Universal Bacterium-Vectored COVID-19 Vaccine Expressing Early SARS-CoV-2 Conserved Proteins Cross-Protects Against Late Variants in Hamsters
Abstract
1. Introduction
2. Methods
2.1. Ethical Statement
2.2. Viruses and Bacteria
2.3. Hamsters
2.4. Proteins, Antibodies, and Heat-Inactivated Bacteria
2.5. Generation of rLVS ΔcapB::RdRp/MN COVID-19 Vaccine Candidate
2.6. Efficacy Study in Hamsters
2.7. Histopathology Assessment
2.8. Virus Assay
2.9. Enzyme-Linked Immunosorbent Assay (ELISA)
2.10. Statistics
3. Results
3.1. Construction and Verification of rLVS ΔcapB:RdRp/MN Vaccine Candidate
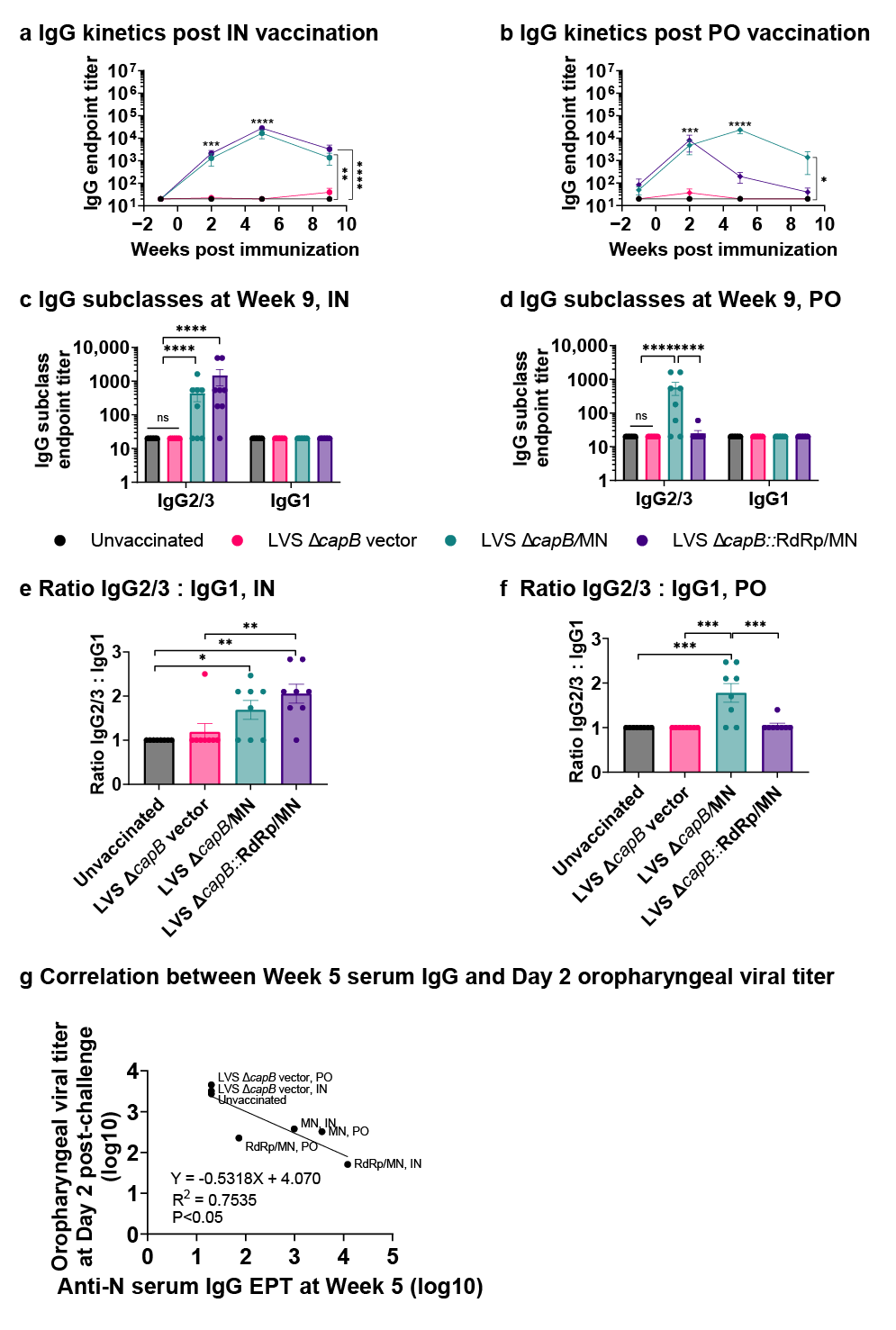
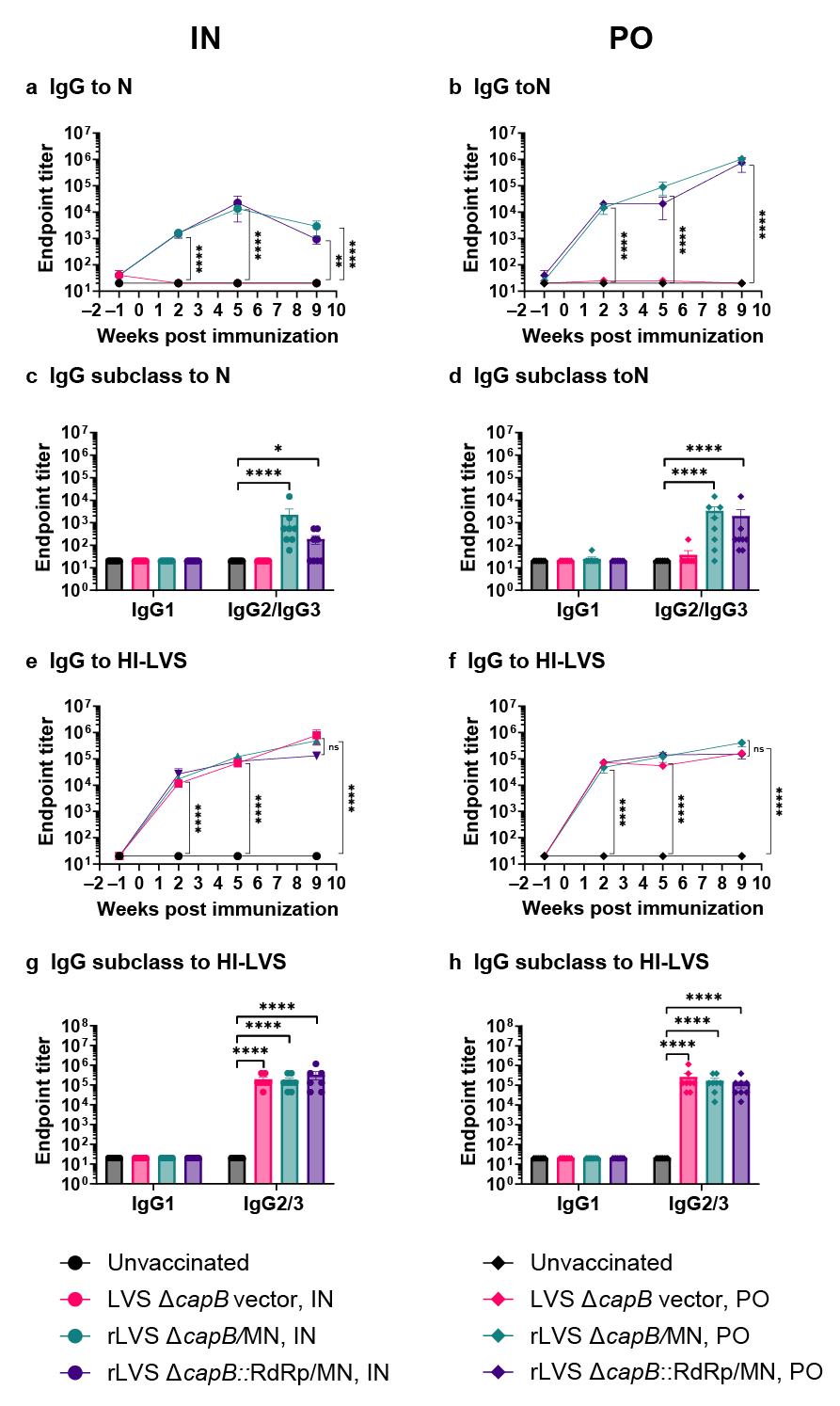
3.2. Study of Vaccine Efficacy Against SARS-CoV-2 Delta Variant Challenge in the Hamster Model—Experiment 1
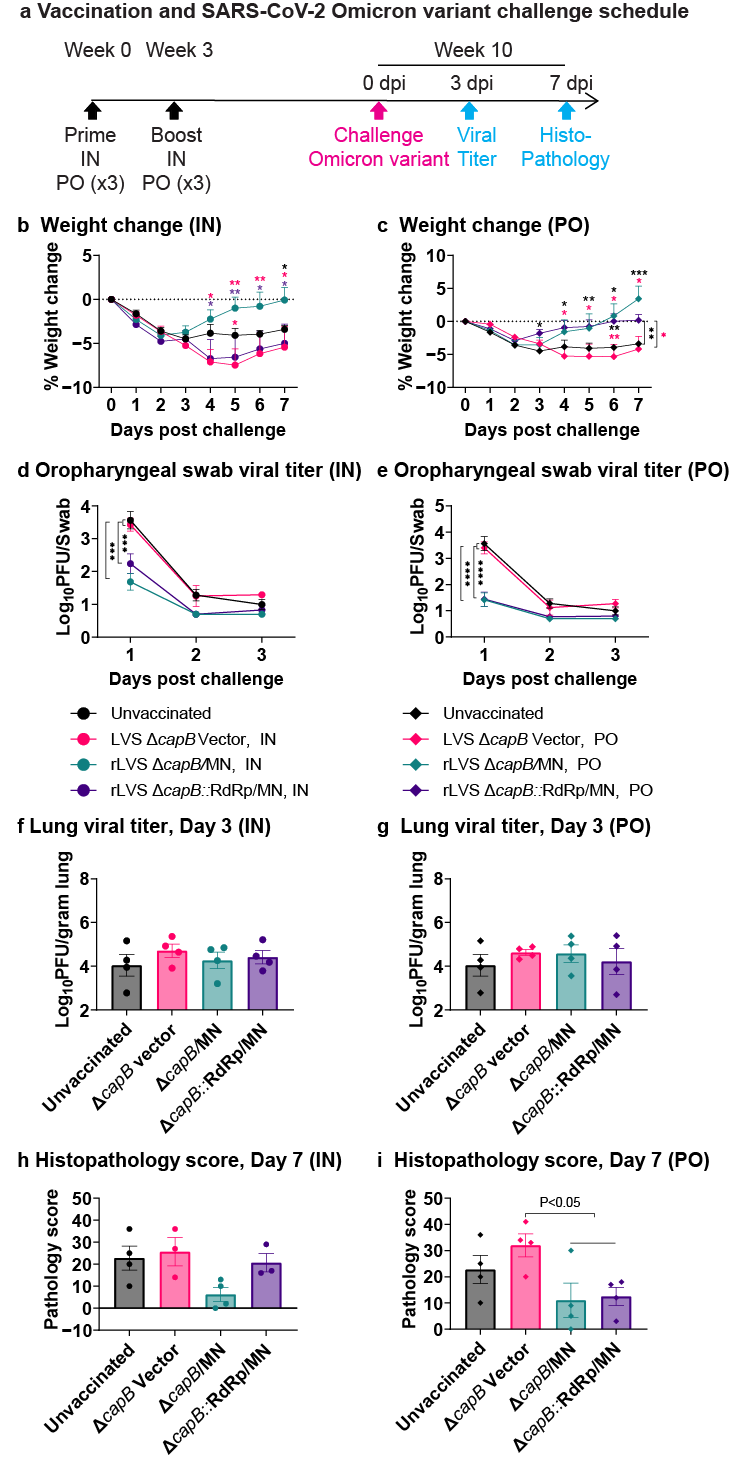
3.3. Study of Vaccine Efficacy Against SARS-CoV-2 Omicron Variant Challenge in the Hamster Model—Experiment 2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Drosten, C.; Gunther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.M.; et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Health Organization 2023 data.who.int, WHO Coronavirus (COVID-19) Dashboard > About [Dashboard]: World Health Organization. 2025. Available online: https://data.who.int/dashboards/covid19/about (accessed on 11 April 2025).
