A Novel Multiepitope Fusion Antigen as a Vaccine Candidate for the Prevention of Enterotoxigenic E. coli-Induced Calf Diarrhea
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Bacterial Strains and Cells
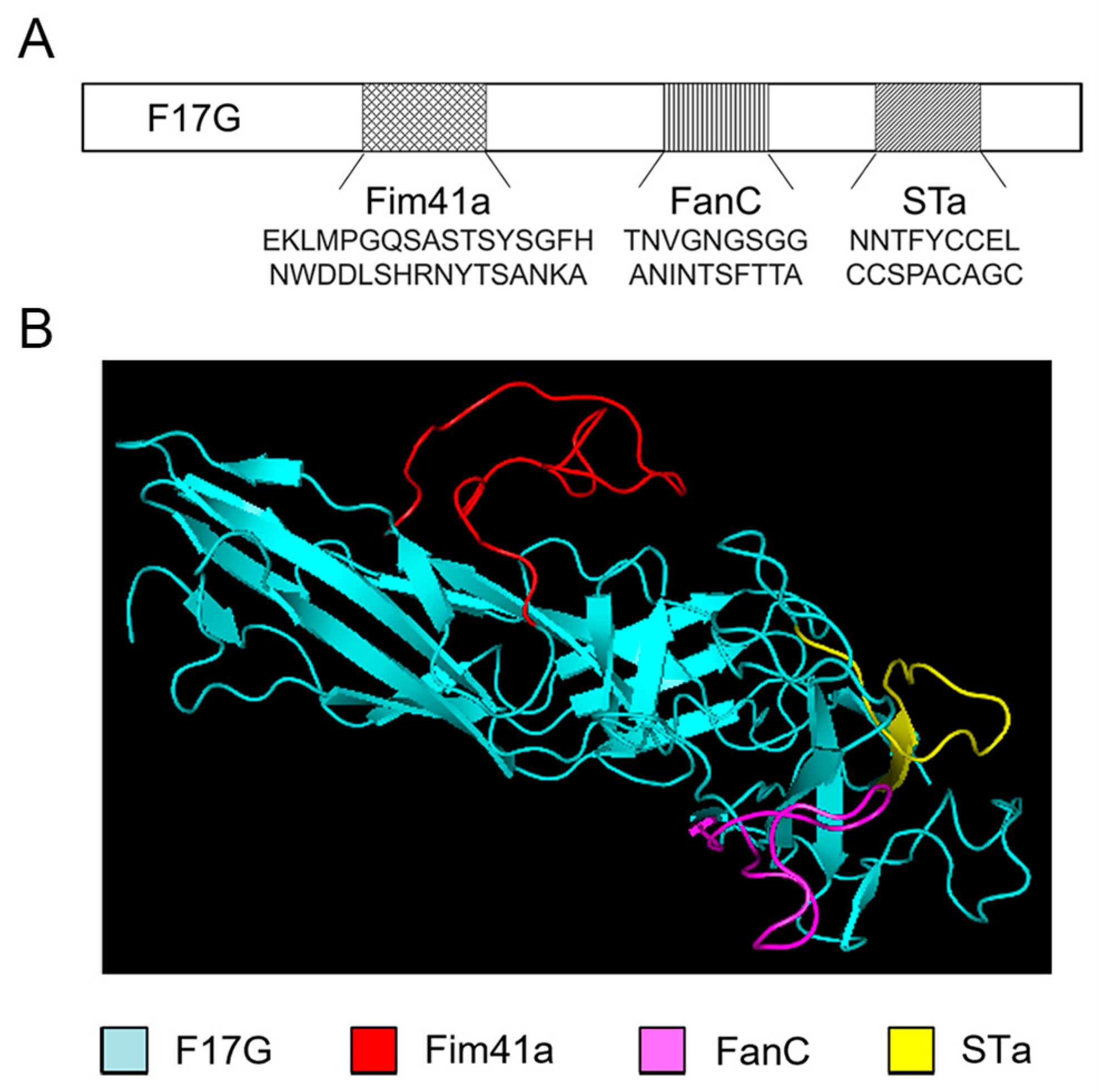
2.3. Cloning of F17G, FanC, Fim41a, and STa Genes and Their Chimeric Gene F17G-Fim41a-FanC-STa
2.4. Expression of the Recombinant Proteins
2.5. Development of Mouse Antiserum
2.6. Antibody Detection with iELISA and WB Assay
2.7. Adherence and Adherence Inhibition Assays
2.8. The Activating Impact and Neutralizing Properties of the Antibody
2.9. Statistical Analysis
3. Results
3.1. Computational Modeling of the MEFA Protein
3.2. Expression of the Recombinant Proteins
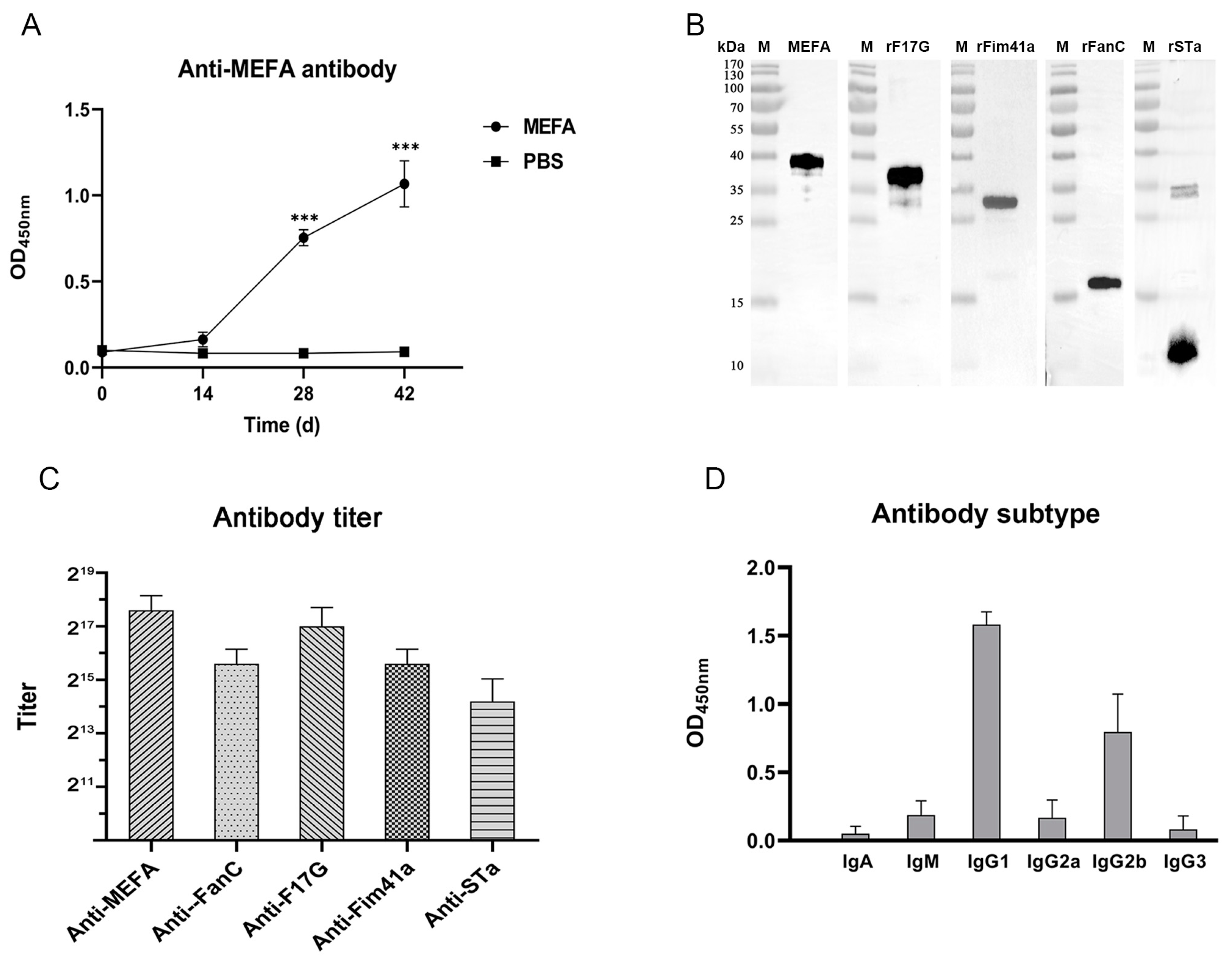
3.3. Antibody Detection with iELISA and WB Assay
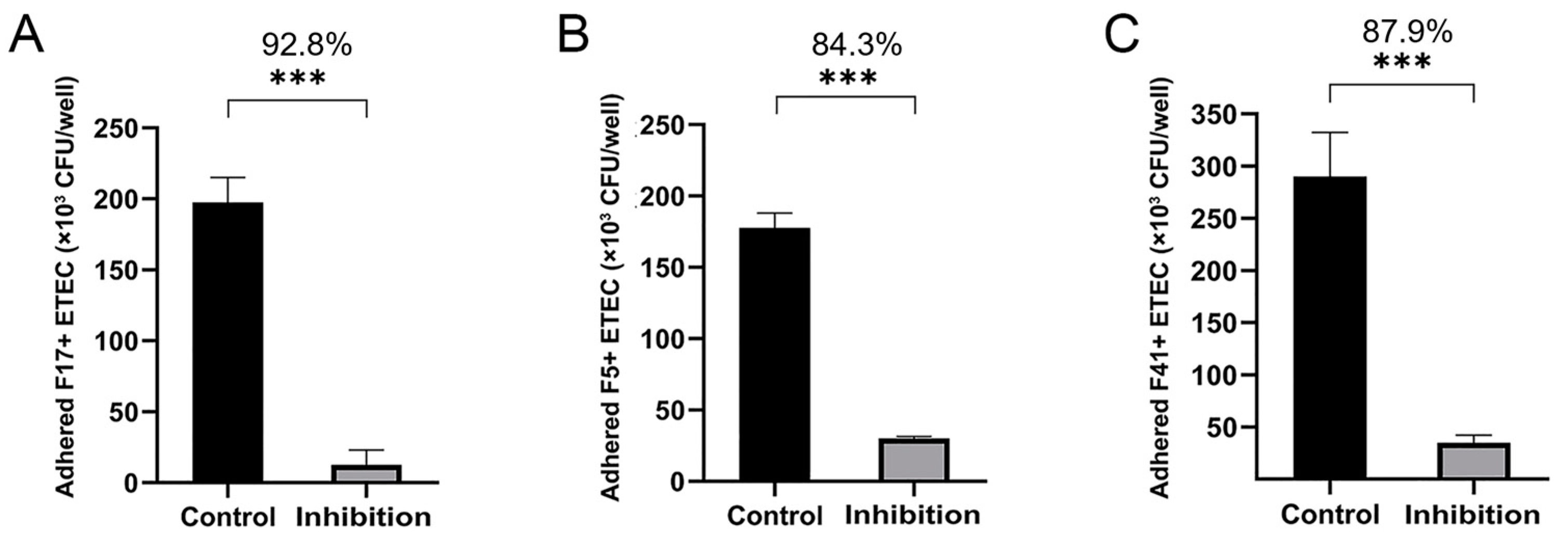
3.4. Adherence and Adherence Inhibition Assay
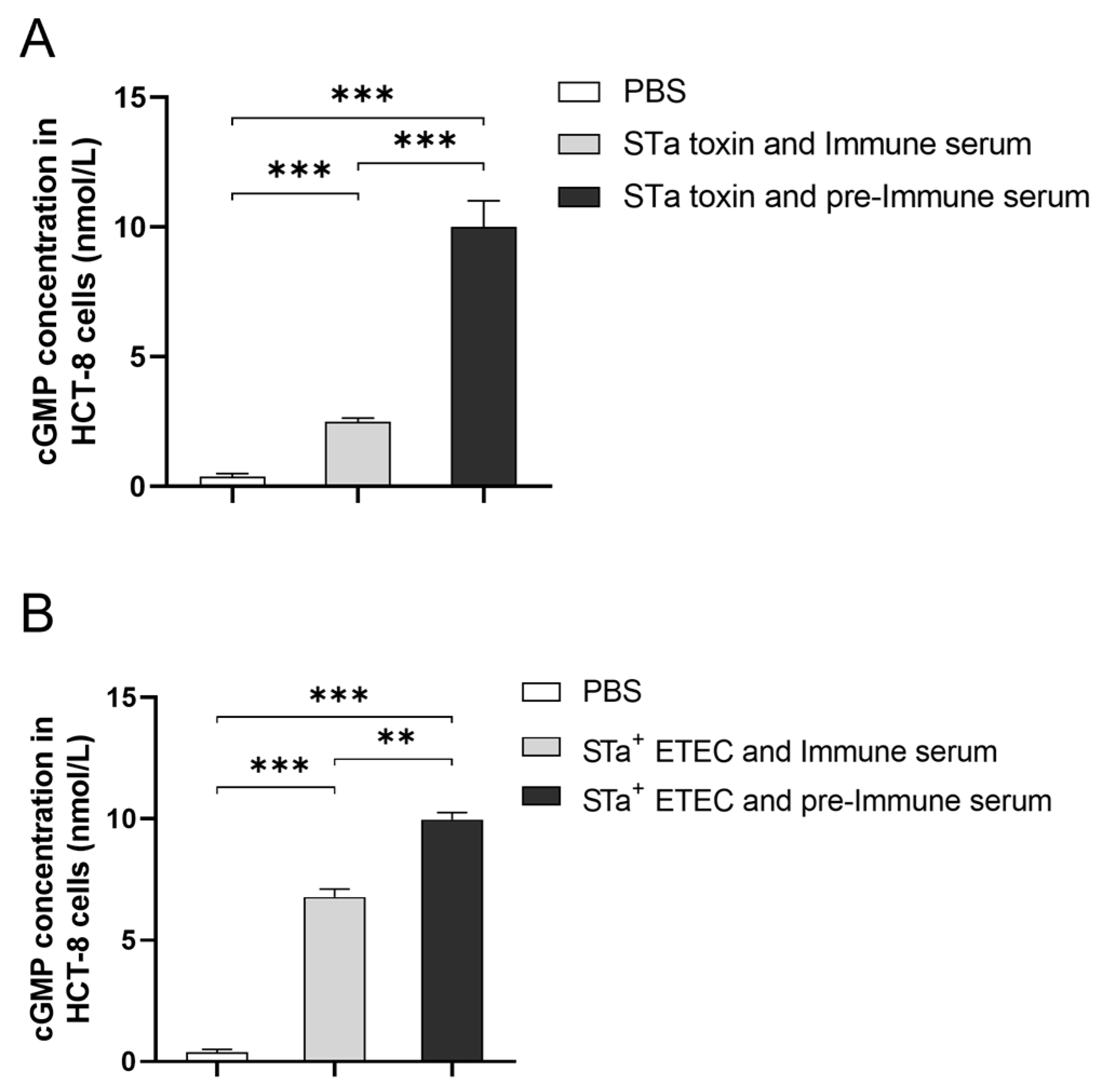
3.5. The Activating Impact and Neutralizing Properties of the Antibody
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meganck, V.; Hoflack, G.; Opsomer, G. Advances in Prevention and Therapy of Neonatal Dairy Calf Diarrhoea: A Systematical Review with Emphasis on Colostrum Management and Fluid Therapy. Acta Vet. Scand. 2014, 56, 75. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.-I.; Han, J.-I.; Wang, C.; Cooper, V.; Schwartz, K.; Engelken, T.; Yoon, K.-J. Case–Control Study of Microbiological Etiology Associated with Calf Diarrhea. Vet. Microbiol. 2013, 166, 375–385. [Google Scholar] [CrossRef]
