Immunoreactivity Analysis of MHC-I Epitopes Derived from the Nucleocapsid Protein of SARS-CoV-2 via Computation and Vaccination
Abstract
1. Introduction
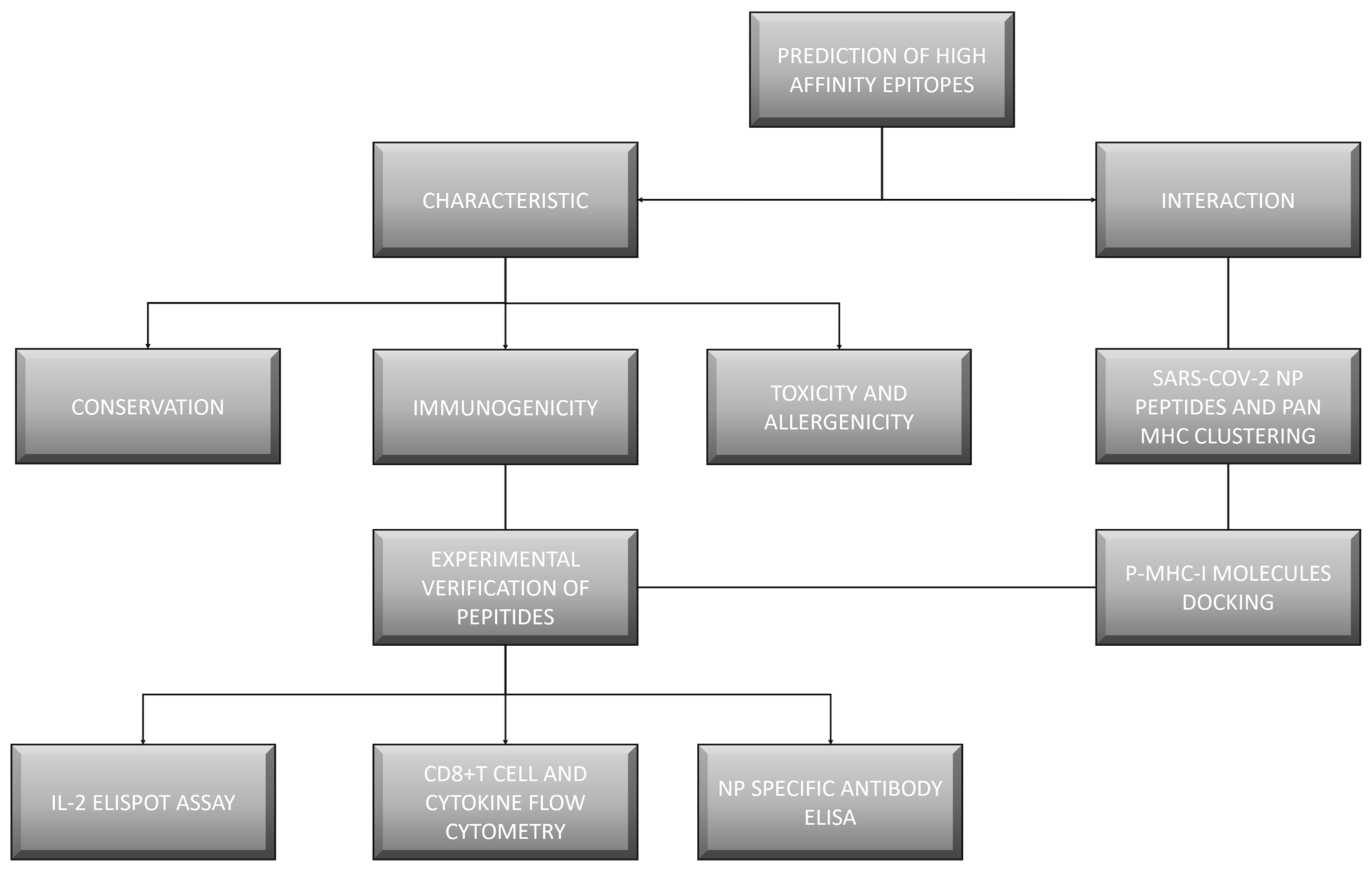
2. Materials and Methods
2.1. SARS-CoV-2 NP Sequence Retrieval
2.2. SARS-CoV-2 NP Pan-MHC-I Epitope Prediction and Screening
2.3. Conservation Analysis
2.4. Immunogenicity Analysis
2.5. Docking of Pan-MHC-I Molecules
2.6. SARS-CoV-2 NP Peptides and Pan-MHC-I Clustering
2.7. Sequence Alignment of SARS-CoV-2 Variants
2.8. Prediction of Peptide Toxicity and Sensitization
2.9. Application of Pan-MHC-I-Restricted SARS-CoV-2 NP Epitopes via a Literature Review
2.10. Vaccine, Animal, and Immunization
2.11. Peptides and ELISpot Assay
2.12. Enzyme-Linked Immunosorbent Assay (ELISA)
2.13. Flow Cytometry
2.14. Statistical Analysis
3. Results
3.1. Affinity Analysis of SARS-CoV-2 NP Epitopes for Mouse H-2 and Major HLA-I Haplotypes
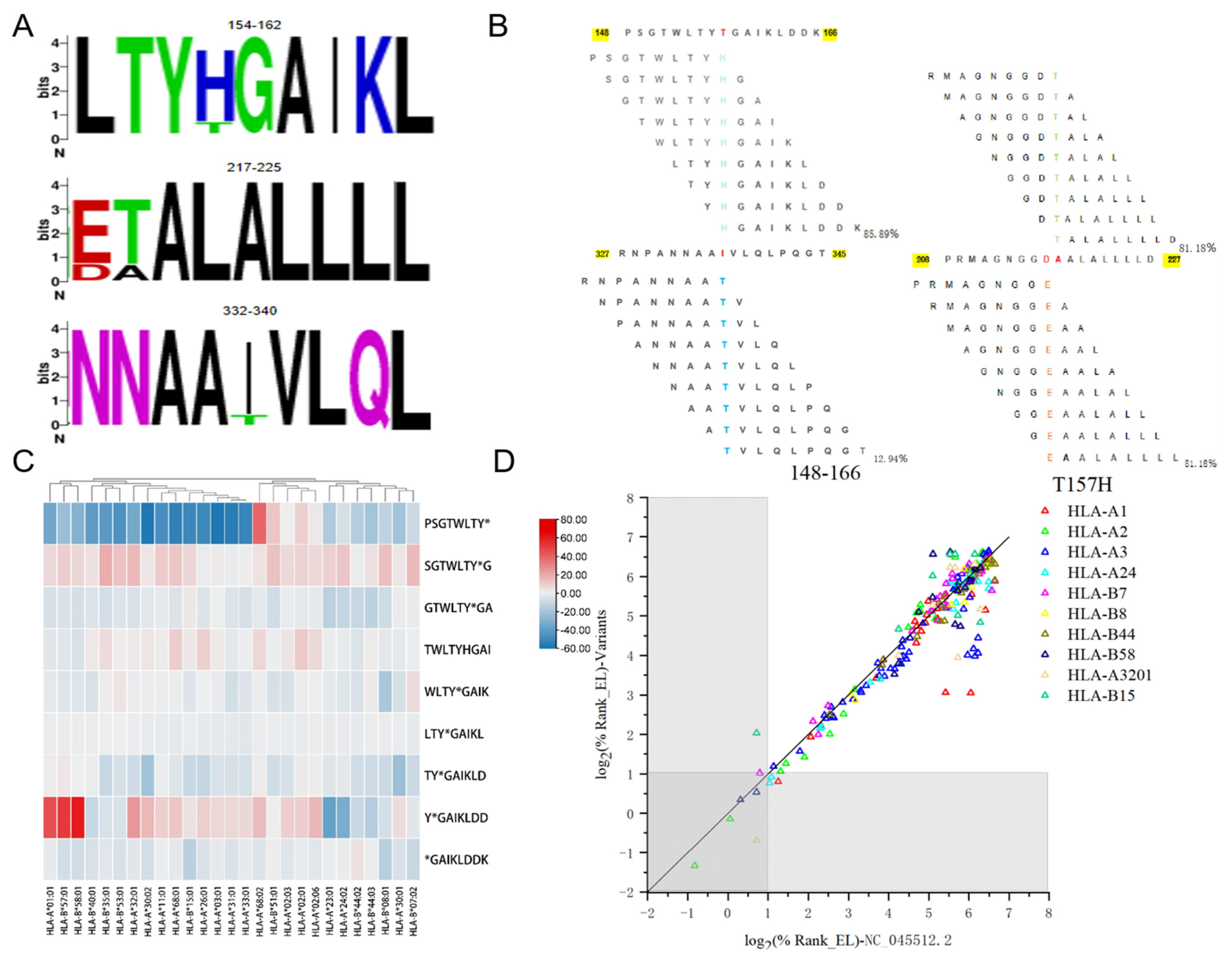
3.2. Conservation Status of SARS-CoV-2 NP 9-mer Dominant Epitopes