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Markov, P.V.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Health Organization 2023 data.who.int, WHO Coronavirus (COVID-19) Dashboard > Variants [Dashboard]: World Health Organization. 2025. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 11 April 2025).
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, Y.; Li, M.; Xiao, H.; Liao, F.; Shen, M.; Ge, W.; Ou, J.; Liu, Y.; Chen, L.; Zhao, Y.; et al. The R203M and D377Y mutations of the nucleocapsid protein promote SARS-CoV-2 infectivity by impairing RIG-I-mediated antiviral signaling. PLoS Pathog. 2025, 21, e1012886. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Uraki, R.; Kiso, M.; Iida, S.; Imai, M.; Takashita, E.; Kuroda, M.; Halfmann, P.J.; Loeber, S.; Maemura, T.; Yamayoshi, S.; et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature 2022, 607, 119–127. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F. SARS-CoV-2 Omicron variant: Recent progress and future perspectives. Signal Transduct. Target. Ther. 2022, 7, 141. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Uraki, R.; Halfmann, P.J.; Iida, S.; Yamayoshi, S.; Furusawa, Y.; Kiso, M.; Ito, M.; Iwatsuki-Horimoto, K.; Mine, S.; Kuroda, M.; et al. Characterization of SARS-CoV-2 Omicron BA.4 and BA.5 isolates in rodents. Nature 2022, 612, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Kakavandi, S.; Zare, I.; VaezJalali, M.; Dadashi, M.; Azarian, M.; Akbari, A.; Zare, I.; VaezJalali, M.; Zalpoor, H. Structural and non-structural proteins in SARS-CoV-2: Potential aspects to COVID-19 treatment or prevention of progression of related diseases. Cell Commun. Signal. 2023, 21, 110. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; COVID-19 Genomics UK Consortium; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yao, Z.; Zhang, L.; Duan, Y.; Tang, X.; Lu, J. Molecular insights into the adaptive evolution of SARS-CoV-2 spike protein. J. Infect. 2024, 88, 106121. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.Y.; Xu, Y.; Gu, Y.; Zeng, D.; Wheeler, B.; Young, H.; Sunny, S.K.; Moore, Z. Effectiveness of Bivalent Boosters against Severe Omicron Infection. N. Engl. J. Med. 2023, 388, 764–766. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liang, Z.; Li, C.; Gong, X.; Ye, G.; Jiang, Y.; Shi, H.; Hussain, A.; Zhao, W.; Li, M.; Tina, Y.; et al. Development of Glycan-masked SARS-CoV-2 RBD vaccines against SARS-related coronaviruses. PLoS Pathog. 2024, 20, e1012599. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sette, A.; Sidney, J.; Crotty, S. T Cell Responses to SARS-CoV-2. Annu Rev Immunol. 2023, 41, 343–373. [Google Scholar] [CrossRef] [PubMed]
- Nesterenko, P.A.; McLaughlin, J.; Tsai, B.L.; Burton Sojo, G.; Cheng, D.; Zhao, D.; Mao, Z.; Bangayan, N.J.; Obusan, M.B.; Su, Y.; et al. HLA-A(*)02:01 restricted T cell receptors against the highly conserved SARS-CoV-2 polymerase cross-react with human coronaviruses. Cell Rep. 2021, 37, 110167. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lopez-Munoz, A.D.; Kosik, I.; Holly, J.; Yewdell, J.W. Cell surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity. Sci Adv. 2022, 8, eabp9770. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Z.; Nomura, N.; Muramoto, Y.; Ekimoto, T.; Uemura, T.; Liu, K.; Yui, M.; Kono, N.; Aoki, J.; Ikeguchi, M.; et al. Structure of SARS-CoV-2 membrane protein essential for virus assembly. Nat. Commun. 