- Nagy, B.; Fekete, P.Z. Enterotoxigenic Escherichia coli in Veterinary Medicine. Int. J. Med. Microbiol. 2005, 295, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Qadri, F.; Svennerholm, A.-M.; Faruque, A.S.G.; Sack, R.B. Enterotoxigenic Escherichia coli in Developing Countries: Epidemiology, Microbiology, Clinical Features, Treatment, and Prevention. Clin. Microbiol. Rev. 2005, 18, 465–483. [Google Scholar] [CrossRef]
- Kopic, S.; Geibel, J.P. Toxin Mediated Diarrhea in the 21 Century: The Pathophysiology of Intestinal Ion Transport in the Course of ETEC, V. Cholerae and Rotavirus Infection. Toxins 2010, 2, 2132–2157. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, J.D. The Whole Shebang: The Gastrointestinal Tract, Escherichia coli Enterotoxins and Secretion. Curr. Issues Mol. Biol. 2012, 14, 71–82. [Google Scholar] [CrossRef]
- Jin, L.Z.; Zhao, X. Intestinal Receptors for Adhesive Fimbriae of Enterotoxigenic Escherichia coli (ETEC) K88 in Swine—A Review. Appl. Microbiol. Biotechnol. 2000, 54, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Guinée, P.A.; Jansen, W.H. Detection of Enterotoxigenicity and Attachment Factors in Escherichia coli Strains of Human, Porcine and Bovine Origin; a Comparative Study. Zentralbl Bakteriol. Orig. A 1979, 243, 245–257. [Google Scholar]
- Schroyen, M.; Stinckens, A.; Verhelst, R.; Niewold, T.; Buys, N. The Search for the Gene Mutations Underlying Enterotoxigenic Escherichia coli F4ab/Ac Susceptibility in Pigs: A Review. Vet. Res. 2012, 43, 70. [Google Scholar] [CrossRef]
- Salhi, I.; Rabti, A.; Dhehibi, A.; Raouafi, N. Sandwich-Based Immunosensor for Dual-Mode Detection of Pathogenic F17–Positive Escherichia coli Strains. Int. J. Mol. Sci. 2022, 23, 6028. [Google Scholar] [CrossRef]
