SARS-CoV-2 Neutralization Assays Used in Clinical Trials: A Narrative Review
Abstract
1. Introduction
2. Current Status of SARS-CoV-2 Vaccines
3. Efficacy of SARS-CoV-2 Vaccines
4. Role of Neutralizing Antibodies
5. Detection Methods for Neutralizing Antibodies
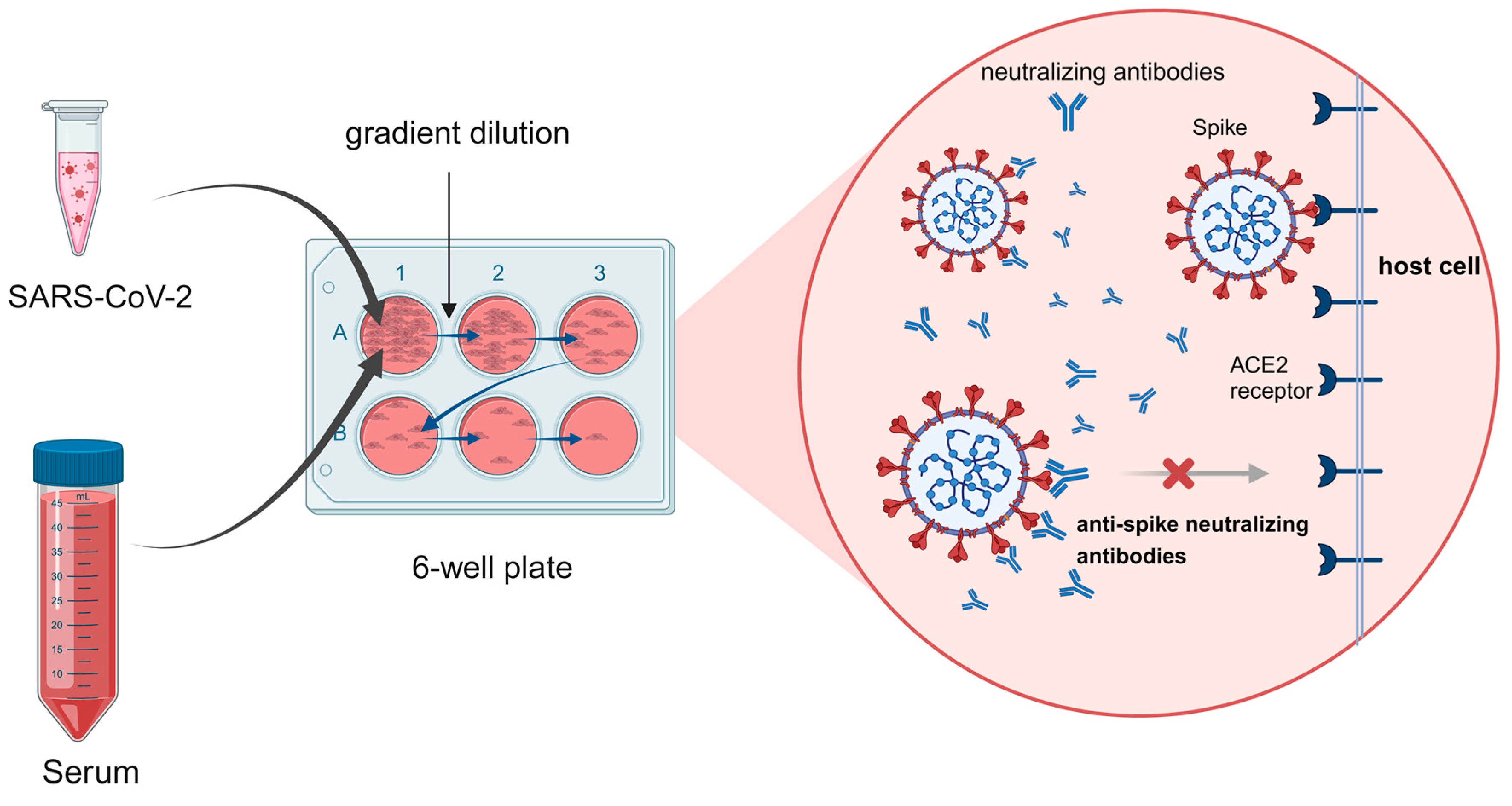
6. Live (Conventional) Virus Neutralization Test (cVNT)
6.1. Cytopathic Effect (CPE)
6.2. Plaque Reduction Neutralization Test (PRNT)
6.3. Recombinant Live Virus
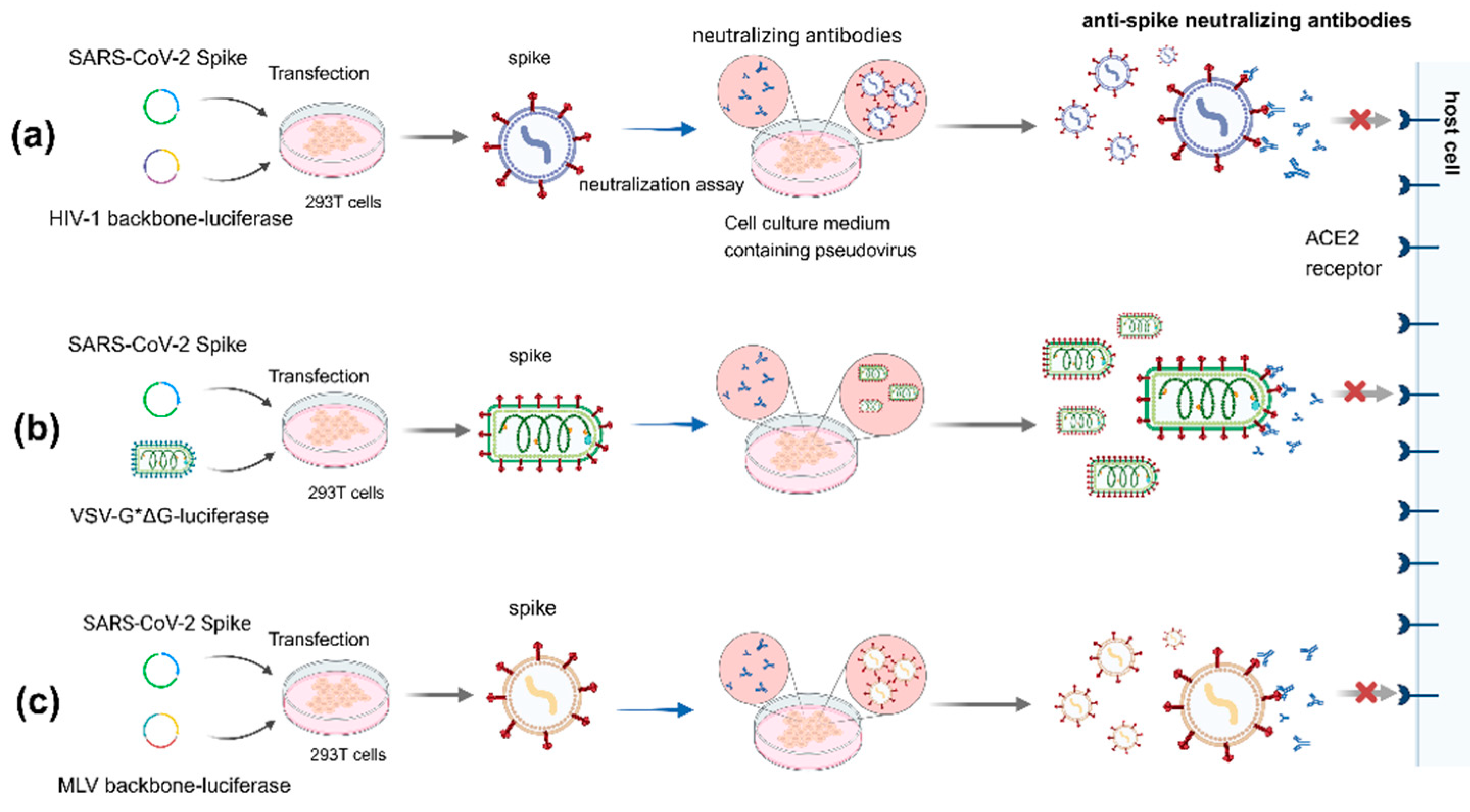
7. Pseudovirus Neutralization Test (pVNT)
7.1. HIV Lentivirus System
7.2. Vesicular Stomatitis Virus (VSV) System
7.3. MLV System
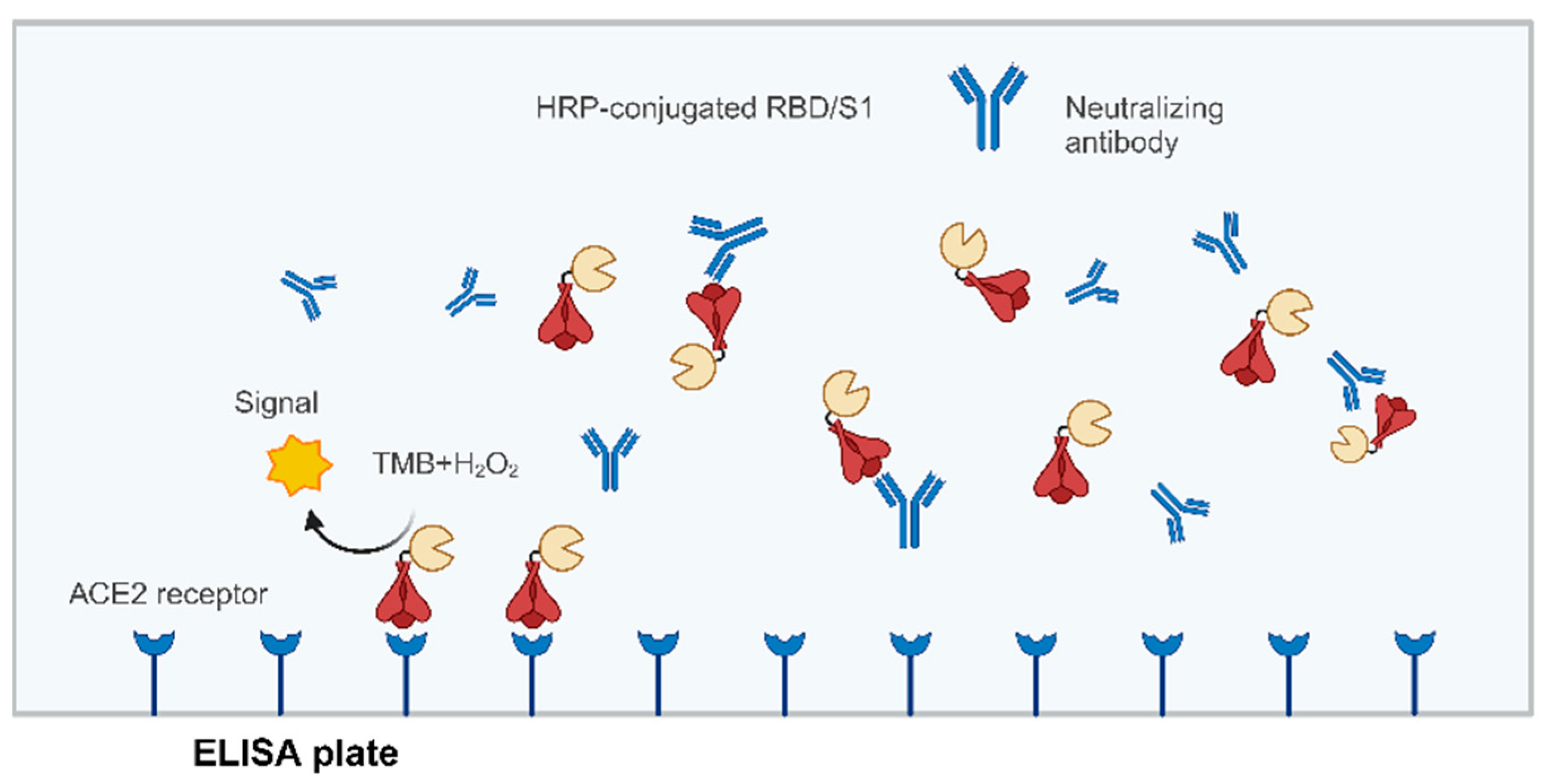
8. Surrogate Virus Neutralization Test (sVNT)
9. Correlation of Neutralizing Antibodies (nAbs) with a Protective Effect
10. Correlation Analysis of Testing Methods
11. Standardization of Neutralizing Antibody Detection Methods
12. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [CrossRef] [PubMed]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Du, L. Effect of Low-Pathogenic Human Coronavirus-Specific Antibodies on SARS-CoV-2. Trends Immunol. 2020, 41, 853–854. [Google Scholar] [CrossRef] [PubMed]
- Stefano, G.B.; Kream, R.M. Convalescent memory T cell immunity in individuals with mild or asymptomatic SARS-CoV-2 infection may result from an evolutionarily adapted immune response to coronavirus and the ‘common cold’. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e929789-1. [Google Scholar] [CrossRef] [PubMed]
- Moss, P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 2022, 23, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Gruell, H.; Vanshylla, K.; Weber, T.; Barnes, C.O.; Kreer, C.; Klein, F. Antibody-mediated neutralization of SARS-CoV-2. Immunity 2022, 55, 925–944. [Google Scholar] [CrossRef] [PubMed]
- WHO. Statement on the Fifteenth Meeting of the IHR (2005) Emergency Committee on the COVID-19 Pandemic. Available online: https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-%282005%29-emergency-committee-regarding-the-coronavirus-disease-%28COVID-19%29-pandemic (accessed on 24 May 2023).
