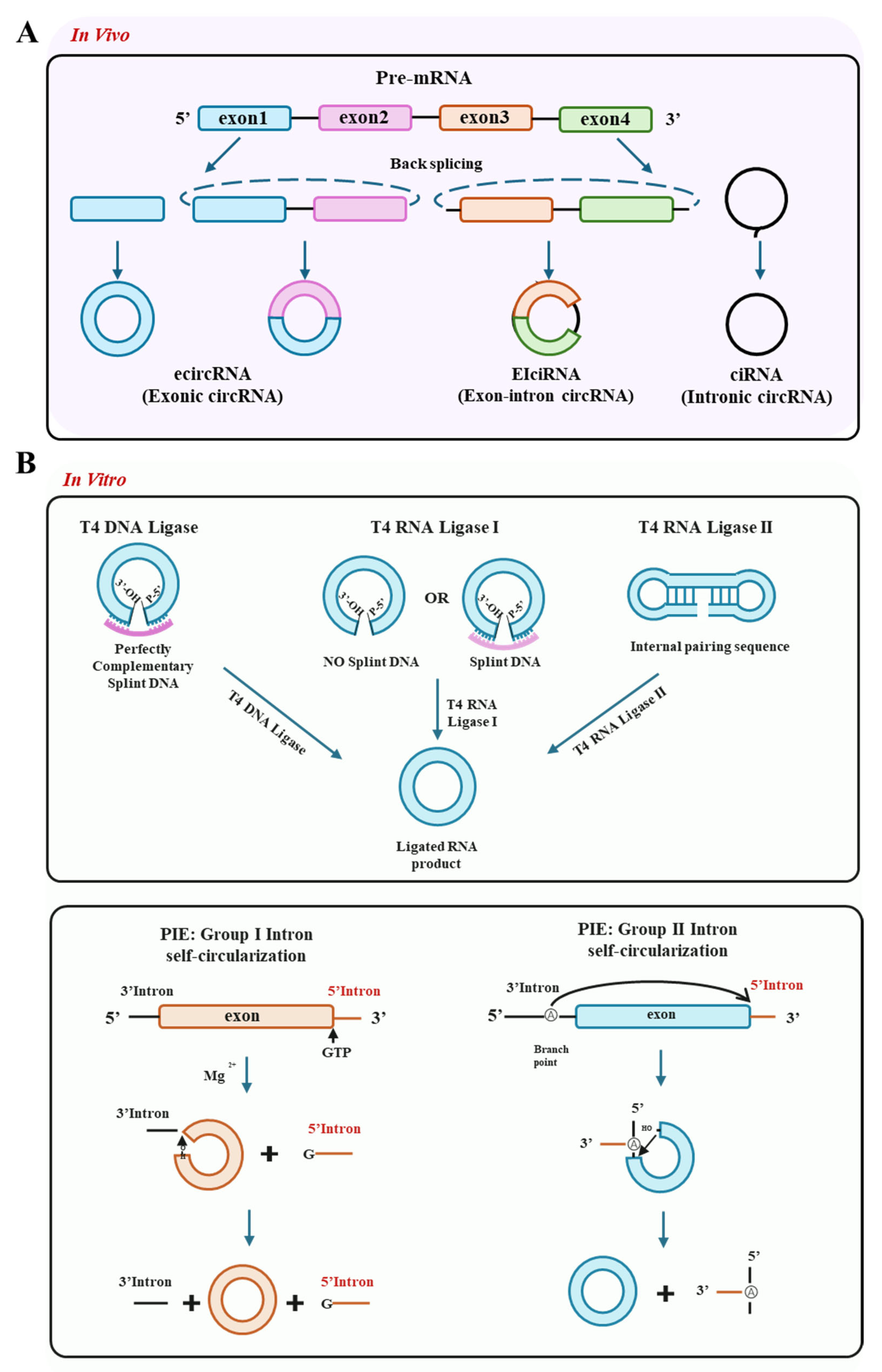
2. Characteristics of circRNA Therapeutics
2.1. Fundamental Properties of circRNA
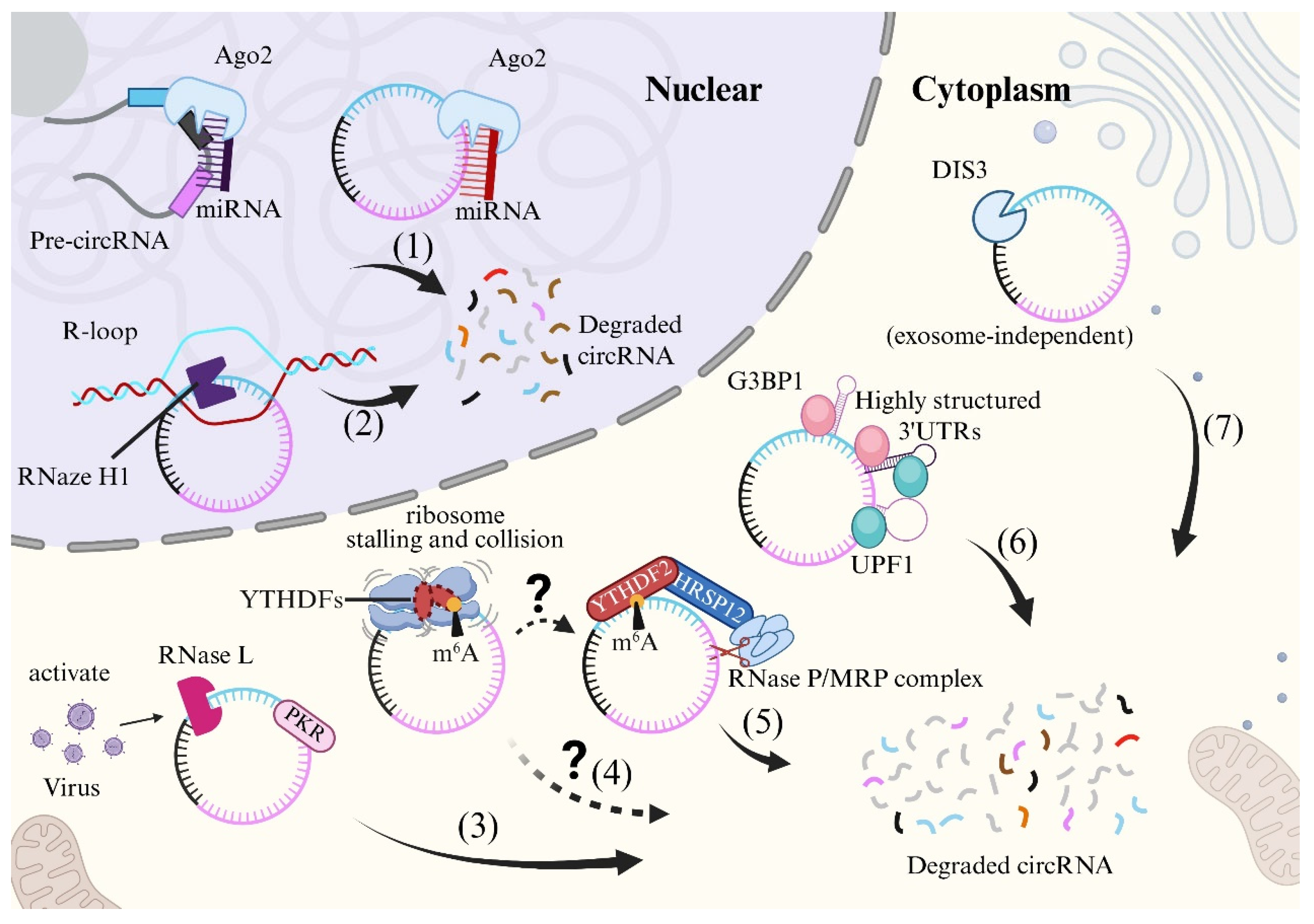
2.2. Stability and Degradation Mechanisms of circRNA
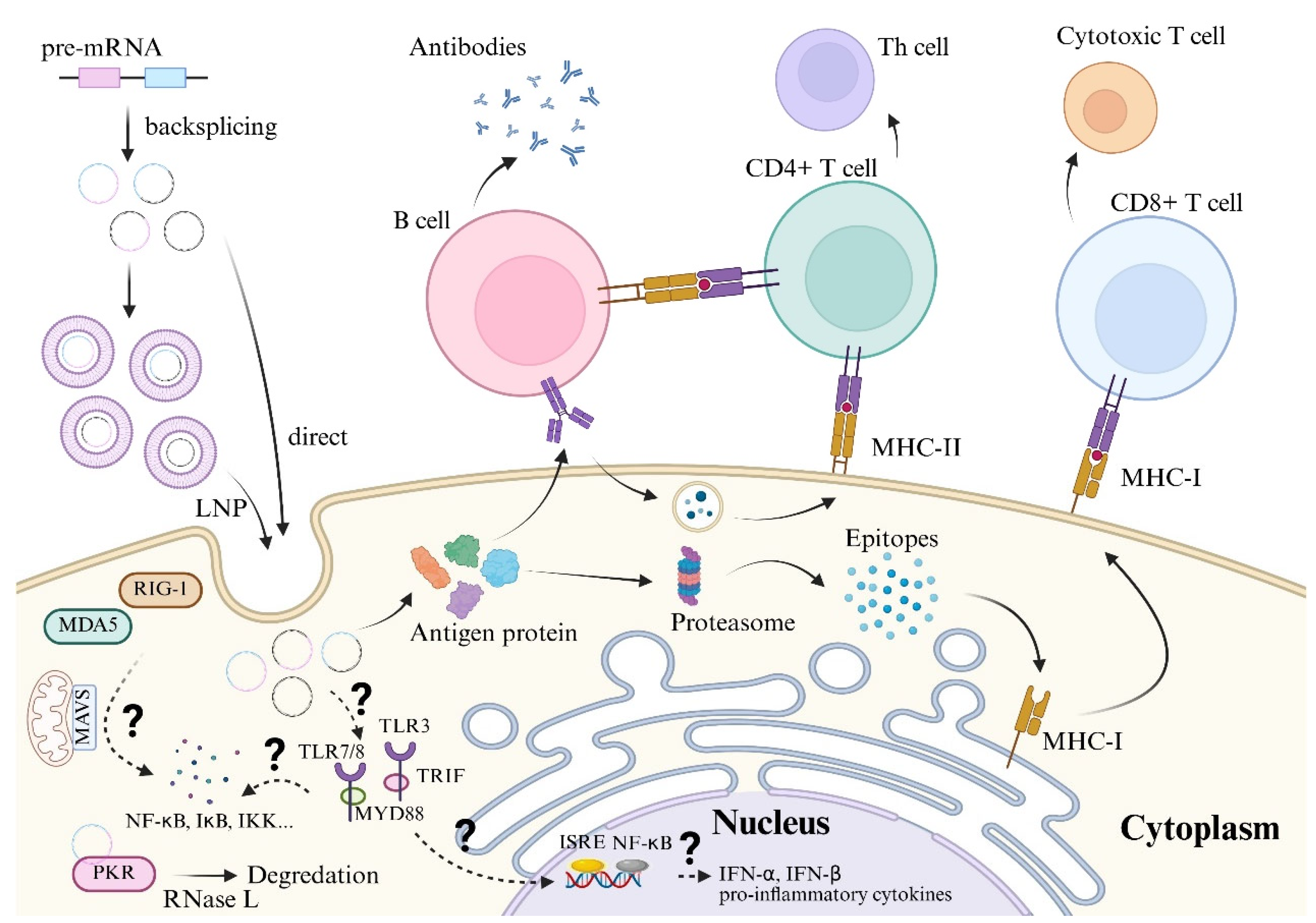
2.3. Immune Responses to circRNA
- (1)
- Sequence and structure: dsRNA regions, GU/U-rich motifs, and imperfect circularization enhance immunogenicity.
- (2)
- Cellular context: Immune cell type (e.g., dendritic cells vs. epithelial cells), subcellular localization, and RNA abundance dictate response magnitude.
- (3)
- Delivery method: Exogenous circRNAs (e.g., synthetic or encapsulated) are more immunogenic than endogenous circRNAs. When circRNAs enter cells (e.g., via exogenous delivery or endogenous release), they can activate immune pathways through multiple mechanisms.
2.3.1. Recognition by Cytosolic Immune Sensors
2.3.2. Endosomal Toll-like Receptor (TLR) Activation
2.3.3. Inflammasome Activation
2.3.4. Adaptive Immune Responses
2.3.5. Immune Evasion Strategies
- (1)
- (2)
- Masking immunostimulatory motifs: Perfectly circularized RNAs lack free ends, reducing RIG-I recognition [56].
2.3.6. Immune Activation as Adjuvants
2.4. Synthesis Strategies, Quality Control, and Delivery Methods of circRNA Vaccines
3. Sequence Design and Optimization Strategies for circRNA Therapeutics
3.1. Optimization of Open Reading Frame Sequences