- Kolenda, R.; Burdukiewicz, M.; Schierack, P. A Systematic Review and Meta-Analysis of the Epidemiology of Pathogenic Escherichia coli of Calves and the Role of Calves as Reservoirs for Human Pathogenic E. coli. Front. Cell Infect. Microbiol. 2015, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.-H.; Kim, S.; Park, J.; Choi, K.-S. Characterization of Virulence Genes in Escherichia coli Strains Isolated from Pre-Weaned Calves in the Republic of Korea. Acta Vet. Scand. 2020, 62, 45. [Google Scholar] [CrossRef] [PubMed]
- Yano, T.; Garcia, M.; Leite, D.S.; Pestana-de-Castro, A.F.; Shenk, M.A. Determination of the Efficiency of K99-F41 Fimbrial Antigen Vaccine in Newborn Calves. Braz. J. Med. Biol. Res. 1995, 28, 651–654. [Google Scholar] [PubMed]
- Viidu, D.-A.; Mõtus, K. Implementation of a Pre-Calving Vaccination Programme against Rotavirus, Coronavirus and Enterotoxigenic Escherichia coli (F5) and Association with Dairy Calf Survival. BMC Vet. Res. 2022, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Xia, P.; Nandre, R.; Zhang, W.; Zhu, G. Review of Newly Identified Functions Associated with the Heat-Labile Toxin of Enterotoxigenic Escherichia coli. Front. Cell. Infect. Microbiol. 2019, 9, 292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sack, D.A. Progress and Hurdles in the Development of Vaccines against Enterotoxigenic Escherichia coli in Humans. Expert Rev. Vaccines 2012, 11, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Li, W.; Wu, X.; Bao, S.; Zeng, J.; Zhao, N.; Luo, M.; Liu, X.; Wang, Y. Immunogenicity and Protective Efficacy of a Recombinant Adenoviral Based Vaccine Expressing Heat-Stable Enterotoxin (STa) and K99 Adhesion Antigen of Enterotoxigenic Escherichia coli in Mice. Mol. Immunol. 2015, 68, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Seo, H.; Upadhyay, I.; Zhang, W. A Polyvalent Adhesin–Toxoid Multiepitope-Fusion-Antigen-Induced Functional Antibodies against Five Enterotoxigenic Escherichia coli Adhesins (CS7, CS12, CS14, CS17, and CS21) but Not Enterotoxins (LT and STa). Microorganisms 2023, 11, 2473. [Google Scholar] [CrossRef] [PubMed]