- WHO. WHO Coronavirus (COVID-19) Dashboard. Available online: https://data.who.int/dashboards/covid19/ (accessed on 12 March 2024).
- Zhang, Y.J.; Zeng, G.; Pan, H.X.; Li, C.G.; Hu, Y.L.; Chu, K.; Han, W.X.; Chen, Z.; Tang, R.; Yin, W.D.; et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021, 21, 181–192. [Google Scholar] [CrossRef]
- Jara, A.; Undurraga, E.A.; Gonzalez, C.; Paredes, F.; Fontecilla, T.; Jara, G.; Pizarro, A.; Acevedo, J.; Leo, K.; Leon, F.; et al. Effectiveness of an Inactivated SARS-CoV-2 Vaccine in Chile. N. Engl. J. Med. 2021, 385, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Tanriover, M.D.; Doganay, H.L.; Akova, M.; Guner, H.R.; Azap, A.; Akhan, S.; Kose, S.; Erdinc, F.S.; Akalin, E.H.; Tabak, O.F.; et al. Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): Interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet 2021, 398, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.L.; Zhang, Y.T.; Wang, Y.X.; Wang, H.; Yang, Y.K.; Gao, G.F.; Tan, W.J.; Wu, G.Z.; Xu, M.; Lou, Z.Y.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021, 21, 39–51. [Google Scholar] [CrossRef]
- Frenck, R.W., Jr.; Klein, N.P.; Kitchin, N.; Gurtman, A.; Absalon, J.; Lockhart, S.; Perez, J.L.; Walter, E.B.; Senders, S.; Bailey, R.; et al. Safety, Immunogenicity, and Efficacy of the BNT162b2 COVID-19 Vaccine in Adolescents. N. Engl. J. Med. 2021, 385, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Muik, A.; Vogler, I.; Derhovanessian, E.; Kranz, L.M.; Vormehr, M.; Quandt, J.; Bidmon, N.; Ulges, A.; Baum, A.; et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 2021, 595, 572–577. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2-Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Chalkias, S.; Harper, C.; Vrbicky, K.; Walsh, S.R.; Essink, B.; Brosz, A.; McGhee, N.; Tomassini, J.E.; Chen, X.; Chang, Y.; et al. A Bivalent Omicron-Containing Booster Vaccine against COVID-19. N. Engl. J. Med. 2022, 387, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Sadoff, J.; Le Gars, M.; Shukarev, G.; Heerwegh, D.; Truyers, C.; de Groot, A.M.; Stoop, J.; Tete, S.; Van Damme, W.; Leroux-Roels, I.; et al. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S COVID-19 Vaccine. N. Engl. J. Med. 2021, 384, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: A pooled analysis of four randomised trials. Lancet 2021, 397, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatullin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G.; et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S.; et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021, 397, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.-C.; Li, Y.-H.; Guan, X.-H.; Hou, L.-H.; Wang, W.-J.; Li, J.-X.; Wu, S.-P.; Wang, B.-S.; Wang, Z.; Wang, L.; et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Zhu, F.-C.; Guan, X.-H.; Li, Y.-H.; Huang, J.-Y.; Jiang, T.; Hou, L.-H.; Li, J.-X.; Yang, B.-F.; Wang, L.; Wang, W.-J.; et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2020, 396, 479–488. [Google Scholar] [CrossRef]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 COVID-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Gao, L.; Tao, L.; Hadinegoro, S.R.; Erkin, M.; Ying, Z.; He, P.; Girsang, R.T.; Vergara, H.; Akram, J.; et al. Efficacy and Safety of the RBD-Dimer-Based COVID-19 Vaccine ZF2001 in Adults. N. Engl. J. Med. 2022, 386, 2097–2111. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, Y.; Dai, L.; Wang, J.; He, P.; Li, C.; Fang, X.; Wang, C.; Zhao, X.; Huang, E.; et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: Two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021, 21, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- Li, M.C.; Wang, H.; Tian, L.L.; Pang, Z.H.; Yang, Q.K.; Huang, T.Q.; Fan, J.F.; Song, L.H.; Tong, Y.G.; Fan, H.H. COVID-19 vaccine development: Milestones, lessons and prospects. Signal Transduct. Target. Ther. 2022, 7, 146. [Google Scholar] [CrossRef] [PubMed]
- Minor, P.D. Live attenuated vaccines: Historical successes and current challenges. Virology 2015, 479, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Pollet, J.; Chen, W.H.; Strych, U. Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv. Drug Deliv. Rev. 2021, 170, 71–82. [Google Scholar] [CrossRef]
- Wu, S.P.; Zhong, G.X.; Zhang, J.; Shuai, L.; Zhang, Z.; Wen, Z.Y.; Wang, B.S.; Zhao, Z.H.; Song, X.H.; Chen, Y.; et al. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020, 11, 4081. [Google Scholar] [CrossRef]
- Chavda, V.P.; Bezbaruah, R.; Athalye, M.; Parikh, P.K.; Chhipa, A.; Patel, S.; Apostolopoulos, V. Replicating Viral Vector-Based Vaccines for COVID-19: Potential Avenue in Vaccination Arena. Viruses-Basel 2022, 14, 759. [Google Scholar] [CrossRef]
- Graham, S.P.; McLean, R.K.; Spencer, A.J.; Belij-Rammerstorfer, S.; Wright, D.; Ulaszewska, M.; Edwards, J.C.; Hayes, J.W.P.; Martini, V.; Thakur, N.; et al. Evaluation of the immunogenicity of prime-boost vaccination with the replication-deficient viral vectored COVID-19 vaccine candidate ChAdOx1 nCoV-19. NPJ Vaccines 2020, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Ikegame, S.; Siddiquey, M.N.A.; Hung, C.-T.; Haas, G.; Brambilla, L.; Oguntuyo, K.Y.; Kowdle, S.; Chiu, H.-P.; Stevens, C.S.; Vilardo, A.E.; et al. Neutralizing activity of Sputnik V vaccine sera against SARS-CoV-2 variants. Nat. Commun. 2021, 12, 4598. [Google Scholar] [CrossRef] [PubMed]
- Matveeva, O.; Ershov, A. Retrospective Cohort Study of the Effectiveness of the Sputnik V and EpiVacCorona Vaccines against the SARS-CoV-2 Delta Variant in Moscow (June–July 2021). Vaccines 2022, 10, 984. [Google Scholar] [CrossRef] [PubMed]
- Ertl, P.F.; Thomsen, L.L. Technical issues in construction of nucleic acid vaccines. Methods 2003, 31, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Lagniton, P.N.P.; Liu, Y.; Xu, R.H. mRNA vaccines for COVID-19: What, why and how. Int. J. Biol. Sci. 2021, 17, 1446–1460. [Google Scholar] [CrossRef] [PubMed]
- Kon, E.; Elia, U.; Peer, D. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr. Opin. Biotechnol. 2022, 73, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Silveira, M.M.; Moreira, G.; Mendonca, M. DNA vaccines against COVID-19: Perspectives and challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes Moderna, Pfizer-BioNTech Bivalent COVID-19 Vaccines for Use as a Booster Dose; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2022.