3.1.1. Codon Usage Bias (CUB) and Optimization
- (1)
- Quantum computing optimizes GC content and minimizes nucleotide repeats.
- (2)
- (3)
- RNNs, including bidirectional LSTM architectures, learn codon usage patterns to recommend optimal codon substitutions (e.g., the ICOR tool for E. coli) [79].
- (4)
- Mixed-integer linear programming balances codon selection with secondary structure constraints [80].
3.1.2. Integrated Algorithmic Platforms
3.2. Selection and Optimization of IRESes
3.2.1. IRES Databases and Resource Platforms
3.2.2. Design Considerations for IRES-ORF Integration
3.3. Optimization Strategies for Group I or II INTRONS and Other Components to Enhance Splicing Efficiency
3.3.1. Design and Optimization of Intronic Ribozymes with High Circularization Efficiency
3.3.2. Optimization of Spacer Sequences in the PIE System
3.3.3. Design and Matching of Homology Arms in the CirCode System
3.4. Design Strategies to Enhance the Stability of circRNA
- (1)
- Avoid incorporating degradation signals, such as specific nucleotide sequences or secondary structures.
- (2)
- Avoid introducing high burden of methylation (e.g., m6A) or other chemical modifications to initiate circRNA degradation.
- (3)
- Engineer hairpin structures or protein-binding motifs into circRNA to generate steric hindrance, thereby blocking degradation by enzymes that could cleave circRNA (e.g., DIS3).