2022, 13, 4399. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Eser, T.M.; Baranov, O.; Huth, M.; Ahmed, M.I.M.; Deak, F.; Held, K.; Lin, L.; Pekayvaz, K.; Leunig, A.; Nicolai, L.; et al. Nucleocapsid-specific T cell responses associate with control of SARS-CoV-2 in the upper airways before seroconversion. Nat. Commun. 2023, 14, 2952. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kundu, R.; Narean, J.S.; Wang, L.; Fenn, J.; Pillay, T.; Fernandez, N.D.; Conibear, E.; Koycheva, A.; Davies, M.; Tolosa-Wright, M.; et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun. 2022, 13, 80. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, Y.; Chen, J.; Chen, S.; Huang, C.; Li, B.; Li, J.; Jin, Z.; Zhang, Q.; Pan, P.; Du, W.; et al. Molecular characterization of SARS-CoV-2 nucleocapsid protein. Front. Cell. Infect. Microbiol. 2024, 14, 1415885. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Lineburg, K.E.; Grant, E.J.; Swaminathan, S.; Chatzileontiadou, D.S.M.; Szeto, C.; Sloane, H.; Panikkar, A.; Raju, J.; Crooks, P.; Rehan, S.; et al. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity 2021, 54, 1055–1065.e5. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ng, O.W.; Chia, A.; Tan, A.T.; Jadi, R.S.; Leong, H.N.; Bertoletti, A.; Tan, Y.J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine 2016, 34, 2008–2014. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021, 34, 108728. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Swadling, L.; Diniz, M.O.; Schmidt, N.M.; Amin, O.E.; Chandran, A.; Shaw, E.; Pade, C.; Gibbons, J.M.; Le Bert, N.; Tan, A.T.; et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2022, 601, 110–117. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jia, Q.; Bielefeldt-Ohmann, H.; Maison, R.M.; Maslesa-Galic, S.; Cooper, S.K.; Bowen, R.A.; Horwitz, M.A. Replicating bacterium-vectored vaccine expressing SARS-CoV-2 Membrane and Nucleocapsid proteins protects against severe COVID-19-like disease in hamsters. NPJ Vaccines 2021, 6, 47. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jia, Q.; Bielefeldt-Ohmann, H.; Maison, R.M.; Hartwig, A.; Maslesa-Galic, S.; Bowen, R.A.; Hartwig, A.; Maison, R.M.; Horwitz, M.A. Oral Administration of Universal Bacterium-Vectored Nucleocapsid-Expressing COVID-19 Vaccine is Efficacious in Hamsters. Microbiol Spectr. 2023, 11, e0503522. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Arieta, C.M.; Xie, Y.J.; Rothenberg, D.A.; Diao, H.; Harjanto, D.; Meda, S.; Marquart, K.; Koenitzer, B.; Sciuto, T.E.; Lobo, A.; et al. The T-cell-directed vaccine BNT162b4 encoding conserved non-spike antigens protects animals from severe SARS-CoV-2 infection. Cell 2023, 186, 2392–2409.e21. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rabdano, S.O.; Ruzanova, E.A.; Vertyachikh, A.E.; Teplykh, V.A.; Emelyanova, A.B.; Rudakov, G.O.; Arakelov, S.A.; Belozerova, N.S.; Lukovenko, A.A.; Rabdano, S.O.; et al. N-protein vaccine is effective against COVID-19: Phase 3, randomized, double-blind, placebo-controlled clinical trial. J. Infect. 2024, 89, 106288. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Bowen, R.; Dillon, B.J.; Maslesa-Galic, S.; Chang, B.T.; Kaidi, A.C.; Horwitz, M.A. Single vector platform vaccine protects against lethal respiratory challenge with Tier 1 select agents of anthrax, plague, and tularemia. Sci. Rep. 2018, 8, 7009. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jia, Q.; Lee, B.Y.; Bowen, R.; Dillon, B.J.; Som, S.M.; Horwitz, M.A. A Francisella tularensis live vaccine strain (LVS) mutant with a deletion in capB, encoding a putative capsular biosynthesis protein, is significantly more attenuated than LVS yet induces potent protective immunity in mice against F. tularensis challenge. Infect Immun. 2010, 78, 4341–4355. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wehrly, T.D.; Chong, A.; Virtaneva, K.; Sturdevant, D.E.; Child, R.; Edwards, J.A.; Porcella, S.F.; Martens, C.; Virtaneva, K.; Sturdevant, D.E.; et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 2009, 11, 1128–1150. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sattler, A.; Angermair, S.; Stockmann, H.; Heim, K.M.; Khadzhynov, D.; Treskatsch, S.; Stockmann, H.; Heim, K.M. SARS-CoV-2-specific T cell responses and correlations with COVID-19 patient predisposition. J. Clin. Investig. 2020, 130, 6477–6489. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Bowen, R.; Lee, B.Y.; Dillon, B.J.; Maslesa-Galic, S.; Horwitz, M.A. Francisella tularensis Live Vaccine Strain deficient in capB and overexpressing the fusion protein of IglA, IglB, and IglC from the bfr promoter induces improved protection against F. tularensis respiratory challenge. Vaccine 2016, 34, 4969–4978. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mlynek, K.D.; Cline, C.R.; Biryukov, S.S.; Toothman, R.G.; Bachert, B.A.; Klimko, C.P.; Shoe, J.L.; Hunter, M.; Hedrick, Z.M.; Dankmeyer, J.L.; et al. The rLVS DeltacapB/iglABC vaccine provides potent protection in Fischer rats against inhalational tularemia caused by various virulent Francisella tularensis strains. Hum. Vaccines Immunother. 2023, 19, 2277083. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tullius, M.V.; Bowen, R.A.; Back, P.S.; Maslesa-Galic, S.; Nava, S.; Horwitz, M.A. LVS DeltacapB-vectored multiantigenic melioidosis vaccines protect against lethal respiratory Burkholderia pseudomallei challenge in highly sensitive BALB/c mice. mBio 2024, 15, e0018624. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Q.; Bielefeldt-Ohmann, H.; Masleša-Galić, S.; Bowen, R.A.; Horwitz, M.A. Universal Bacterium-Vectored COVID-19 Vaccine Expressing Early SARS-CoV-2 Conserved Proteins Cross-Protects Against Late Variants in Hamsters. Vaccines 2025, 13, 633. https://doi.org/10.3390/vaccines13060633
Jia Q, Bielefeldt-Ohmann H, Masleša-Galić S, Bowen RA, Horwitz MA. Universal Bacterium-Vectored COVID-19 Vaccine Expressing Early SARS-CoV-2 Conserved Proteins Cross-Protects Against Late Variants in Hamsters. Vaccines. 2025; 13(6):633. https://doi.org/10.3390/vaccines13060633
Chicago/Turabian StyleJia, Qingmei, Helle Bielefeldt-Ohmann, Saša Masleša-Galić, Richard A. Bowen, and Marcus A. Horwitz. 2025. "Universal Bacterium-Vectored COVID-19 Vaccine Expressing Early SARS-CoV-2 Conserved Proteins Cross-Protects Against Late Variants in Hamsters" Vaccines 13, no. 6: 633. https://doi.org/10.3390/vaccines13060633
APA StyleJia, Q., Bielefeldt-Ohmann, H., Masleša-Galić, S., Bowen, R. A., & Horwitz, M. A. (2025). Universal Bacterium-Vectored COVID-19 Vaccine Expressing Early SARS-CoV-2 Conserved Proteins Cross-Protects Against Late Variants in Hamsters. Vaccines, 13(6), 633. https://doi.org/10.3390/vaccines13060633