- Chalkias, S.; Eder, F.; Essink, B.; Khetan, S.; Nestorova, B.; Feng, J.; Chen, X.; Chang, Y.; Zhou, H.; Montefiori, D.; et al. Safety, immunogenicity and antibody persistence of a bivalent Beta-containing booster vaccine against COVID-19: A phase 2/3 trial. Nat. Med. 2022, 28, 2388–2397. [Google Scholar] [CrossRef] [PubMed]
- Link-Gelles, R.; Ciesla, A.A.; Fleming-Dutra, K.E.; Smith, Z.R.; Britton, A.; Wiegand, R.E.; Miller, J.D.; Accorsi, E.K.; Schrag, S.J.; Verani, J.R.; et al. Effectiveness of Bivalent mRNA Vaccines in Preventing Symptomatic SARS-CoV-2 Infection–Increasing Community Access to Testing Program, United States, September–November 2022. Mmwr-Morb. Mortal. Wkly. Rep. 2022, 71, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- WHO. Vaccine Efficacy, Effectiveness and Protection. Available online: https://www.who.int/news-room/feature-stories/detail/vaccine-efficacy-effectiveness-and-protection (accessed on 7 May 2023).
- Suthar, M.S.; Zimmerman, M.G.; Kauffman, R.C.; Mantus, G.; Linderman, S.L.; Hudson, W.H.; Vanderheiden, A.; Nyhoff, L.; Davis, C.W.; Adekunle, O.; et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep. Med. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Gilbert, P.B.; DeGruttola, V.G.; Hudgens, M.G.; Self, S.G.; Hammer, S.M.; Corey, L. What constitutes efficacy for a human immunodeficiency virus vaccine that ameliorates viremia: Issues involving surrogate end points in phase 3 trials. J. Infect. Dis. 2003, 188, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. A correlate of protection for SARS-CoV-2 vaccines is urgently needed. Nat. Med. 2021, 27, 1147–1148. [Google Scholar] [CrossRef] [PubMed]
- Klasse, P.J.; Sattentau, Q.J. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 2002, 83, 2091–2108. [Google Scholar] [CrossRef] [PubMed]
- Dörner, T.; Radbruch, A. Antibodies and B cell memory in viral immunity. Immunity 2007, 27, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.K.; Ackerman, M.E. Increasing the Clinical Potential and Applications of Anti-HIV Antibodies. Front. Immunol. 2017, 8, 1655. [Google Scholar] [CrossRef] [PubMed]
- Morales-Nunez, J.J.; Munoz-Valle, J.F.; Torres-Hernandez, P.C.; Hernandez-Bello, J. Overview of Neutralizing Antibodies and Their Potential in COVID-19. Vaccines 2021, 9, 1376. [Google Scholar] [CrossRef] [PubMed]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Stephens, D.S.; McElrath, M.J. COVID-19 and the Path to Immunity. JAMA 2020, 324, 1279–1281. [Google Scholar] [CrossRef] [PubMed]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Earle, K.A.; Ambrosino, D.M.; Fiore-Gartland, A.; Goldblatt, D.; Gilbert, P.B.; Siber, G.R.; Dull, P.; Plotkin, S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine 2021, 39, 4423–4428. [Google Scholar] [CrossRef]
- Bewley, K.R.; Coombes, N.S.; Gagnon, L.; McInroy, L.; Baker, N.; Shaik, I.; St-Jean, J.R.; St-Amant, N.; Buttigieg, K.R.; Humphries, H.E.; et al. Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat. Protoc. 2021, 16, 3114–3140. [Google Scholar] [CrossRef] [PubMed]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly Conserved Protective Epitopes on Influenza B Viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.H.; Li, Q.Q.; Wu, J.J.; Zhao, C.Y.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.L.; Qin, H.Y.; Wang, M.; et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.; Tiu, C.; Hu, Z.; Chen, V.C.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Case, J.B.; Rothlauf, P.W.; Chen, R.E.; Liu, Z.; Zhao, H.; Kim, A.S.; Bloyet, L.M.; Zeng, Q.; Tahan, S.; Droit, L.; et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe 2020, 28, 475–485.e475. [Google Scholar] [CrossRef] [PubMed]
- Kaminskyy, V.; Zhivotovsky, B. To kill or be killed: How viruses interact with the cell death machinery. J. Intern Med. 2010, 267, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Muruato, A.E.; Fontes-Garfias, C.R.; Ren, P.; Garcia-Blanco, M.A.; Menachery, V.D.; Xie, X.P.; Shi, P.Y. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020, 11, 4059. [Google Scholar] [CrossRef] [PubMed]
- Frische, A.; Brooks, P.T.; Gybel-Brask, M.; Sækmose, S.G.; Jensen, B.A.; Mikkelsen, S.; Bruun, M.T.; Boding, L.; Strandh, C.P.; Jørgensen, C.S.; et al. Optimization and evaluation of a live virus SARS-CoV-2 neutralization assay. PLoS ONE 2022, 17, e0272298. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S. Revisiting the concept of a cytopathic viral infection. PLoS Pathog. 2017, 13, e1006409. [Google Scholar] [CrossRef]
- Traggiai, E.; Becker, S.; Subbarao, K.; Kolesnikova, L.; Uematsu, Y.; Gismondo, M.R.; Murphy, B.R.; Rappuoli, R.; Lanzavecchia, A. An efficient method to make human monoclonal antibodies from memory B cells: Potent neutralization of SARS coronavirus. Nat. Med. 2004, 10, 871–875. [Google Scholar] [CrossRef]
- Perera, R.A.; Mok, C.K.; Tsang, O.T.; Lv, H.; Ko, R.L.; Wu, N.C.; Yuan, M.; Leung, W.S.; Chan, J.M.; Chik, T.S.; et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro. Surveill. 2020, 25, 2000421. [Google Scholar] [CrossRef] [PubMed]