3.3. Immunogenicity of SARS-CoV-2 NP 9-mer Peptides
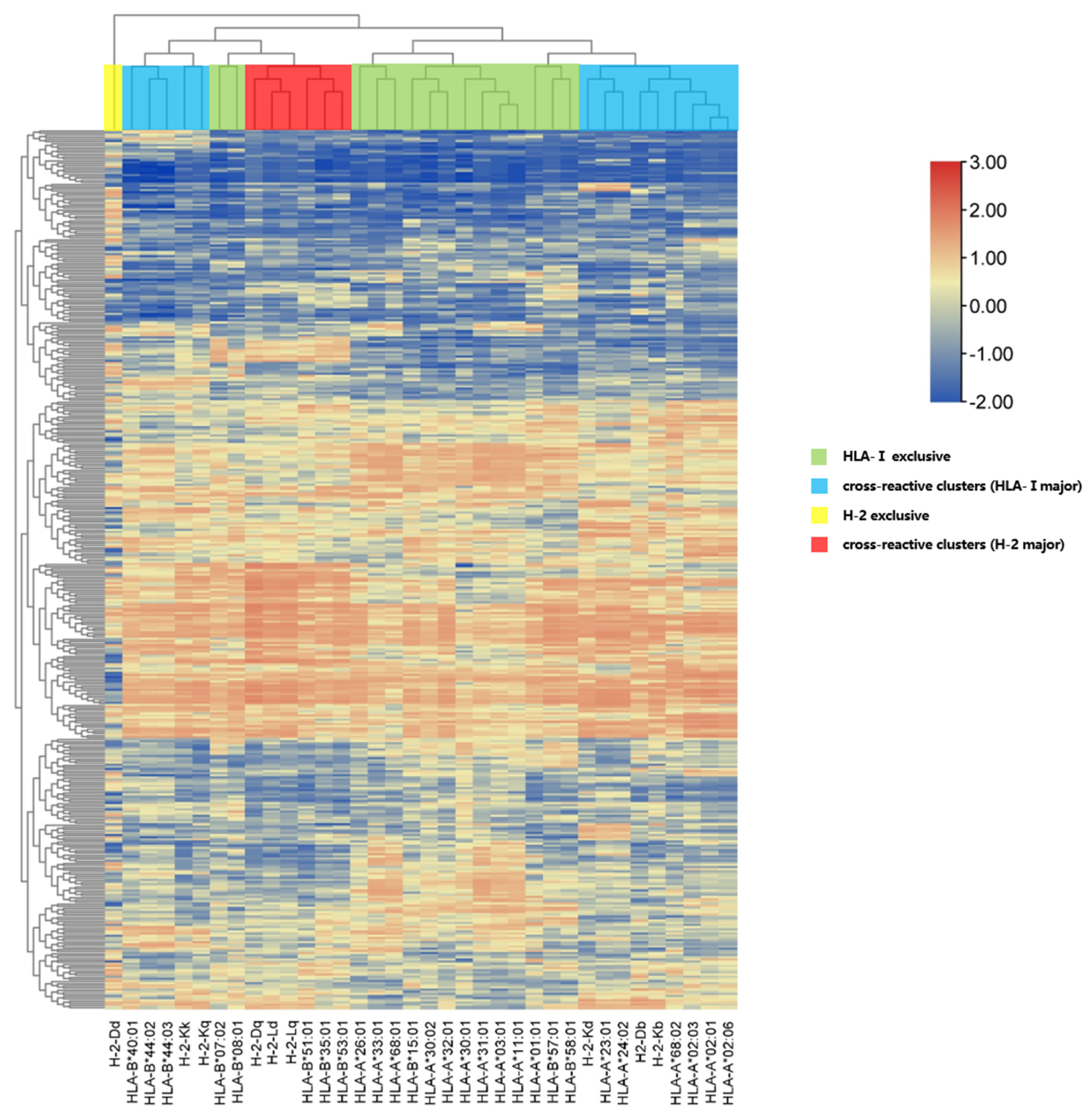
3.4. Interactions Between Pan-MHC-I Molecules and SARS-CoV-2 NP 9-mer Peptides via Hierarchical Clustering
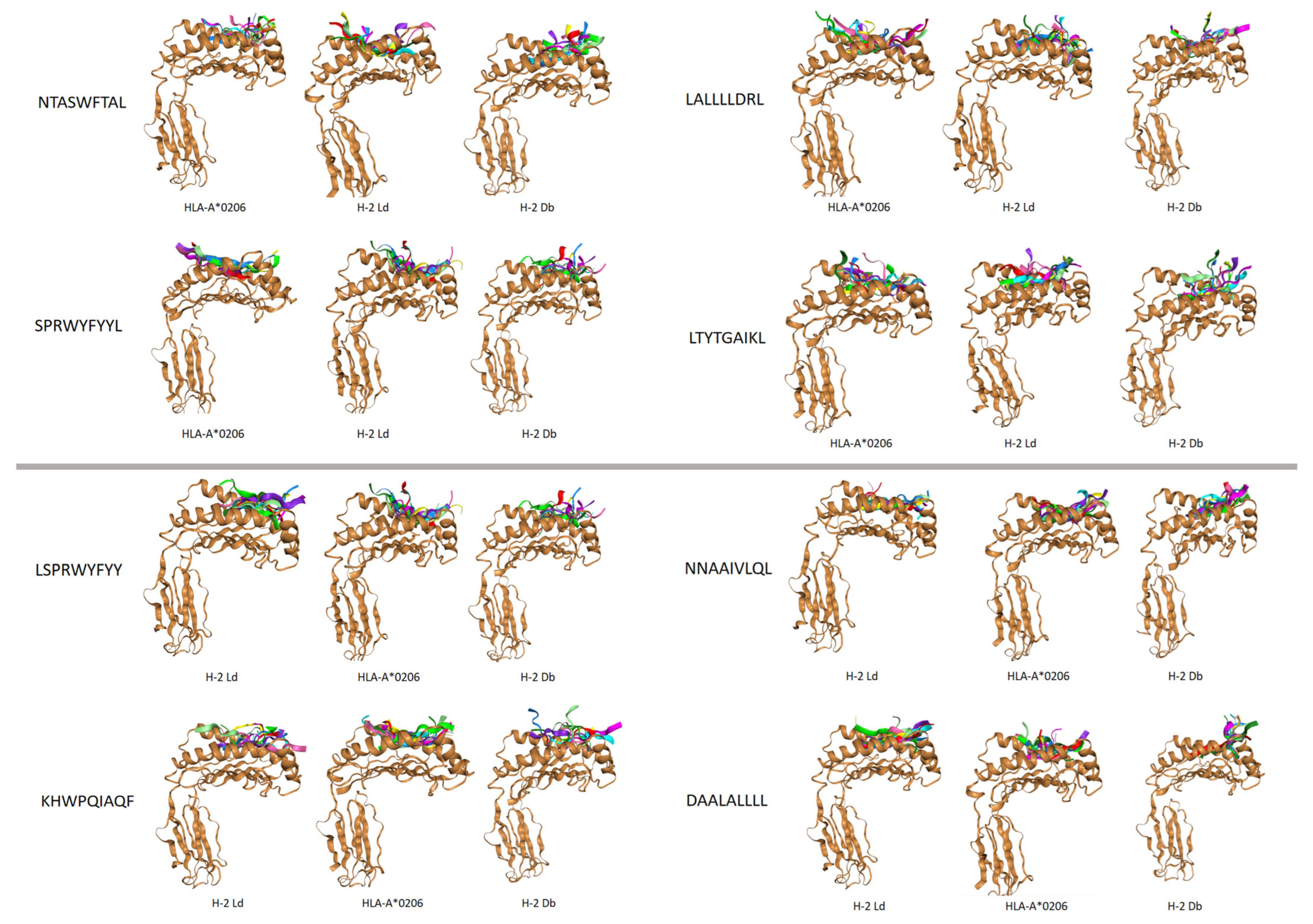
3.5. Docking of Pan-MHC-I Molecules with Preferred Epitopes
3.6. Multiple Sequence Alignment with 84 SARS-CoV-2 Mutant Strains
3.7. Differences in the Immunoreaction Among SARS-CoV-2 and Its Variants
3.8. Toxicity and Sensitization Analysis of SARS-CoV-2 NP Epitopes
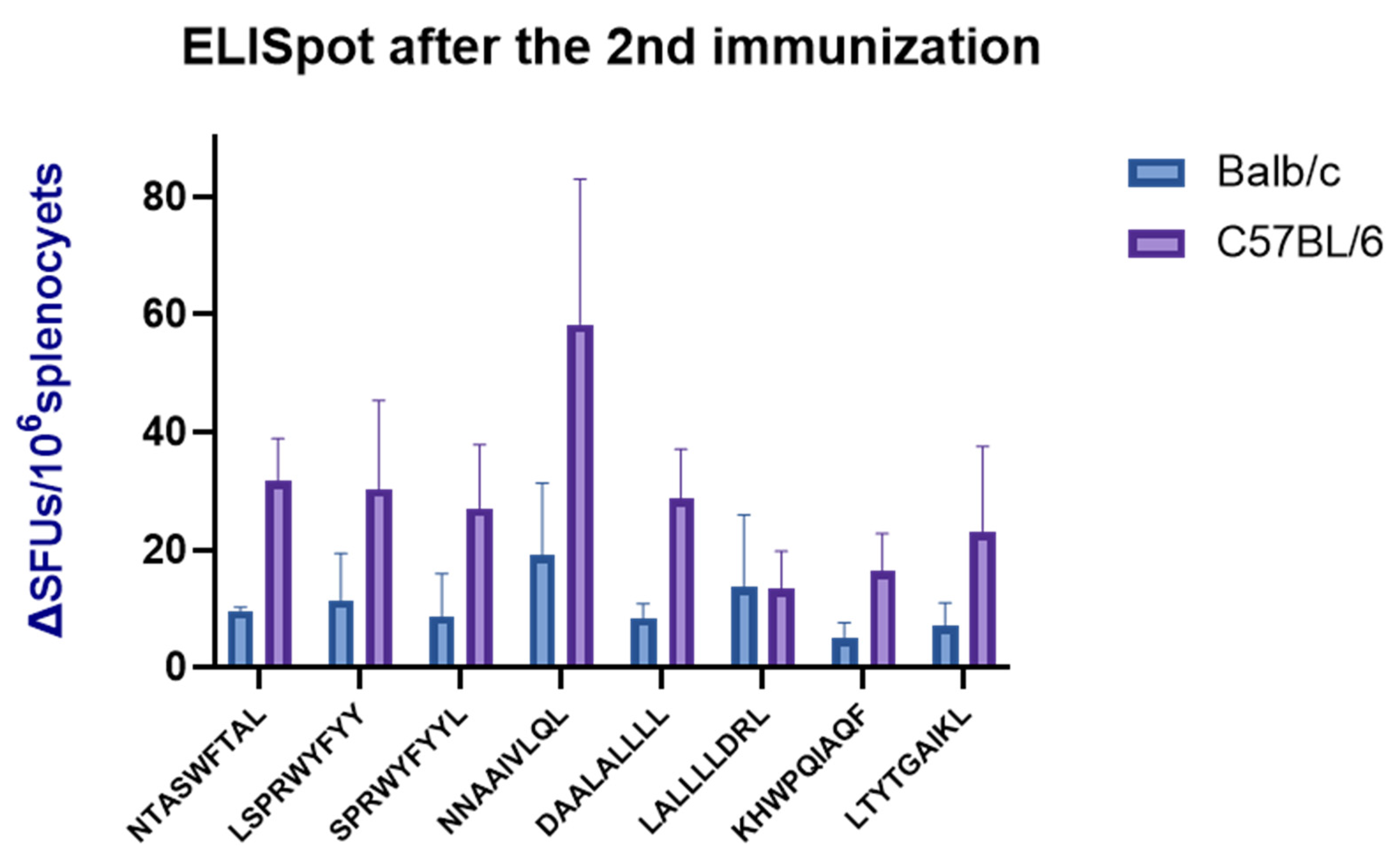
3.9. ELISpot Validation of the SARS-CoV-2 NP Epitopes
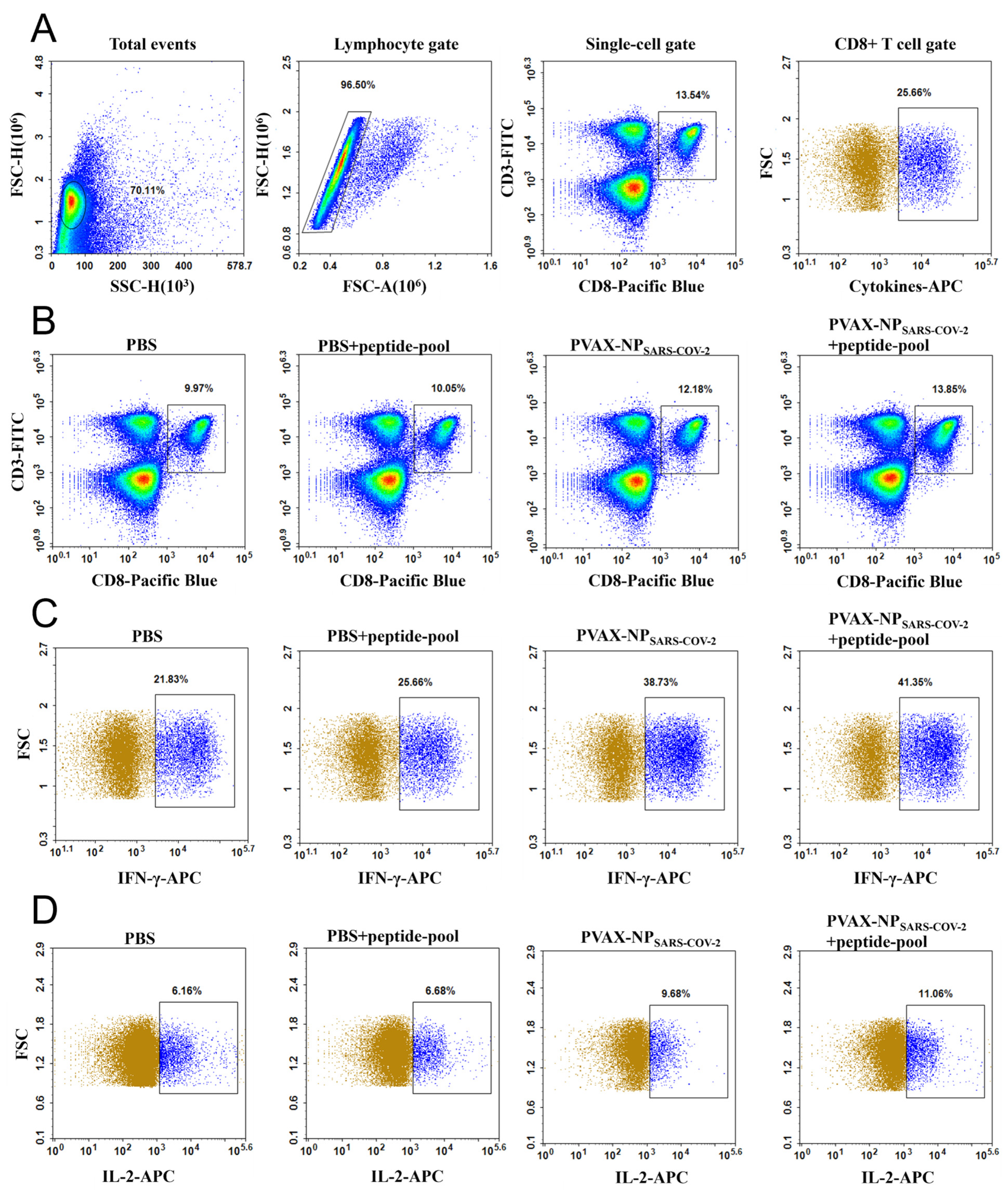
3.10. SARS-CoV-2 NP Epitopes Enhanced the Secretion of IFN-γ and IL-2
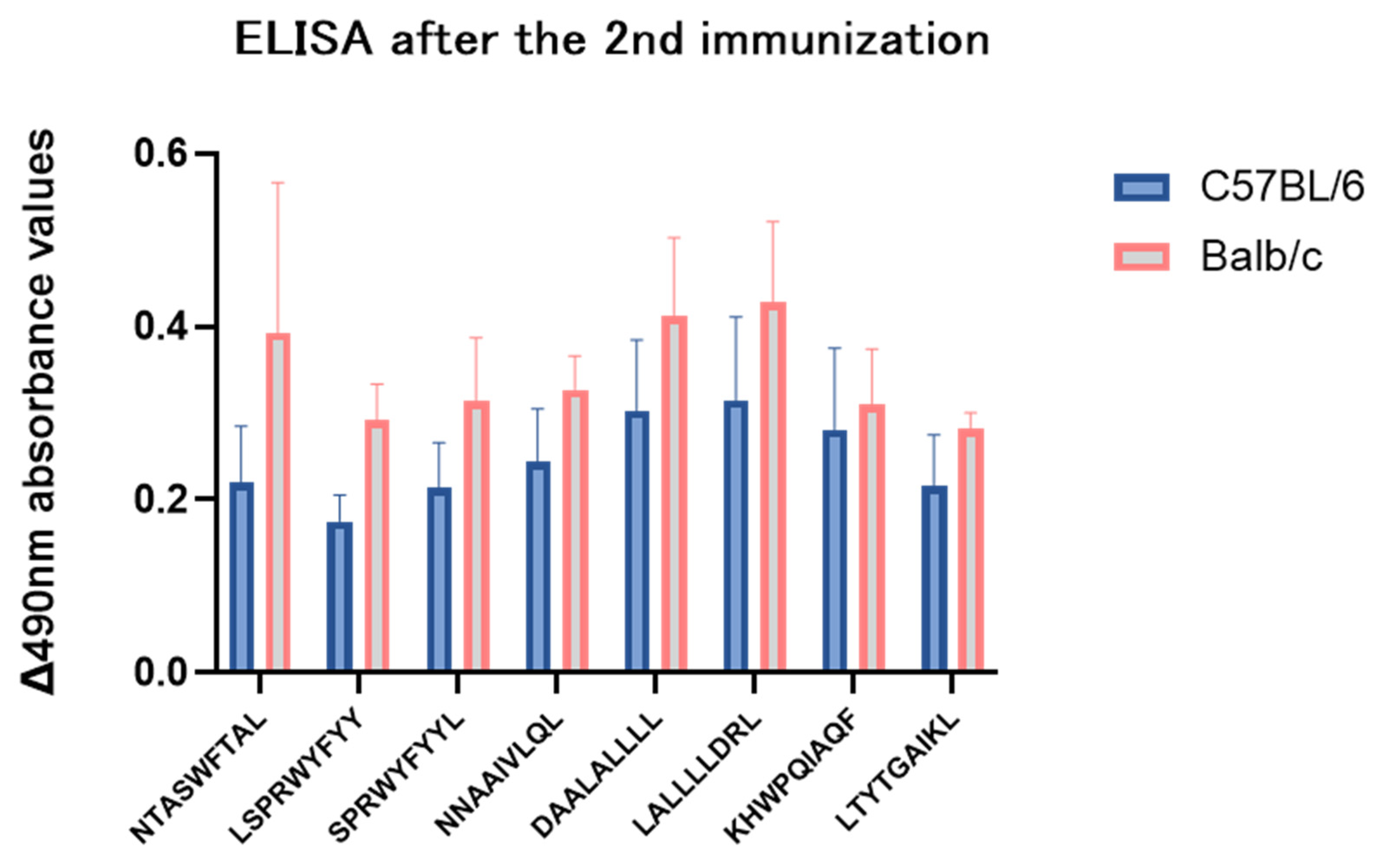
3.11. The Preferred Epitopes Induced Humoral Immune Responses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Bio-Medica Atenei Parm. 2020, 91, 157–160. [Google Scholar] [CrossRef]