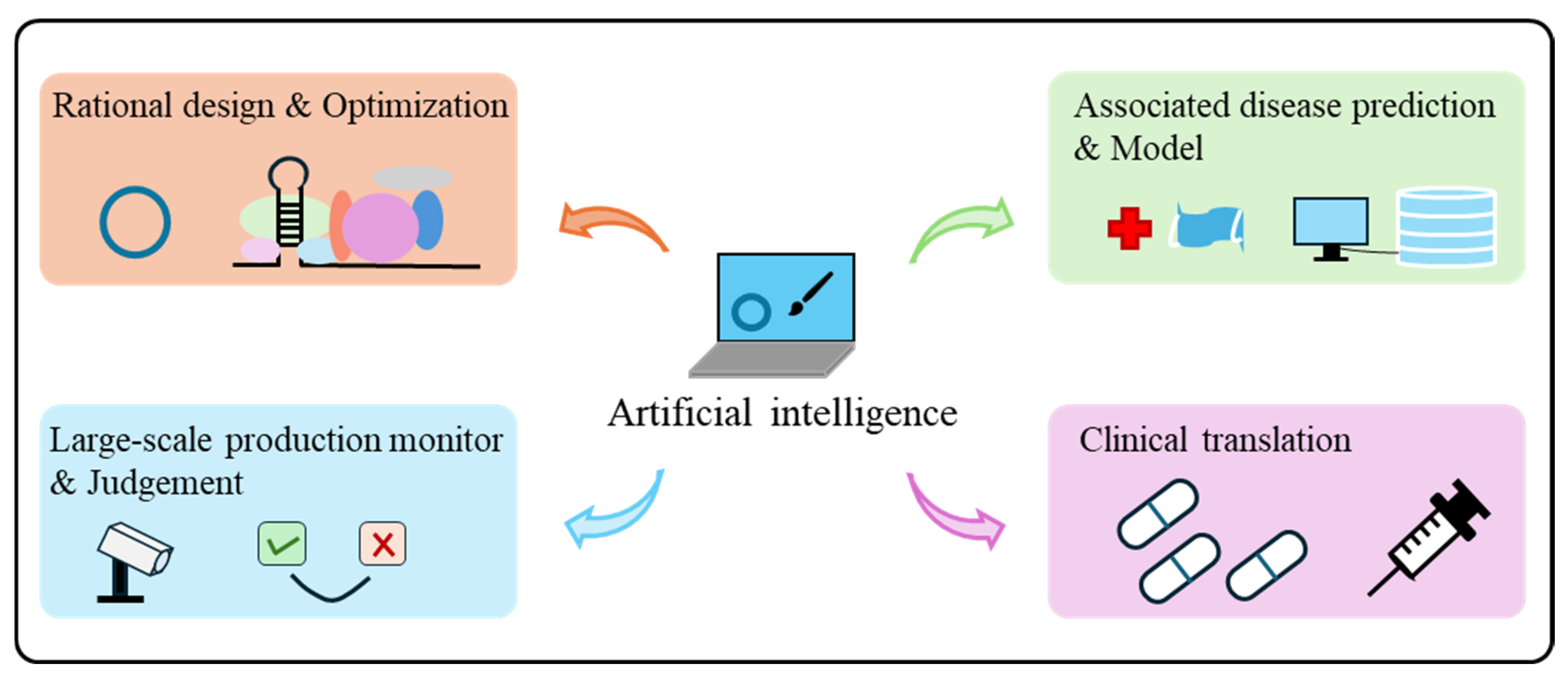
4. Artificial Intelligence (AI) in Advancing circRNA Vaccines
4.1. The Rational Design
4.2. Model Construction and Databases
4.3. Manufacturing
4.4. Clinical Translation
5. Recent Advances in circRNA Therapeutics
5.1. CircRNA Vaccines for Viral Infectious Diseases
5.1.1. SARS-CoV-2 and Variants
5.1.2. Monkeypox Virus (MPXV)
5.1.3. Zika Virus (ZIKV)
5.1.4. Rabies Virus
5.1.5. Influenza Virus
5.2. CircRNA Vaccines for Cancer Therapy
5.2.1. High-Efficiency Vaccine Platforms Inducing Antitumor Immune Responses
5.2.2. Targeting the Immunosuppressive Tumor Microenvironment
5.2.3. Tumor-Specific circRNAs as Neoantigen Sources
5.2.4. Synergistic Combination Therapies
5.3. CircRNA Therapeutics for Autoimmune and Metabolic Disorders
5.4. Advances in Other Protein Replacement Therapies via circRNA-Encoded Functional Proteins
6. Clinical Trials of circRNA-Based Therapeutics
7. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iwasaki, A.; Omer, S.B. Why and How Vaccines Work. Cell 2020, 183, 290–295. [Google Scholar] [CrossRef]
- Niu, D.; Wu, Y.; Lian, J. Circular RNA vaccine in disease prevention and treatment. Signal Transduct. Target. Ther. 2023, 8, 341. [Google Scholar] [CrossRef]
- Baden, L.R.; Sahly, H.M.E.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.-H.; Zheng, L.; Wang, Z. mRNA therapeutics: New vaccination and beyond. Fundam. Res. 2023, 3, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. Why rings of RNA could be the next blockbuster drug. Nature 2023, 622, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Hsu, M.-T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- Arnberg, A.C.; Van Ommen, G.J.B.; Grivell, L.A.; Van Bruggen, E.F.J.; Borst, P. Some yeast mitochondrial RNAs are circular. Cell 1980, 19, 313–319. [Google Scholar] [CrossRef]
- Kos, A.; Dijkema, R.; Arnberg, A.C.; van der Meide, P.H.; Schellekens, H. The hepatitis delta (δ) virus possesses a circular RNA. Nature 1986, 323, 558–560. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Chen, C.Y.; Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995, 268, 415–417. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015, 21, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e27. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef]
- Beaudry, D.; Perreault, J.P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res 1995, 23, 3064–3066. [Google Scholar] [CrossRef]
- Moore, M.J.; Sharp, P.A. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science 1992, 256, 992–997. [Google Scholar] [CrossRef]
- Wesselhoeft, R.A.; Kowalski, P.S.; Anderson, D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018, 9, 2629. [Google Scholar] [CrossRef]
- Puttaraju, M.; Been, M.D. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 1992, 20, 5357–5364. [Google Scholar] [CrossRef]
- Wei, Y.H.; Fan, X.J.; Mao, M.W. A Review on Circular RNA Translation and Its Implications in Disease. Methods Mol. Biol. 2025, 2883, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. Not just a sponge: New functions of circular RNAs discovered. Sci. China Life Sci. 2015, 58, 407–408. [Google Scholar] [CrossRef] [PubMed]
- Misir, S.; Wu, N.; Yang, B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022, 29, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Li, J.; Gao, X.; Zhang, Z.; Lai, Y.; Lin, X.; Lin, B.; Ma, M.; Liang, X.; Li, X.; Lv, W.; et al. CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502–5p/KRAS and IGF2BP2/Myc axes. Mol. Cancer 2021, 20, 138. [Google Scholar] [CrossRef]
- Silenzi, V.; D’Ambra, E.; Santini, T.; D’Uva, S.; Setti, A.; Salvi, N.; Nicoletti, C.; Scarfò, R.; Cordella, F.; Mongiardi, B.; et al. A tripartite circRNA/mRNA/miRNA interaction regulates glutamatergic signaling in the mouse brain. Cell Rep. 2024, 43, 114766. [Google Scholar] [CrossRef]
- Boo, S.H.; Shin, M.-K.; Hwang, H.J.; Hwang, H.; Chang, S.; Kim, T.; Baek, D.; Kim, Y.K. Circular RNAs trigger nonsense-mediated mRNA decay. Mol. Cell 2024, 84, 4862–4877.e4867. [Google Scholar] [CrossRef]
- Rossi, F.; Beltran, M.; Damizia, M.; Grelloni, C.; Colantoni, A.; Setti, A.; Di Timoteo, G.; Dattilo, D.; Centrón-Broco, A.; Nicoletti, C.; et al. Circular RNA ZNF609/CKAP5 mRNA interaction regulates microtubule dynamics and tumorigenicity. Mol. Cell 2022, 82, 75–89.e79. [Google Scholar] [CrossRef]
- Xu, X.; Peng, Q.; Ren, Z.; Han, Y.; Jiang, X.; Wu, Z.; Tan, S.; Yang, W.; Oyang, L.; Luo, X.; et al. CircRNF13 enhances IGF2BP1 phase separation-mediated ITGB1 mRNA stabilization in an m6A-dependent manner to promote oral cancer cisplatin chemoresistance. Mol. Cancer 2025, 24, 36. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Xu, C.; Liu, J.; Chen, J.; Li, G.; Huang, B.; Pan, Y.; Zhang, Y.; Wei, Q.; et al. Circular RNA circGlis3 protects against islet β-cell dysfunction and apoptosis in obesity. Nat. Commun. 2023, 14, 351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Liu, J.; Deng, H.; Ma, R.; Liao, J.-Y.; Liang, H.; Hu, J.; Li, J.; Guo, Z.; Cai, J.; et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 2020, 183, 76–93.e22. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zuber, P.K.; Xiao, H.; Li, X.; Gordiyenko, Y.; Ramakrishnan, V. Efficient circular RNA synthesis for potent rolling circle translation. Nat. Biomed. Eng. 2025, 9, 1062–1074. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.-K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2023, 41, 262–272. [Google Scholar] [CrossRef]
- Fan, X.; Yang, Y.; Chen, C.; Wang, Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 2022, 13, 3751. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Z. Cellular functions and biomedical applications of circular RNAs. Acta Biochim. Biophys. Sin. 2025, 57, 157–168. [Google Scholar] [CrossRef]
- Sun, M.; Yang, Y. Biological functions and applications of circRNAs—Next generation of RNA-based therapy. J. Mol. Cell Biol. 2023, 15, mjad031. [Google Scholar] [CrossRef]
- Liu, C.-X.; Chen, L.-L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Chen, L.-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Loan Young, T.; Chang Wang, K.; James Varley, A.; Li, B. Clinical delivery of circular RNA: Lessons learned from RNA drug development. Adv. Drug Deliv. Rev. 2023, 197, 114826. [Google Scholar] [CrossRef] [PubMed]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef]