- Coates, H.V.; Alling, D.W.; Chanock, R.M. An antigenic analysis of respiratory syncytial virus isolates by a plaque reduction neutralization test. Am. J. Epidemiol. 1966, 83, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Lau, E.H.Y.; Tsang, O.T.Y.; Hui, D.S.C.; Kwan, M.Y.W.; Chan, W.H.; Chiu, S.S.; Ko, R.L.W.; Chan, K.H.; Cheng, S.M.S.; Perera, R.; et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat. Commun. 2021, 12, 63. [Google Scholar] [CrossRef]
- Kisch, A.L.; Johnson, K.M. A plaque assay for respiratory syncytial virus. Proceedings of the Society for Experimental Biology and Medicine. Soc. Exp. Biol. Med. 1963, 112, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Dulbecco, R. Production of Plaques in Monolayer Tissue Cultures by Single Particles of an Animal Virus. Proc. Natl. Acad. Sci. USA 1952, 38, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Septisetyani, E.P.; Prasetyaningrum, P.W.; Anam, K.; Santoso, A. SARS-CoV-2 Antibody Neutralization Assay Platforms Based on Epitopes Sources: Live Virus, Pseudovirus, and Recombinant S Glycoprotein RBD. Immune Netw. 2021, 21, e39. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Fujino, M.; Yoshida, N.; Kimura, K.; Zhou, J.; Motegi, Y.; Komase, K.; Nakayama, T. Development of a new neutralization test for measles virus. J. Virol. Methods 2007, 142, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Rostad, C.A.; Currier, M.C.; Moore, M.L. Fluorescent and Bioluminescent Reporter Myxoviruses. Viruses 2016, 8, 214. [Google Scholar] [CrossRef]
- Zhang, F.Q.; Perez-Martin, E.; Juleff, N.; Charleston, B.; Seago, J. A replication-competent foot-and-mouth disease virus expressing a luciferase reporter. J. Virol. Methods 2017, 247, 38–44. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Liu, J.; Bailey, A.L.; Plante, K.S.; Plante, J.A.; Zou, J.; Xia, H.; Bopp, N.E.; Aguilar, P.V.; et al. A trans-complementation system for SARS-CoV-2 recapitulates authentic viral replication without virulence. Cell 2021, 184, 2229–2238.e13. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Kurhade, C.; Chang, H.C.; Hu, Y.; Meza, J.A.; Beaver, D.; Trinh, K.; Omlid, J.; Elghetany, B.; Desai, R.; et al. An Integrated Research-Clinical BSL-2 Platform for a Live SARS-CoV-2 Neutralization Assay. Viruses 2023, 15, 1855. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Xia, H.; Shi, P.Y.; Xie, X.; Ren, P. A Single-Round Infection Fluorescent SARS-CoV-2 Neutralization Test for COVID-19 Serological Testing at a Biosafety Level-2 Laboratory. Viruses 2022, 14, 1211. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Xia, H.; Zou, J.; Weaver, S.C.; Swanson, K.A.; Cai, H.; Cutler, M.; Cooper, D.; Muik, A.; et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature 2021, 596, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Xia, H.; Zhang, X.; Zou, J.; Fontes-Garfias, C.R.; Weaver, S.C.; Swanson, K.A.; Cai, H.; Sarkar, R.; et al. BNT162b2-Elicited Neutralization against New SARS-CoV-2 Spike Variants. N. Engl. J. Med. 2021, 385, 472–474. [Google Scholar] [CrossRef] [PubMed]
- Falsey, A.R.; Frenck, R.W., Jr.; Walsh, E.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Bailey, R.; Swanson, K.A.; Xu, X.; et al. SARS-CoV-2 Neutralization with BNT162b2 Vaccine Dose 3. N. Engl. J. Med. 2021, 385, 1627–1629. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, F.M.; Sher, L.D.; Sabharwal, C.; Gurtman, A.; Xu, X.; Kitchin, N.; Lockhart, S.; Riesenberg, R.; Sexter, J.M.; Czajka, H.; et al. Evaluation of BNT162b2 COVID-19 Vaccine in Children Younger than 5 Years of Age. N. Engl. J. Med. 2023, 388, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Chiem, K.; Vasquez, D.M.; Park, J.G.; Platt, R.N.; Anderson, T.; Walter, M.R.; Kobie, J.J.; Ye, C.; Martínez-Sobrido, L.; Chamari, K.; et al. Generation and Characterization of recombinant SARS-CoV-2 expressing reporter genes. J. Virol. 2021, 95, e02209-20. [Google Scholar] [CrossRef] [PubMed]
- Lichtenegger, S.; Saiger, S.; Hardt, M.; Kulnik, S.; Wagner, G.E.; Kleinhappl, B.; Assig, K.; Zauner, A.; Ober, M.; Kimpel, J.; et al. Development of a Rapid Live SARS-CoV-2 Neutralization Assay Based on a qPCR Readout. J. Clin. Microbiol. 2022, 60, e0037622. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Wang, Z.; Cao, S.; Huang, W.; Yuan, L.; Huang, Y.J.; Zheng, Y.; Chen, J.; Ying, B.; et al. Safety and superior immunogenicity of heterologous boosting with an RBD-based SARS-CoV-2 mRNA vaccine in Chinese adults. Cell Res. 2022, 32, 777–780. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Zhang, Z.; Wu, J.; Zhang, J.; Hu, H.; Zhu, T.; Zhang, J.; Luo, L.; Fan, P.; et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type-5 vector-based COVID-19 vaccine (Ad5-nCoV) in adults: Preliminary report of an open-label and randomised phase 1 clinical trial. Lancet Infect. Dis. 2021, 21, 1654–1664. [Google Scholar] [CrossRef] [PubMed]