- Hashish, E.A.; Zhang, C.; Ruan, X.; Knudsen, D.E.; Chase, C.C.; Isaacson, R.E.; Zhou, G.; Zhang, W. A Multiepitope Fusion Antigen Elicits Neutralizing Antibodies against Enterotoxigenic Escherichia coli and Homologous Bovine Viral Diarrhea Virus In Vitro. Clin. Vaccine Immunol. 2013, 20, 1076–1083. [Google Scholar] [CrossRef]
- Clifford, J.N.; Høie, M.H.; Deleuran, S.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-3.0: Improved B-cell Epitope Prediction Using Protein Language Models. Protein Sci. 2022, 31, e4497. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. Prediction Methods for B-Cell Epitopes. In Immunoinformatics; Flower, D.R., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2007; Volume 409, pp. 387–394. ISBN 978-1-58829-699-3. [Google Scholar]
- Ruan, X.; Robertson, D.C.; Nataro, J.P.; Clements, J.D.; Zhang, W. Characterization of Heat-Stable (STa) Toxoids of Enterotoxigenic Escherichia coli Fused to Double Mutant Heat-Labile Toxin Peptide in Inducing Neutralizing Anti-STa Antibodies. Infect. Immun. 2014, 82, 1823–1832. [Google Scholar] [CrossRef] [PubMed]
- Stefańska, B.; Katzer, F.; Golińska, B.; Sobolewska, P.; Smulski, S.; Frankiewicz, A.; Nowak, W. Different Methods of Eubiotic Feed Additive Provision Affect the Health, Performance, Fermentation, and Metabolic Status of Dairy Calves during the Preweaning Period. BMC Vet. Res. 2022, 18, 138. [Google Scholar] [CrossRef] [PubMed]