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011, 30, 4414–4422. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.-L.; Lei, Y.-N.; Liu, X.-Q.; Xue, W.; Zhang, Y.; Nan, F.; Gao, X.; Zhang, J.; Wei, J.; et al. Linking circular intronic RNA degradation and function in transcription by RNase H1. Sci. China Life Sci. 2021, 64, 1795–1809. [Google Scholar] [CrossRef]
- Liu, C.-X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.-K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.; et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177, 865–880.e821. [Google Scholar] [CrossRef]
- Murakami, S.; Olarerin-George, A.O.; Liu, J.F.; Zaccara, S.; Hawley, B.; Jaffrey, S.R. m(6)A alters ribosome dynamics to initiate mRNA degradation. Cell 2025, 188, 3728–3743. [Google Scholar] [CrossRef]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507.e498. [Google Scholar] [CrossRef]
- Guo, Y.; Wei, X.; Peng, Y. Structure-Mediated Degradation of CircRNAs. Trends Cell Biol. 2020, 30, 501–503. [Google Scholar] [CrossRef]
- Fischer, J.W.; Busa, V.F.; Shao, Y.; Leung, A.K.L. Structure-Mediated RNA Decay by UPF1 and G3BP1. Mol. Cell 2020, 78, 70–84.e76. [Google Scholar] [CrossRef]
- Tao, X.; Zhai, S.-N.; Liu, C.-X.; Huang, Y.; Wei, J.; Guo, Y.-L.; Liu, X.-Q.; Li, X.; Yang, L.; Chen, L.-L. Degradation of circular RNA by the ribonuclease DIS3. Mol. Cell 2025, 85, 1674–1685. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yu, H.; Han, F.; Lai, X.; Ye, K.; Lei, S.; Mai, M.; Lai, M.; Zhang, H. Tumor-suppressive circRHOBTB3 is excreted out of cells via exosome to sustain colorectal cancer cell fitness. Mol. Cancer 2022, 21, 46. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Kim, M.V.; Chen, X.; Batista, P.J.; Aoyama, S.; Wilusz, J.E.; Iwasaki, A.; Chang, H.Y. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol. Cell 2017, 67, 228–238.e225. [Google Scholar] [CrossRef] [PubMed]
- Wesselhoeft, R.A.; Kowalski, P.S.; Parker-Hale, F.C.; Huang, Y.; Bisaria, N.; Anderson, D.G. RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo. Mol. Cell 2019, 74, 508–520.e504. [Google Scholar] [CrossRef]
- Breuer, J.; Barth, P.; Noe, Y.; Shalamova, L.; Goesmann, A.; Weber, F.; Rossbach, O. What goes around comes around: Artificial circular RNAs bypass cellular antiviral responses. Mol. Ther. Nucleic Acids 2022, 28, 623–635. [Google Scholar] [CrossRef]
- Wei, H.; Chen, C.; Zhang, K.; Li, Z.; Wei, T.; Tang, C.; Yang, Y.; Wang, Z. A flexible, efficient, and scalable platform to produce circular RNAs as new therapeutics. bioRxiv 2022. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Shen, T.; Wang, Q.; Zhou, S.; Yang, S.; Liao, S.; Su, T.; Mei, L.; Zhang, B.; et al. Small circular RNAs as vaccines for cancer immunotherapy. Nat. Biomed. Eng. 2025, 9, 249–267. [Google Scholar] [CrossRef]
- Wilusz, J.E. Circle the Wagons: Circular RNAs Control Innate Immunity. Cell 2019, 177, 797–799. [Google Scholar] [CrossRef]
- Pereira, M.; Durso, D.F.; Bryant, C.E.; Kurt-Jones, E.A.; Silverman, N.; Golenbock, D.T.; Gazzinelli, R.T. The IRAK4 scaffold integrates TLR4-driven TRIF and MYD88 signaling pathways. Cell Rep. 2022, 40, 111225. [Google Scholar] [CrossRef]
- Paik, S.; Kim, J.K.; Silwal, P.; Sasakawa, C.; Jo, E.-K. An update on the regulatory mechanisms of NLRP3 inflammasome activation. Cell. Mol. Immunol. 2021, 18, 1141–1160. [Google Scholar] [CrossRef]
- Saw, P.E.; Song, E. circRNA Vaccines. In RNA Therapeutics in Human Diseases; Saw, P.E., Song, E., Eds.; Springer Nature: Singapore, 2025; pp. 639–662. [Google Scholar]
- Amaya, L.; Grigoryan, L.; Li, Z.; Lee, A.; Wender, P.A.; Pulendran, B.; Chang, H.Y. Circular RNA vaccine induces potent T cell responses. Proc. Natl. Acad. Sci. USA 2023, 120, e2302191120. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 2019, 76, 96–109.e109. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Hou, Q.; Zhao, Y.; Zhu, J.; Zhai, M.; Li, D.; Li, Y.; Liu, C.; Li, N.; Cao, Y.; et al. Clean-PIE: A novel strategy for efficiently constructing precise circRNA with thoroughly minimized immunogenicity to direct potent and durable protein expression. bioRxiv 2022. [Google Scholar] [CrossRef]
- Wang, L.; Dong, C.; Zhang, W.; Ma, X.; Rou, W.; Yang, K.; Cui, T.; Qi, S.; Yang, L.; Xie, J.; et al. Developing an enhanced chimeric permuted intron-exon system for circular RNA therapeutics. Theranostics 2024, 14, 5869–5882. [Google Scholar] [CrossRef]
- Guillen-Cuevas, K.; Lu, X.; Birtwistle, M.R.; Husson, S.M. Purifying circular RNA by ultrafiltration. bioRxiv 2024. [Google Scholar] [CrossRef]
- Huang, K.; Li, N.; Li, Y.; Zhu, J.; Fan, Q.; Yang, J.; Gao, Y.; Liu, Y.; Gao, S.; Zhao, P.; et al. Circular mRNA Vaccine against SARS-COV-2 Variants Enabled by Degradable Lipid Nanoparticles. ACS Appl. Mater. Interfaces 2025, 17, 4699–4710. [Google Scholar] [CrossRef]
- Jia, Y.; Xu, L.; Leng, S.; Sun, Y.; Huang, X.; Wang, Y.; Ren, H.; Li, G.; Bai, Y.; Zhang, Z.; et al. Nose-to-Brain Delivery of Circular RNA SCMH1-Loaded Lipid Nanoparticles for Ischemic Stroke Therapy. Adv. Mater. 2025, 37, e2500598. [Google Scholar] [CrossRef]
- Yang, J.; Zhu, J.; Sun, J.; Chen, Y.; Du, Y.; Tan, Y.; Wu, L.; Zhai, M.; Wei, L.; Li, N.; et al. Intratumoral delivered novel circular mRNA encoding cytokines for immune modulation and cancer therapy. Mol. Ther.-Nucleic Acids 2022, 30, 184–197. [Google Scholar] [CrossRef]
- Gong, Z.; Hu, W.; Zhou, C.; Guo, J.; Yang, L.; Wang, B. Recent advances and perspectives on the development of circular RNA cancer vaccines. npj Vaccines 2025, 10, 41. [Google Scholar] [CrossRef]
- Cao, X.; Cai, Z.; Zhang, J.; Zhao, F. Engineering circular RNA medicines. Nat. Rev. Bioeng. 2025, 3, 270–287. [Google Scholar] [CrossRef]
- Technical guidelines for pharmaceutical research on mRNA vaccines for the prevention of 2019 novel coronavirus (Trial). 2020. Available online: https://www.nmpa.gov.cn/zhuanti/yqyjzxd/yqyjxd/20200814230916157.html (accessed on 25 July 2025).