- Ansarah-Sobrinho, C.; Nelson, S.; Jost, C.A.; Whitehead, S.S.; Pierson, T.C. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology 2008, 381, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Song, Z.; Qi, Y.; Feng, X.; Xu, N.; Sun, Y.; Wu, X.; Yao, X.; Mao, Q.; Li, X.; et al. Molecular determinants of enterovirus 71 viral entry: Cleft around GLN-172 on VP1 protein interacts with variable region on scavenge receptor B 2. J. Biol. Chem. 2012, 287, 6406–6420. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.; Weisblum, Y.; Muecksch, F.; Hoffmann, H.H.; Michailidis, E.; Lorenzi, J.C.C.; Mendoza, P.; Rutkowska, M.; Bednarski, E.; Gaebler, C.; et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020, 217, e20201181. [Google Scholar] [CrossRef] [PubMed]
- Lillico, S.; Vasey, D.; King, T.; Whitelaw, B. Lentiviral transgenesis in livestock. Transgenic. Res. 2011, 20, 441–442. [Google Scholar] [CrossRef] [PubMed]
- Banga Ndzouboukou, J.L.; Zhang, Y.D.; Fan, X.L. Recent Developments in SARS-CoV-2 Neutralizing Antibody Detection Methods. Curr. Med. Sci. 2021, 41, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Ledgerwood, J.E.; Coates, E.E.; Yamshchikov, G.; Saunders, J.G.; Holman, L.; Enama, M.E.; DeZure, A.; Lynch, R.M.; Gordon, I.; Plummer, S.; et al. Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin. Exp. Immunol. 2015, 182, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Gao, Q.; He, C.; Huang, A.; Tang, N.; Wang, K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. 2020, 7, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.W.; Tai, C.H.; Hsu, Y.M.; Cheng, D.; Hung, S.J.; Chai, K.M.; Wang, Y.F.; Wang, J.R. Assessing the application of a pseudovirus system for emerging SARS-CoV-2 and re-emerging avian influenza virus H5 subtypes in vaccine development. Biomed. J. 2020, 43, 375–387. [Google Scholar] [CrossRef]
- Crawford, K.H.D.; Eguia, R.; Dingens, A.S.; Loes, A.N.; Malone, K.D.; Wolf, C.R.; Chu, H.Y.; Tortorici, M.A.; Veesler, D.; Murphy, M.; et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 2020, 12, 513. [Google Scholar] [CrossRef]
- Kumar, M.; Keller, B.; Makalou, N.; Sutton, R.E. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001, 12, 1893–1905. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Huang, B.; A, R.; Li, W.; Wang, W.; Deng, Y.; Tan, W. Development and effectiveness of pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro. Biosaf. Health 2020, 2, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Lichty, B.D.; Power, A.T.; Stojdl, D.F.; Bell, J.C. Vesicular stomatitis virus: Re-inventing the bullet. Trends Mol. Med. 2004, 10, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Fukushi, S.; Watanabe, R.; Taguchi, F. Pseudotyped vesicular stomatitis virus for analysis of virus entry mediated by SARS coronavirus spike proteins. Methods Mol. Biol. 2008, 454, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Whitt, M.A. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J. Virol. Methods 2010, 169, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Witte, O.N.; Baltimore, D. Mechanism of formation of pseudotypes between vesicular stomatitis virus and murine leukemia virus. Cell 1977, 11, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Zettl, F.; Meister, T.L.; Vollmer, T.; Fischer, B.; Steinmann, J.; Krawczyk, A.; V’Kovski, P.; Todt, D.; Steinmann, E.; Pfaender, S.; et al. Rapid Quantification of SARS-CoV-2-Neutralizing Antibodies Using Propagation-Defective Vesicular Stomatitis Virus Pseudotypes. Vaccines 2020, 8, 386. [Google Scholar] [CrossRef]
- Li, Q.; Liu, Q.; Huang, W.; Li, X.; Wang, Y. Current status on the development of pseudoviruses for enveloped viruses. Rev. Med. Virol. 2018, 28, e1963. [Google Scholar] [CrossRef] [PubMed]
- Fan, H. Leukemogenesis by Moloney murine leukemia virus: A multistep process. Trends Microbiol. 1997, 5, 74–82. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef]
- Zheng, Y.; Larragoite, E.T.; Williams, E.S.C.P.; Lama, J.; Cisneros, I.; Delgado, J.C.; Slev, P.; Rychert, J.; Innis, E.A.; Coiras, M.; et al. Neutralization assay with SARS-CoV-1 and SARS-CoV-2 spike pseudotyped murine leukemia virions. Virol. J. 2021, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Dowd, K.A.; Manhart, C.J.; Ledgerwood, J.E.; Durbin, A.P.; Whitehead, S.S.; Pierson, T.C. Mechanism and Significance of Cell Type-Dependent Neutralization of Flaviviruses. J. Virol. 2014, 88, 7210–7220. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-T.; Han, Y.-J.; Wu, G.-H.; Huang, K.-Y.A.; Huang, P.-N. Overview of Neutralization Assays and International Standard for Detecting SARS-CoV-2 Neutralizing Antibody. Viruses 2022, 14, 1560. [Google Scholar] [CrossRef]