- Ravi, V.; Saxena, S.; Panda, P.S. Basic virology of SARS-CoV 2. Indian J. Med. Microbiol. 2022, 40, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Xu, J.; Zhang, L.; Shu, T.; Wu, D.; Huang, M.; Ren, Y.; Li, X.; Geng, Q.; Xu, Y.; et al. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci. China Life Sci. 2020, 63, 1413–1416. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Ye, F.; Cheng, M.L.; Feng, Y.; Deng, Y.Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52, 971–977.e973. [Google Scholar] [CrossRef]
- Xiang, F.; Wang, X.; He, X.; Peng, Z.; Yang, B.; Zhang, J.; Zhou, Q.; Ye, H.; Ma, Y.; Li, H.; et al. Antibody Detection and Dynamic Characteristics in Patients With Coronavirus Disease 2019. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 1930–1934. [Google Scholar] [CrossRef]
- Pratheek, B.M.; Saha, S.; Maiti, P.K.; Chattopadhyay, S.; Chattopadhyay, S. Immune regulation and evasion of Mammalian host cell immunity during viral infection. Indian J. Virol. Off. Organ Indian Virol. Soc. 2013, 24, 1–15. [Google Scholar] [CrossRef]
- Sieker, F.; May, A.; Zacharias, M. Predicting affinity and specificity of antigenic peptide binding to major histocompatibility class I molecules. Curr. Protein Pept. Sci. 2009, 10, 286–296. [Google Scholar] [CrossRef]
- Blum, J.S.; Wearsch, P.A.; Cresswell, P. Pathways of antigen processing. Annu. Rev. Immunol. 2013, 31, 443–473. [Google Scholar] [CrossRef]
- Pratheek, B.M.; Suryawanshi, A.R.; Chattopadhyay, S.; Chattopadhyay, S. In silico analysis of MHC-I restricted epitopes of Chikungunya virus proteins: Implication in understanding anti-CHIKV CD8(+) T cell response and advancement of epitope based immunotherapy for CHIKV infection. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 31, 118–126. [Google Scholar] [CrossRef]
- Khan, M.A.; Hossain, M.U.; Rakib-Uz-Zaman, S.M.; Morshed, M.N. Epitope-based peptide vaccine design and target site depiction against Ebola viruses: An immunoinformatics study. Scand. J. Immunol. 2015, 82, 25–34. [Google Scholar] [CrossRef]