- Evaluation of the quality, safety and efficacy of messenger RNA vaccines for the prevention of infectious diseases: Regulatory considerations. 2021. Available online: https://www.who.int/publications/m/item/evaluation-of-the-quality-safety-and-efficacy-of-messenger-rna-vaccines-for-the-prevention-of-infectious-diseases-regulatory-considerations (accessed on 25 July 2025).
- Analytical procedures for mRNA vaccine quality. 2022. Available online: https://www.uspnf.com/notices/analytical-procedures-mrna-vaccines-20220210 (accessed on 25 July 2025).
- Gong, H.; Wen, J.; Luo, R.; Feng, Y.; Guo, J.; Fu, H.; Zhou, X. Integrated mRNA sequence optimization using deep learning. Brief. Bioinform. 2023, 24, bbad001. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Noroozizadeh, S.; Moayedpour, S.; Kogler-Anele, L.; Xue, Z.; Zheng, D.; Montoya, F.U.; Agarwal, V.; Bar-Joseph, Z.; Jager, S. mRNA-LM: Full-length integrated SLM for mRNA analysis. Nucleic Acids Res. 2025, 53, gkaf044. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Jain, A.; Mauro, E.; LeShane, K.; Densmore, D. ICOR: Improving codon optimization with recurrent neural networks. BMC Bioinform. 2023, 24, 132. [Google Scholar] [CrossRef]
- Şen, A.; Kargar, K.; Akgün, E.; Pınar, M.Ç. Codon optimization: A mathematical programing approach. Bioinformatics 2020, 36, 4012–4020. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, L.; Lin, A.; Xu, C.; Li, Z.; Liu, K.; Liu, B.; Ma, X.; Zhao, F.; Jiang, H.; et al. Algorithm for optimized mRNA design improves stability and immunogenicity. Nature 2023, 621, 396–403. [Google Scholar] [CrossRef]
- Xu, C.; Pu, C.; Chen, R.; Wang, W.; Jiang, F.; Deng, C.; Zhai, D.; Chen, Y.; Hu, W.; Zhang, Y.; et al. circDesign Algorithm for Designing Synthetic Circular RNA. bioRxiv 2025. preprint. [Google Scholar]
- Kieft, J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008, 33, 274–283. [Google Scholar] [CrossRef]
- Weingarten-Gabbay, S.; Elias-Kirma, S.; Nir, R.; Gritsenko, A.A.; Stern-Ginossar, N.; Yakhini, Z.; Weinberger, A.; Segal, E. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 2016, 351, aad4939. [Google Scholar] [CrossRef]
- Chen, C.K.; Cheng, R.; Demeter, J.; Chen, J.; Weingarten-Gabbay, S.; Jiang, L.; Snyder, M.P.; Weissman, J.S.; Segal, E.; Jackson, P.K.; et al. Structured elements drive extensive circular RNA translation. Mol. Cell 2021, 81, 4300–4318.e4313. [Google Scholar] [CrossRef] [PubMed]
- Bonnal, S.; Boutonnet, C.; Prado-Lourenco, L.; Vagner, S. IRESdb: The Internal Ribosome Entry Site database. Nucleic Acids Res. 2003, 31, 427–428. [Google Scholar] [CrossRef] [PubMed]
- Mokrejs, M.; Vopalensky, V.; Kolenaty, O.; Masek, T.; Feketova, Z.; Sekyrova, P.; Skaloudova, B.; Kriz, V.; Pospisek, M. IRESite: The database of experimentally verified IRES structures (www.iresite.org). Nucleic Acids Res. 2006, 34, D125–D130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Y.; Wang, C.; Zhang, H.; Zhang, H.; Jiang, B.; Guo, X.; Song, X. IRESbase: A Comprehensive Database of Experimentally Validated Internal Ribosome Entry Sites. Genom. Proteom. Bioinform. 2020, 18, 129–139. [Google Scholar] [CrossRef]
- Abe, N.; Matsumoto, K.; Nishihara, M.; Nakano, Y.; Shibata, A.; Maruyama, H.; Shuto, S.; Matsuda, A.; Yoshida, M.; Ito, Y.; et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 2015, 5, 16435. [Google Scholar] [CrossRef]
- Busa, V.F.; Leung, A.K.L. Thrown for a (stem) loop: How RNA structure impacts circular RNA regulation and function. Methods 2021, 196, 56–67. [Google Scholar] [CrossRef]
- Khodadoust, M.S.; Olsson, N.; Wagar, L.E.; Haabeth, O.A.; Chen, B.; Swaminathan, K.; Rawson, K.; Liu, C.L.; Steiner, D.; Lund, P.; et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 2017, 543, 723–727. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, L.; Wang, W.; Tang, Y.; Wang, Q.; An, J.; Xu, H.; Ge, Y.; Zhu, H.; Wang, H.; et al. Improving the Circularization Efficiency, Stability and Translatability of Circular RNA by circDesign. bioRxiv 2023. [Google Scholar] [CrossRef]
- Peng, L.; Li, H.; Yuan, S.; Meng, T.; Chen, Y.; Fu, X.; Cao, D. metaCDA: A Novel Framework for CircRNA-Driven Drug Discovery Utilizing Adaptive Aggregation and Meta-Knowledge Learning. J. Chem. Inf. Model. 2025, 65, 2129–2144. [Google Scholar] [CrossRef]
- Wang, S.; Xiong, Y.; Zhang, Y.; Wang, H.; Chen, M.; Li, J.; Luo, P.; Luo, Y.-H.; Hecht, M.; Frey, B.; et al. TCCIA: A comprehensive resource for exploring CircRNA in cancer immunotherapy. J. Immunother. Cancer 2024, 12, e008040. [Google Scholar] [CrossRef]
- Chu, Y.; Zhang, Y.; Wang, Q.; Zhang, L.; Wang, X.; Wang, Y.; Salahub, D.R.; Xu, Q.; Wang, J.; Jiang, X.; et al. A transformer-based model to predict peptide–HLA class I binding and optimize mutated peptides for vaccine design. Nat. Mach. Intell. 2022, 4, 300–311. [Google Scholar] [CrossRef]
- Chuwdhury, G.S.; Guo, Y.; Chiang, C.L.; Lam, K.O.; Kam, N.W.; Liu, Z.; Dai, W. ImmuneMirror: A machine learning-based integrative pipeline and web server for neoantigen prediction. Brief. Bioinform. 2024, 25, bbae024. [Google Scholar] [CrossRef] [PubMed]
- Madern, M.F.; Yang, S.; Witteveen, O.; Segeren, H.A.; Bauer, M.; Tanenbaum, M.E. Long-term imaging of individual ribosomes reveals ribosome cooperativity in mRNA translation. Cell 2025, 188, 1896–1911.e1824. [Google Scholar] [CrossRef] [PubMed]
- Vartak, A.; Sucheck, S.J. Recent Advances in Subunit Vaccine Carriers. Vaccines 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cordón, P.J.; Montoya, M.; Reis, A.L.; Dixon, L.K. African swine fever: A re-emerging viral disease threatening the global pig industry. Vet. J. 2018, 233, 41–48. [Google Scholar] [CrossRef]