- Byrnes, J.R.; Zhou, X.X.; Lui, I.; Elledge, S.K.; Glasgow, J.E.; Lim, S.A.; Loudermilk, R.P.; Chiu, C.Y.; Wang, T.T.; Wilson, M.R.; et al. Competitive SARS-CoV-2 Serology Reveals Most Antibodies Targeting the Spike Receptor-Binding Domain Compete for ACE2 Binding. Msphere 2020, 5, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Kostin, N.N.; Bobik, T.V.; Skryabin, G.A.; Simonova, M.A.; Knorre, V.D.; Abrikosova, V.A.; Mokrushina, Y.A.; Smirnov, I.V.; Aleshenko, N.L.; Kruglova, N.A.; et al. An ELISA Platform for the Quantitative Analysis of SARS-CoV-2 RBD-neutralizing Antibodies As an Alternative to Monitoring of the Virus-Neutralizing Activity. Acta Naturae 2022, 14, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Kohmer, N.; Ruhl, C.; Ciesek, S.; Rabenau, H.F. Utility of Different Surrogate Enzyme-Linked Immunosorbent Assays (sELISAs) for Detection of SARS-CoV-2 Neutralizing Antibodies. J. Clin. Med. 2021, 10, 2128. [Google Scholar] [CrossRef] [PubMed]
- Girl, P.; Zwirglmaier, K.; von Buttlar, H.; Wölfel, R.; Müller, K. Evaluation of Two Rapid Lateral Flow Tests and Two Surrogate ELISAs for the Detection of SARS-CoV-2 Specific Neutralizing Antibodies. Front. Med. 2022, 9, 820151. [Google Scholar] [CrossRef] [PubMed]
- Shi, A.C.; Ren, P. SARS-CoV-2 serology testing: Progress and challenges. J. Immunol. Methods 2021, 494, 113060. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Muruato, A.E.; Zhang, X.; Lokugamage, K.G.; Fontes-Garfias, C.R.; Zou, J.; Liu, J.; Ren, P.; Balakrishnan, M.; Cihlar, T.; et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020, 11, 5214. [Google Scholar] [CrossRef] [PubMed]
- Condor Capcha, J.M.; Lambert, G.; Dykxhoorn, D.M.; Salerno, A.G.; Hare, J.M.; Whitt, M.A.; Pahwa, S.; Jayaweera, D.T.; Shehadeh, L.A. Generation of SARS-CoV-2 Spike Pseudotyped Virus for Viral Entry and Neutralization Assays: A 1-Week Protocol. Front. Cardiovasc. Med. 2020, 7, 618651. [Google Scholar] [CrossRef] [PubMed]
- von Rhein, C.; Scholz, T.; Henss, L.; Kronstein-Wiedemann, R.; Schwarz, T.; Rodionov, R.N.; Corman, V.M.; Tonn, T.; Schnierle, B.S. Comparison of potency assays to assess SARS-CoV-2 neutralizing antibody capacity in COVID-19 convalescent plasma. J. Virol. Methods 2021, 288, 114031. [Google Scholar] [CrossRef] [PubMed]
- Herrlein, M.-L.; Hein, S.; Zahn, T.; Mhedhbi, I.; Raupach, J.; Husria, Y.; Benz, N.I.; Eisert, J.; Bender, D.; Haberger, V.; et al. Comparative Investigation of Methods for Analysis of SARS-CoV-2-Spike-Specific Antisera. Viruses 2022, 14, 410. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mao, Q.; Wu, X.; He, Q.; Bian, L.; Bai, Y.; Wang, Z.; Wang, Q.; Zhang, J.; Liang, Z.; et al. Considerations for the Feasibility of Neutralizing Antibodies as a Surrogate Endpoint for COVID-19 Vaccines. Front. Immunol. 2022, 13, 814365. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.B.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Benkeser, D.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022, 375, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.W.; Mak, L.; Leung, G.M.; Cowling, B.J.; Peiris, M. Comparative immunogenicity of mRNA and inactivated vaccines against COVID-19. Lancet Microbe 2021, 2, e423. [Google Scholar] [CrossRef] [PubMed]
- Bergwerk, M.; Gonen, T.; Lustig, Y.; Amit, S.; Lipsitch, M.; Cohen, C.; Mandelboim, M.; Levin, E.G.; Rubin, C.; Indenbaum, V.; et al. COVID-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021, 385, 1474–1484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jia, Y.; Ji, Y.; Cong, X.; Liu, Y.; Yang, R.; Kong, X.; Shi, Y.; Zhu, L.; Wang, Z.; et al. Inactivated vaccines against SARS-CoV-2: Neutralizing antibody titers in vaccine recipients. Front. Microbiol. 2022, 13, 816778. [Google Scholar] [CrossRef]
- Levin, E.G.; Lustig, Y.; Cohen, C.; Fluss, R.; Indenbaum, V.; Amit, S.; Doolman, R.; Asraf, K.; Mendelson, E.; Ziv, A.; et al. Waning Immune Humoral Response to BNT162b2 COVID-19 Vaccine over 6 Months. N. Engl. J. Med. 2021, 385, e84. [Google Scholar] [CrossRef]
- Israel, A.; Shenhar, Y.; Green, I.; Merzon, E.; Golan-Cohen, A.; Schäffer, A.A.; Ruppin, E.; Vinker, S.; Magen, E. Large-Scale Study of Antibody Titer Decay following BNT162b2 mRNA Vaccine or SARS-CoV-2 Infection. Vaccines 2021, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Tang, H.; McDanal, C.; Wagh, K.; Fischer, W.; Theiler, J.; Yoon, H.; Li, D.; Haynes, B.F.; Sanders, K.O.; et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe 2021, 29, 529–539.e3. [Google Scholar] [CrossRef] [PubMed]
- Edara, V.V.; Norwood, C.; Floyd, K.; Lai, L.; Davis-Gardner, M.E.; Hudson, W.H.; Mantus, G.; Nyhoff, L.E.; Adelman, M.W.; Fineman, R.; et al. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe 2021, 29, 516–521.e3. [Google Scholar] [CrossRef] [PubMed]
- Rössler, A.; Riepler, L.; Bante, D.; von Laer, D.; Kimpel, J. SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons. N. Engl. J. Med. 2022, 386, 698–700. [Google Scholar] [CrossRef] [PubMed]
- Altarawneh, H.N.; Chemaitelly, H.; Ayoub, H.H.; Hasan, M.R.; Coyle, P.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Al-Kanaani, Z.; Al-Kuwari, E.; et al. Protective Effect of Previous SARS-CoV-2 Infection against Omicron BA.4 and BA.5 Subvariants. N. Engl. J. Med. 2022, 387, 1620–1621. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. New Omicron-specific vaccines offer similar protection to existing boosters. Nature 2022, 609, 232–233. [Google Scholar] [CrossRef]