- Rappuoli, R.; Bottomley, M.J.; D’Oro, U.; Finco, O.; De Gregorio, E. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J. Exp. Med. 2016, 213, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Padilla, J.; Lafuente, E.M.; Reche, P.A. Computer-Aided Design of an Epitope-Based Vaccine against Epstein-Barr Virus. J. Immunol. Res. 2017, 2017, 9363750. [Google Scholar] [CrossRef] [PubMed]
- Francini, G.; Scardino, A.; Kosmatopoulos, K.; Lemonnier, F.A.; Campoccia, G.; Sabatino, M.; Pozzessere, D.; Petrioli, R.; Lozzi, L.; Neri, P.; et al. High-affinity HLA-A(*)02.01 peptides from parathyroid hormone-related protein generate in vitro and in vivo antitumor CTL response without autoimmune side effects. J. Immunol. 2002, 169, 4840–4849. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.; Kaushal, N.; Manish, M.; Subbarao, N.; Shakirova, V.; Martynova, E.; Liu, R.; Hamza, S.; Rizvanov, A.A.; Khaiboullina, S.F.; et al. Identification of conserved immunogenic peptides of SARS-CoV-2 nucleocapsid protein. J. Biomol. Struct. Dyn. 2023, 1–17. [Google Scholar] [CrossRef]