- Berzosa, M.; Nemeskalova, A.; Zúñiga-Ripa, A.; Salvador-Bescós, M.; Larrañeta, E.; Donnelly, R.F.; Gamazo, C.; Irache, J.M. Immune Response after Skin Delivery of a Recombinant Heat-Labile Enterotoxin B Subunit of Enterotoxigenic Escherichia coli in Mice. Pharmaceutics 2022, 14, 239. [Google Scholar] [CrossRef] [PubMed]
- Sukwa, N.; Mubanga, C.; Hatyoka, L.M.; Chilyabanyama, O.N.; Chibuye, M.; Mundia, S.; Munyinda, M.; Kamuti, E.; Siyambango, M.; Badiozzaman, S.; et al. Safety, Tolerability, and Immunogenicity of an Oral Inactivated ETEC Vaccine (ETVAX®) with dmLT Adjuvant in Healthy Adults and Children in Zambia: An Age Descending Randomised, Placebo-Controlled Trial. Vaccine 2023, 41, 6884–6894. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Xu, Y.; Li, X.; Yin, J.; Li, G.; Zhao, H.; Li, S.; Li, J.; Wang, L. Protective Efficacy of a Recombinant Enterotoxin Antigen in a Maternal Immunization Model and the Inhibition of Specific Maternal Antibodies to Neonatal Oral Vaccination. J. Reprod. Immunol. 2023, 157, 103946. [Google Scholar] [CrossRef] [PubMed]
- Durel, L.; Rose, C.; Bainbridge, T.; Roubert, J.; Dressel, K.-U.; Bennemann, J.; Rückner, A.; Vahlenkamp, T.; Maillard, R. Immune Response of Mature Cows Subjected to Annual Booster Vaccination against Neonatal Calf Diarrhoea with Two Different Commercial Vaccines: A Non-Inferiority Study. Livest. Sci. 2017, 204, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhu, J.; Zhu, G. Fimbriae of animal-originated enterotoxigenic Escherichia coli—A review. Wei Sheng Wu Xue Bao 2012, 52, 679–686. [Google Scholar] [PubMed]
- Liu, Y.; Maciel, M.; O’Dowd, A.; Poole, S.T.; Rollenhagen, J.E.; Etobayeva, I.V.; Savarino, S.J. Development and Comparison of a Panel of Modified CS17 Fimbrial Tip Adhesin Proteins as Components for an Adhesin-Based Vaccine against Enterotoxigenic Escherichia coli. Microorganisms 2021, 9, 1646. [Google Scholar] [CrossRef] [PubMed]