- Knezevic, I.; Mattiuzzo, G.; Page, M.; Minor, P.; Griffiths, E.; Nuebling, M.; Moorthy, V. WHO International Standard for evaluation of the antibody response to COVID-19 vaccines: Call for urgent action by the scientific community. Lancet Microbe 2022, 3, e235–e240. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Mao, Q.; Tan, D.; Liu, J.; Zhang, X.; Li, L.; Liu, M.; Wang, Z.; Cheng, F.; Cui, B.; et al. Establishment of national standard for anti-SARS-Cov-2 neutralizing antibody in China: The first National Standard calibration traceability to the WHO International Standard. Front. Immunol. 2023, 14, 1107639. [Google Scholar] [CrossRef]
- Lerdsamran, H.; Anusorntanawat, R.; Sangsiriwut, K.; Sawadpongpan, S.; Prasertsopon, J.; Thinpan, N.; Intalapaporn, P.; Techasuwanna, R.; Okada, P.; Puthavathana, P. Higher correlation between neutralizing antibodies and surrogate neutralizing or binding antibodies in COVID-19 patients than vaccine recipients. PLoS ONE 2024, 19, e0298033. [Google Scholar] [CrossRef]
- Pieri, M.; Infantino, M.; Manfredi, M.; Nuccetelli, M.; Grossi, V.; Lari, B.; Tomassetti, F.; Sarubbi, S.; Russo, E.; Amedei, A.; et al. Performance evaluation of four surrogate Virus Neutralization Tests (sVNTs) in comparison to the in vivo gold standard test. FBL 2022, 27, 74. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Chmielewska, A.M.; Czarnota, A.; Bieńkowska-Szewczyk, K.; Grzyb, K. Immune response against SARS-CoV-2 variants: The role of neutralization assays. NPJ Vaccines 2021, 6, 142. [Google Scholar] [CrossRef] [PubMed]
- CEPI. CEPI Establishes Global Network of Laboratories to Centralise Assessment of COVID-19 Vaccine Candidates. Available online: https://cepi.net/news_cepi/cepi-establishes-global-network-of-laboratories-to-centralise-assessment-of-COVID-19-vaccine-candidates/ (accessed on 19 May 2023).
- WHO. First WHO International Standard for Anti-SARS-CoV-2 Immunoglobulin (Human) NIBSC Code: 20/136 Instructions for Use. Available online: https://www.nibsc.org/documents/ifu/20-136.pdf (accessed on 21 May 2023).
- WHO. First WHO International Reference Panel for Anti-SARS-CoV-2 Immunoglubulin NIBSC Code: 20/268 Instructions for Use. Available online: https://www.nibsc.org/documents/ifu/20-268.pdf (accessed on 19 May 2023).
- Kristiansen, P.A.; Page, M.; Bernasconi, V.; Mattiuzzo, G.; Dull, P.; Makar, K.; Plotkin, S.; Knezevic, I. WHO International Standard for anti-SARS-CoV-2 immunoglobulin. Lancet 2021, 397, 1347–1348. [Google Scholar] [CrossRef]
- Khoury, D.S.; Schlub, T.E.; Cromer, D.; Steain, M.; Fong, Y.; Gilbert, P.B.; Subbarao, K.; Triccas, J.A.; Kent, S.J.; Davenport, M.P. Correlates of Protection, Thresholds of Protection, and Immunobridging among Persons with SARS-CoV-2 Infection. Emerg. Infect. Dis. 2023, 29, 381–388. [Google Scholar] [CrossRef]
| Vaccine Type | Name | Manufacturer | Immunogenic Detection Method | Immune Indexes | References |
|---|---|---|---|---|---|
| Inactivated vaccines | CoronaVac | Sinovac |
|
| [12,13,14] |
| |||||
| |||||
| |||||
| BBIBP-CorV | Sinopharm/BIBP |
|
| [15,16] | |
| |||||
| mRNA vaccines | BNT162b2 | Pfizer-BioNTech |
|
| [17,18,19,20] |
| |||||
| mRNA-1273 | Moderna |
|
| [21,22] | |
| |||||
| |||||
| mRNA-1273.214 | Moderna |
|
| [23] | |
| |||||
| Adenovirus vaccines | Ad26.COV2.S | Janssen–Cilag InternationalNV |
|
| [15,24] |
| |||||
| |||||
| AZD1222/Vaxzevria | AstraZeneca |
|
| [25,26,27] | |
| |||||
| Sputnik V | Gamaleya Nat. Center of Epidem. and Microbiol. |
|
| [28,29] | |
| |||||
| |||||
| Ad5-nCoV/Convidecia | CanSinoBIO |
|
| [30,31] | |
| |||||
| |||||
| |||||
| |||||
| |||||
| Subunit vaccines | NVX-CoV2373/Nuvaxovid | Novavax |
|
| [32,33] |
| |||||
| ZF2001 | Zhifei Longcom |
|
| [34,35] | |
| |||||
| |||||
|
| Method Classification | Method Name | Advantages | Disadvantages |
|---|---|---|---|
| cVNT | CPE |
|
|
| PRNT | |||
| Recombinant live virus |
|
| |
| pVNT | HIV system |
|
|
| VSV system | |||
| MLV system | |||
| sVNT | ELISA |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Huang, W.; Xiang, H.; Nie, J. SARS-CoV-2 Neutralization Assays Used in Clinical Trials: A Narrative Review. Vaccines 2024, 12, 554. https://doi.org/10.3390/vaccines12050554
Sun Y, Huang W, Xiang H, Nie J. SARS-CoV-2 Neutralization Assays Used in Clinical Trials: A Narrative Review. Vaccines. 2024; 12(5):554. https://doi.org/10.3390/vaccines12050554
Chicago/Turabian StyleSun, Yeqing, Weijin Huang, Hongyu Xiang, and Jianhui Nie. 2024. "SARS-CoV-2 Neutralization Assays Used in Clinical Trials: A Narrative Review" Vaccines 12, no. 5: 554. https://doi.org/10.3390/vaccines12050554
APA StyleSun, Y., Huang, W., Xiang, H., & Nie, J. (2024). SARS-CoV-2 Neutralization Assays Used in Clinical Trials: A Narrative Review. Vaccines, 12(5), 554. https://doi.org/10.3390/vaccines12050554