- Jin, X.; Ding, Y.; Sun, S.; Wang, X.; Zhou, Z.; Liu, X.; Li, M.; Chen, X.; Shen, A.; Wu, Y.; et al. Screening HLA-A-restricted T cell epitopes of SARS-CoV-2 and the induction of CD8(+) T cell responses in HLA-A transgenic mice. Cell. Mol. Immunol. 2021, 18, 2588–2608. [Google Scholar] [CrossRef]
- Hu, C.; Shen, M.; Han, X.; Chen, Q.; Li, L.; Chen, S.; Zhang, J.; Gao, F.; Wang, W.; Wang, Y.; et al. Identification of cross-reactive CD8(+) T cell receptors with high functional avidity to a SARS-CoV-2 immunodominant epitope and its natural mutant variants. Genes Dis. 2022, 9, 216–229. [Google Scholar] [CrossRef]
- Rigo, M.M.; Fasoulis, R.; Conev, A.; Hall-Swan, S.; Antunes, D.A.; Kavraki, L.E. SARS-Arena: Sequence and Structure-Guided Selection of Conserved Peptides from SARS-related Coronaviruses for Novel Vaccine Development. Front. Immunol. 2022, 13, 931155. [Google Scholar] [CrossRef]
- Kumar, J.; Qureshi, R.; Sagurthi, S.R.; Qureshi, I.A. Designing of Nucleocapsid Protein Based Novel Multi-epitope Vaccine Against SARS-COV-2 Using Immunoinformatics Approach. Int. J. Pept. Res. Ther. 2021, 27, 941–956. [Google Scholar] [CrossRef]