- Bihannic, M.; Ghanbarpour, R.; Auvray, F.; Cavalié, L.; Châtre, P.; Boury, M.; Brugère, H.; Madec, J.-Y.; Oswald, E. Identification and Detection of Three New F17 Fimbrial Variants in Escherichia coli Strains Isolated from Cattle. Vet. Res. 2014, 45, 76. [Google Scholar] [CrossRef]
- Bi, Y.; Yang, C.; Diao, Q.; Tu, Y. Effects of Dietary Supplementation with Two Alternatives to Antibiotics on Intestinal Microbiota of Preweaned Calves Challenged with Escherichia coli K99. Sci. Rep. 2017, 7, 5439. [Google Scholar] [CrossRef]
- Duan, Q.; Pang, S.; Wu, W.; Jiang, B.; Zhang, W.; Liu, S.; Wang, X.; Pan, Z.; Zhu, G. A Multivalent Vaccine Candidate Targeting Enterotoxigenic Escherichia coli Fimbriae for Broadly Protecting against Porcine Post-Weaning Diarrhea. Vet. Res. 2020, 51, 93. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Jiang, L.; McIntyre, M.E.; Petrova, E.; Cheng, S.X. Calcimimetic Acts on Enteric Neuronal CaSR to Reverse Cholera Toxin-Induced Intestinal Electrolyte Secretion. Sci. Rep. 2018, 8, 7851. [Google Scholar] [CrossRef] [PubMed]
- Elhelw, H.A.; El Fadeel, M.R.A.; El-Sergany, E.; Allam, A.; Elbayoumy, M.K.; El-Kattan, A.M.; El-kholy, A.A.-M. Preparation and Field Study of Combined Vaccine against Clostridium Perfringens Type A and Bovine Viral Diarrhea Virus in Camels. Clin. Exp. Vaccine Res. 2022, 11, 30. [Google Scholar] [CrossRef]
- Katsoulos, P.D.; Karatzia, M.A.; Dovas, C.I.; Filioussis, G.; Papadopoulos, E.; Kiossis, E.; Arsenopoulos, K.; Papadopoulos, T.; Boscos, C.; Karatzias, H. Evaluation of the In-Field Efficacy of Oregano Essential Oil Administration on the Control of Neonatal Diarrhea Syndrome in Calves. Res. Vet. Sci. 2017, 115, 478–483. [Google Scholar] [CrossRef] [PubMed]