- You, H.; Jones, M.K.; Gordon, C.A.; Arganda, A.E.; Cai, P.; Al-Wassiti, H.; Pouton, C.W.; McManus, D.P. The mRNA Vaccine Technology Era and the Future Control of Parasitic Infections. Clin. Microbiol. Rev. 2023, 36, e00241–00221. [Google Scholar] [CrossRef]
- Zhang, C.; Maruggi, G.; Shan, H.; Li, J. Advances in mRNA Vaccines for Infectious Diseases. Front. Immunol. 2019, 10, 594. [Google Scholar] [CrossRef]
- Chung, J.Y.; Thone, M.N.; Kwon, Y.J. COVID-19 vaccines: The status and perspectives in delivery points of view. Adv. Drug Deliv. Rev. 2021, 170, 1–25. [Google Scholar] [CrossRef]
- Ge, Q.; Chen, P.; Cheng, Y.; Xiao, Y. The long road for vaccine development with difficulties and hopes. Emerg. Microbes Infect. 2024, 13, 2396886. [Google Scholar] [CrossRef]
- Zhang, Z.; Fu, Y.; Ju, X.; Zhang, F.; Zhang, P.; He, M. Advances in Engineering Circular RNA Vaccines. Pathogens 2024, 13, 692. [Google Scholar] [CrossRef]
- Zhou, J.; Ye, T.; Yang, Y.; Li, E.; Zhang, K.; Wang, Y.; Chen, S.; Hu, J.; Zhang, K.; Liu, F.; et al. Circular RNA vaccines against monkeypox virus provide potent protection against vaccinia virus infection in mice. Mol. Ther. J. Am. Soc. Gene Ther. 2024, 32, 1779–1789. [Google Scholar] [CrossRef]
- Yue, X.; Zhong, C.; Cao, R.; Liu, S.; Qin, Z.; Liu, L.; Zhai, Y.; Luo, W.; Lian, Y.; Zhang, M.; et al. CircRNA based multivalent neuraminidase vaccine induces broad protection against influenza viruses in mice. npj Vaccines 2024, 9, 170. [Google Scholar] [CrossRef]
- Liu, X.; Li, Z.; Li, X.; Wu, W.; Jiang, H.; Zheng, Y.; Zhou, J.; Ye, X.; Lu, J.; Wang, W.; et al. A single-dose circular RNA vaccine prevents Zika virus infection without enhancing dengue severity in mice. Nat. Commun. 2024, 15, 8932. [Google Scholar] [CrossRef]
- Wang, F.; Cai, G.; Wang, Y.; Zhuang, Q.; Cai, Z.; Li, Y.; Gao, S.; Li, F.; Zhang, C.; Zhao, B.; et al. Circular RNA-based neoantigen vaccine for hepatocellular carcinoma immunotherapy. MedComm 2024, 5, e667. [Google Scholar] [CrossRef] [PubMed]
- Singh, O.N.; Berry, U.; Joshi, G.; Asuru, T.R.; Chandrasekar, K.; Narayanan, S.; Srivastava, P.; Tiwari, M.; Chattopadhyay, S.; Mehdi, F.; et al. Immunogenicity and protection efficacy of self-amplifying and circular mRNA vaccines for SARS-CoV-2. bioRxiv 2024. [Google Scholar] [CrossRef]
- Xu, S.; Xu, Y.; Solek, N.C.; Chen, J.; Gong, F.; Varley, A.J.; Golubovic, A.; Pan, A.; Dong, S.; Zheng, G.; et al. Tumor-Tailored Ionizable Lipid Nanoparticles Facilitate IL-12 Circular RNA Delivery for Enhanced Lung Cancer Immunotherapy. Adv. Mater. 2024, 36, e2400307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ma, B.; Li, B.; Li, Z.; Gao, M.; Zhao, H.; Peng, R.; Hu, J.; Wang, Y.; You, W.; et al. Cardiolipin-mimic lipid nanoparticles without antibody modification delivered senolytic in-vivo CAR-T therapy for inflamm-aging. bioRxiv 2024. [Google Scholar] [CrossRef]
- Hu, Q.; Zhao, H.; Zhou, K.; Hua, X.; Zhang, X. Scarless circular mRNA-based CAR-T cell therapy elicits superior anti-tumor efficacy. bioRxiv 2024. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Liu, C.; Jiang, Y.; Wang, Z.; Guo, Z.; Li, X. A single dose of VEGF-A circular RNA sustains in situ long-term expression of protein to accelerate diabetic wound healing. J. Control. Release Off. J. Control. Release Soc. 2024, 373, 319–335. [Google Scholar] [CrossRef]
- Tong, M.; Palmer, N.; Dailamy, A.; Kumar, A.; Khaliq, H.; Han, S.; Finburgh, E.; Wing, M.; Hong, C.; Xiang, Y.; et al. Robust genome and cell engineering via in vitro and in situ circularized RNAs. Nat. Biomed. Eng. 2025, 9, 109–126. [Google Scholar] [CrossRef]
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744. [Google Scholar] [CrossRef]
- Seephetdee, C.; Bhukhai, K.; Buasri, N.; Leelukkanaveera, P.; Lerdwattanasombat, P.; Manopwisedjaroen, S.; Phueakphud, N.; Kuhaudomlarp, S.; Olmedillas, E.; Saphire, E.O.; et al. A circular mRNA vaccine prototype producing VFLIP-X spike confers a broad neutralization of SARS-CoV-2 variants by mouse sera. Antivir. Res. 2022, 204, 105370. [Google Scholar] [CrossRef]
- Huang, D.; Zhu, X.; Ye, S.; Zhang, J.; Liao, J.; Zhang, N.; Zeng, X.; Wang, J.; Yang, B.; Zhang, Y.; et al. Tumour circular RNAs elicit anti-tumour immunity by encoding cryptic peptides. Nature 2024, 625, 593–602. [Google Scholar] [CrossRef]
- Wan, J.; Wang, Z.; Wang, L.; Wu, L.; Zhang, C.; Zhou, M.; Fu, Z.F.; Zhao, L. Circular RNA vaccines with long-term lymph node-targeting delivery stability after lyophilization induce potent and persistent immune responses. Mbio 2024, 15, e01775–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, Y.; Zhong, J.; Wu, X.; Leng, Z.; Liu, M.; Wang, Y.; Wang, Y.; Yang, X.; Huang, N.; et al. Lnc-H19-derived protein shapes the immunosuppressive microenvironment of glioblastoma. Cell Rep. Med. 2024, 5, 101806. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Manoharan, T.; Liu, B.; Cheng, C.Z.M.; En Siew, B.; Cheong, W.-K.; Lee, K.Y.; Tan, I.J.-W.; Lieske, B.; Tan, K.-K.; et al. Circular RNA as a source of neoantigens for cancer vaccines. J. Immunother. Cancer 2024, 12, e008402. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hu, Y.; Li, J.; He, J.; Yu, G.; Wang, J.; Lin, X. Intranasal prime-boost RNA vaccination elicits potent T cell response for lung cancer therapy. Signal Transduct. Target. Ther. 2025, 10, 101. [Google Scholar] [CrossRef]
- Cai, Z.; Wuri, Q.; Song, Y.; Qu, X.; Hu, H.; Cao, S.; Wu, H.; Wu, J.; Wang, C.; Yu, X.; et al. CircRNA-loaded DC vaccine in combination with low-dose gemcitabine induced potent anti-tumor immunity in pancreatic cancer model. Cancer Immunol. Immunother. 2025, 74, 68. [Google Scholar] [CrossRef]
- Guo, S.-K.; Liu, C.-X.; Xu, Y.-F.; Wang, X.; Nan, F.; Huang, Y.; Li, S.; Nan, S.; Li, L.; Kon, E.; et al. Therapeutic application of circular RNA aptamers in a mouse model of psoriasis. Nat. Biotechnol. 2024, 43, 236–246. [Google Scholar] [CrossRef]
- Shi, R.; Jin, Y.; Zhao, S.; Yuan, H.; Shi, J.; Zhao, H. Hypoxic ADSC-derived exosomes enhance wound healing in diabetic mice via delivery of circ-Snhg11 and induction of M2-like macrophage polarization. Biomed. Pharmacother. 2022, 153, 113463. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Chen, L.Y.; Zhang, M.; Wang, C.; Du, X.L.; Ye, S.L.; Li, X.Q.; Chen, H.; Hu, N. Exosomes derived from BMSCs enhance diabetic wound healing through circ-Snhg11 delivery. Diabetol. Metab. Syndr. 2024, 16, 37. [Google Scholar] [CrossRef] [PubMed]
- Hu, N.; Cai, Z.W.; Jiang, X.D.; Wang, C.; Tang, T.; Xu, T.Z.; Chen, H.; Li, X.Q.; Du, X.L.; Cui, W.G. Hypoxia-pretreated ADSC-derived exosome-embedded hydrogels promote angiogenesis and accelerate diabetic wound healing. Acta Biomater. 2023, 157, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.F.; Jia, P.F.; Zhao, S.M.; Yuan, H.X.; Shi, J.H.; Zhao, H. Upregulation of circ-IGF1R increased therapeutic effect of hypoxia-pretreated ADSC-derived extracellular vesicle by regulating miR-503-5p/HK2/VEGFA axis. J. Cell. Mol. Med. 2024, 28, e18471. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.L.; Chu, Z.Q.; Wang, Z.H.; Li, Q.K.; Meng, S.; Lu, Y.; Ma, K.; Cui, S.N.; Hu, W.Z.; Zhang, W.H.; et al. circCDK13-loaded small extracellular vesicles accelerate healing in preclinical diabetic wound models. Nat. Commun. 2024, 15, 3904. [Google Scholar] [CrossRef]
- Wang, X.; Jian, W.; Luo, Q.; Fang, L. CircSEMA4B inhibits the progression of breast cancer by encoding a novel protein SEMA4B-211aa and regulating AKT phosphorylation. Cell Death Dis. 2022, 13, 794. [Google Scholar] [CrossRef]
- Li, H.; Peng, K.; Yang, K.; Ma, W.; Qi, S.; Yu, X.; He, J.; Lin, X.; Yu, G. Circular RNA cancer vaccines drive immunity in hard-to-treat malignancies. Theranostics 2022, 12, 6422–6436. [Google Scholar] [CrossRef]
- Yu, H.; Wen, Y.; Yu, W.; Lu, L.; Yang, Y.; Liu, C.; Hu, Z.; Fang, Z.; Huang, S. Optimized circular RNA vaccines for superior cancer immunotherapy. Theranostics 2025, 15, 1420–1438. [Google Scholar] [CrossRef]
- Zaiou, M. The Emerging Role and Promise of Circular RNAs in Obesity and Related Metabolic Disorders. Cells 2020, 9, 1473. [Google Scholar] [CrossRef]
- Zhai, X.Y.; Zhang, Y.F.; Xin, S.Y.; Cao, P.F.; Lu, J.H. Insights into the Involvement of Circular RNAs in Autoimmune Diseases. Front. Immunol. 2021, 12, 622316. [Google Scholar] [CrossRef]
- Hunter, T.L.; Bao, Y.; Zhang, Y.; Matsuda, D.; Riener, R.; Wang, A.; Li, J.J.; Soldevila, F.; Chu, D.S.H.; Nguyen, D.P.; et al. In vivo CAR T cell generation to treat cancer and autoimmune disease. Science 2025, 388, 1311–1317. [Google Scholar] [CrossRef]
- Wang, S.; Wu, C.W.; Wang, J.W.; Yuan, F.; Hou, Y.F.; Cao, T.T.; Xu, L.S.; Qian, L.; Xia, Y.B.; Xu, L.; et al. Hsa_circ_0002301 inhibits ferroptosis in gastric cancer by encoding the de novo protein HECTD1-463aa. J. Transl. Med. 2025, 23, 250. [Google Scholar] [CrossRef]
- Guan, C.H.; Gao, J.J.; Zou, X.L.; Shi, W.J.; Hao, Y.H.; Ge, Y.F.; Xu, Z.Q.; Yang, C.R.; Bi, S.W.; Jiang, X.M.; et al. A Novel 167-Amino Acid Protein Encoded by CircPCSK6 Inhibits Intrahepatic Cholangiocarcinoma Progression via IKBα Ubiquitination. Adv. Sci. 2025, 12, 2409173. [Google Scholar] [CrossRef]
- Peng, D.Z.; Wei, C.; Jing, B.Y.; Yu, R.Z.; Zhang, Z.Y.; Han, L. A novel protein encoded by circCOPA inhibits the malignant phenotype of glioblastoma cells and increases their sensitivity to temozolomide by disrupting the NONO-SFPQ complex. Cell Death Dis. 2024, 15, 616. [Google Scholar] [CrossRef]

| Vaccine Type | Profile | Advantages | Disadvantages |
|---|---|---|---|
| Inactivated | Pathogen inactivated in entirety |
|
|
| Live-attenuated | Live pathogen with attenuated virulence |
|
|
| Subunit | Specific pathogen components (protein/polysaccharide/VLP) |
|
|
| Viral Vector | Antigen gene delivered via harmless viral vector |
|
|
| DNA | Antigen gene inserted into plasmid DNA vector |
|
|
| mRNA | Antigen-encoding mRNA encapsulated in lipid nanoparticles |
|
|
| circRNA | Synthetic circRNA encapsulated in lipid nanoparticles |
|
|
| CircRNA | Expressed Protein | Disease/Pathogen | Application Field | Mechanism/Function | Reference |
|---|---|---|---|---|---|
| circRNA RBD-Delta | Spike trimeric RBD (Delta/Omicron) | COVID-19 | Vaccine prevention | Induces neutralizing antibodies and Th1-skewed cellular immunity, providing broad-spectrum protection against variants | [115] |
| circRNA VFLIP-X | VFLIP-X spike protein (K417N mutations) | SARS-CoV-2 variants | Vaccine prevention | Elicits broad neutralizing antibodies and balanced Th1/Th2 responses against multiple VOCs/VOIs | [116] |
| circFAM53B | Cryptic peptides (ALFRLTNRA/RTAHYGTGR) | Breast cancer, melanoma | Universal cancer vaccine | Encodes tumor-specific antigens (TSAs) via HLA-I/II dual presentation, activating CD8+/CD4+ T cells | [117] |
| circRNA EDIII-Fc + NS1 | EDIII-Fc and NS1 proteins | Zika virus | Vaccine prevention | Induces neutralizing antibodies and germinal center (GC) reactions, avoiding DENV ADE effect with single-dose efficacy | [107] |
| circRNA-G | Rabies virus glycoprotein G (RABV-G) | Rabies | Vaccine prevention | Enhances humoral immunity (high IgG/neutralizing antibodies) and lymph node-targeted delivery for prolonged antigen expression | [118] |
| circRNA-NA (N1/N2/IBV) | Neuraminidase (N1, N2, influenza B) | Influenza | Vaccine prevention | Induces broad neutralizing antibodies and Th1-skewed immunity against H1N1/H3N2/Victoria/Yamagata strains | [106] |