| Primers | Sequences (5′–3′) |
|---|---|
| F17G-F | CTTTAAGAAGGAGATATACATATGGCGGCAGTTTCATTTATTGGTGC |
| F17G-R | GGCCGCAAGCTTGTCGACCTGATAGGAAAATGTAAATG |
| FanC-F | CTTTAAGAAGGAGATATACATATGAATACAGGTACTATTAAC |
| FanC-R | TGCGGCCGCAAGCTTGTCGACCATATAAGTGACTAAGAAGG |
| Fim41a-F | CTTTAAGAAGGAGATATACATATGGCTGCTGATTGGACGGAAGG |
| Fim41a-R | TGCGGCCGCAAGCTTGTCGACACTATAAATAACGGTGATAG |
| STa-F | CTTTAAGAAGGAGATATACATATGATGAAAAAGCTAATGTTGGC |
| STa-R | TGCGGCCGCAAGCTTGTCGACATAACATCCAGCACAGGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Yuan, X.; He, Y.; Chen, Y.; Hu, C.; Chen, J.; Zhang, L.; Chen, X.; Guo, A. A Novel Multiepitope Fusion Antigen as a Vaccine Candidate for the Prevention of Enterotoxigenic E. coli-Induced Calf Diarrhea. Vaccines 2024, 12, 457. https://doi.org/10.3390/vaccines12050457
Zhang H, Yuan X, He Y, Chen Y, Hu C, Chen J, Zhang L, Chen X, Guo A. A Novel Multiepitope Fusion Antigen as a Vaccine Candidate for the Prevention of Enterotoxigenic E. coli-Induced Calf Diarrhea. Vaccines. 2024; 12(5):457. https://doi.org/10.3390/vaccines12050457
Chicago/Turabian StyleZhang, Haoyun, Xinwei Yuan, Yanfei He, Yingyu Chen, Changmin Hu, Jianguo Chen, Lei Zhang, Xi Chen, and Aizhen Guo. 2024. "A Novel Multiepitope Fusion Antigen as a Vaccine Candidate for the Prevention of Enterotoxigenic E. coli-Induced Calf Diarrhea" Vaccines 12, no. 5: 457. https://doi.org/10.3390/vaccines12050457
APA StyleZhang, H., Yuan, X., He, Y., Chen, Y., Hu, C., Chen, J., Zhang, L., Chen, X., & Guo, A. (2024). A Novel Multiepitope Fusion Antigen as a Vaccine Candidate for the Prevention of Enterotoxigenic E. coli-Induced Calf Diarrhea. Vaccines, 12(5), 457. https://doi.org/10.3390/vaccines12050457