| cirA29L/cirA35R/cirB6R/cirM1R | A29L, A35R, B6R, M1R proteins | Monkeypox | Vaccine prevention | Elicits neutralizing antibodies and T cell responses via multivalent antigens, reducing tissue viral load | [105] |
| H19-IRP | H19-IRP (protein encoded by lncRNA) | Glioblastoma | Cancer immunotherapy | Activates CCL2/Galectin-9 transcription to recruit MDSCs/TAMs; triggers T cell responses as a TAA | [119] |
| circRNA-PTPN2 | PTPN2 (neoantigen) | Hepatocellular carcinoma | Neoantigen vaccine | Activates DC maturation and T cell responses via circRNA-LNP delivery, enhancing tumor cell targeting | [108] |
| circRAPGEF5, circMYH9 | Tumor-specific cryptic peptides | Colorectal cancer | Liquid biopsy-driven therapy | Presented via HLA-A*11:01, inducing T cell-mediated tumor organoid clearance | [120] |
| circRNA-LNP | SIIINFEKL (OVA antigen) | Lung cancer | Mucosal immunotherapy | Enhances antigen-specific T cell responses via cDC1s and alveolar macrophages, reducing systemic toxicity | [121] |
| circRNA-FS (FAPα/survivin) | FAPα, survivin | Pancreatic cancer | Chemo-immunotherapy combination | Enhances DC vaccine antigen expression, induces ICD, synergizes with gemcitabine to suppress Tregs | [122] |
| Small circRNA (<300 nt) (e.g., circRNA-SIINFEKL) | Peptide antigens (e.g., SIINFEKL) | Low-immunogenic tumors (e.g., melanoma) | Long-term immune memory induction | High stability (half-life > 7 days), low PKR activation, synergizes with immune checkpoint inhibitors | [59] |
| circRNA SCAR | - | Non-alcoholic steatohepatitis | Metabolic disease/Liver disease | Binds ATP5B and inhibits mitochondrial permeability transition pore (mPTP) opening, reduces mitochondrial ROS output, alleviates fibroblast activation and inflammation | [35] |
| circPOLR2A (EPIC) | - | Psoriasis | Immune modulation/inflammation control | Stabilizes PKR binding to inhibit its activity, attenuates IFN-α signaling and dsRNA-mediated inflammatory responses | [123] |
| circ-Snhg11 | - | Diabetes mellitus | Wound healing | Inhibits hyperglycemia-induced endothelial damage via miR-144-3p/HIF-1α axis, induces M2-like macrophage polarization | [124] |
| circ-Snhg11 | - | Diabetes mellitus | Diabetic wound healing | Enhances SLC7A11/GPX4-mediated anti-ferroptosis signaling through miR-144-3p sponge effect, promotes angiogenesis | [125] |
| circ-Snhg11 | - | Diabetes mellitus | Angiogenesis | Activates miR-144-3p/NFE2L2/HIF1α pathway to suppress oxidative stress and improve endothelial function with vascular regeneration | [126] |
| circ-IGF1R | - | Diabetes mellitus | Diabetic foot ulcer | Upregulates HK2 and VEGFA expression via miR-503-5p sponge adsorption, enhances angiogenesis while reducing apoptosis | [127] |
| circCDK13 | - | Diabetes mellitus | Wound healing/regenerative medicine | Interacts with IGF2BP3 in m6A-dependent manner to enhance CD44 and c-MYC expression, promoting cutaneous cell proliferation/migration | [128] |
| VEGF-A circRNA | VEGF-A | Diabetes mellitus | Diabetic foot ulcer | Achieves sustained VEGF-A protein expression via lipid nanoparticle delivery to promote angiogenesis | [113] |
| IL-12 circRNA | IL-12 | Lung cancer | Immunotherapy | Delivers circRNA-encoded IL-12 through H1L1A1B3 LNPs, activates immune response, increases CD8+ T cell infiltration, inhibits tumor growth | [113] |
| circSEMA4B | SEMA4B-211aa | Breast cancer | Cancer therapy | Encodes SEMA4B-211aa to suppress PI3K/AKT pathway via miR-330-3p/PDCD4 axis-mediated inhibition of AKT phosphorylation | [129] |
| Study Title | Year | Location | Sponsor | Study Status | Study ID | Data From |
|---|---|---|---|---|---|---|
| A Study to Evealuate Safety and Immunogenicity of TI-0010 SARS-CoV-2 Vaccine in Healthy Adults | 2023 | China | National Drug Clinical Trial Institute of the Second Affiliated Hospital of Bengbu Medical College | RECRUITING | NCT06205524 | clinicaltrials.gov |
| A Single Arm Clinical Study of Dendritic Cell Vaccine Loaded With CircRNA Encoding Cryptic Peptide for Patients With HER2-negative Advanced Breast Cancer | 2024 | China | Sun Yat-Sen Memorial Hospital of Sun Yat-Sen University | NOT_YET_RECRUITING | NCT06530082 | clinicaltrials.gov |
| First-in-Human Pilot Study of Epicardial CircRNA-HM2002 Injection in CABG for Ischemic Heart Failure | 2024 | China | Ruijin Hospital | ACTIVE_NOT_RECRUITING | NCT06621576 | clinicaltrials.gov |
| HM2002 Injection | 2025 | China | Shanghai CirCode Biomed Co., Ltd, Shanghai, China. | UNKONW | CXSL2400740 | Center for Drug Evaluation of NMPA |
| Study of CircRNA Treatment in Patients with Radiation Induced Xerostomia-1 (RXRG001) | 2025 | China | RiboX Therapeutics Ltd, Shanghai, China. | RECRUITING | NCT06714253 | clinicaltrials.gov |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.-Y.; Zhu, F.-M.; Zhang, Y.-J.; Wei, H.Y. Harnessing the Loop: The Perspective of Circular RNA in Modern Therapeutics. Vaccines 2025, 13, 821. https://doi.org/10.3390/vaccines13080821
Zhao Y-Y, Zhu F-M, Zhang Y-J, Wei HY. Harnessing the Loop: The Perspective of Circular RNA in Modern Therapeutics. Vaccines. 2025; 13(8):821. https://doi.org/10.3390/vaccines13080821
Chicago/Turabian StyleZhao, Yang-Yang, Fu-Ming Zhu, Yong-Juan Zhang, and Huanhuan Y. Wei. 2025. "Harnessing the Loop: The Perspective of Circular RNA in Modern Therapeutics" Vaccines 13, no. 8: 821. https://doi.org/10.3390/vaccines13080821
APA StyleZhao, Y.-Y., Zhu, F.-M., Zhang, Y.-J., & Wei, H. Y. (2025). Harnessing the Loop: The Perspective of Circular RNA in Modern Therapeutics. Vaccines, 13(8), 821. https://doi.org/10.3390/vaccines13080821