- Rakib, A.; Sami, S.A.; Islam, M.A.; Ahmed, S.; Faiz, F.B.; Khanam, B.H.; Marma, K.K.S.; Rahman, M.; Uddin, M.M.N.; Nainu, F.; et al. Epitope-Based Immunoinformatics Approach on Nucleocapsid Protein of Severe Acute Respiratory Syndrome-Coronavirus-2. Molecules 2020, 25, 5088. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, D.; Li, M.; Liu, M.; Yao, S.; Zhan, J.; Liu, W.J.; Gao, G.F. A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8(+) T Cell Epitope from SARS-CoV-2—Hubei Province, China, 2021. China CDC Wkly. 2022, 4, 83–87. [Google Scholar] [CrossRef]
- Kim, Y.; Ponomarenko, J.; Zhu, Z.; Tamang, D.; Wang, P.; Greenbaum, J.; Lundegaard, C.; Sette, A.; Lund, O.; Bourne, P.E.; et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012, 40, W525–W530. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Sidney, J.; Pinilla, C.; Sette, A.; Peters, B. Derivation of an amino acid similarity matrix for peptide: MHC binding and its application as a Bayesian prior. BMC Bioinform. 2009, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Rammensee, H.; Bachmann, J.; Emmerich, N.P.; Bachor, O.A.; Stevanović, S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics 1999, 50, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Reche, P.A.; Glutting, J.P.; Reinherz, E.L. Prediction of MHC class I binding peptides using profile motifs. Hum. Immunol. 2002, 63, 701–709. [Google Scholar] [CrossRef]
- Reche, P.A.; Glutting, J.P.; Zhang, H.; Reinherz, E.L. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics 2004, 56, 405–419. [Google Scholar] [CrossRef]
- Feltkamp, M.C.; Vierboom, M.P.; Kast, W.M.; Melief, C.J. Efficient MHC class I-peptide binding is required but does not ensure MHC class I-restricted immunogenicity. Mol. Immunol. 1994, 31, 1391–1401. [Google Scholar] [CrossRef]
- Soria-Guerra, R.E.; Nieto-Gomez, R.; Govea-Alonso, D.O.; Rosales-Mendoza, S. An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. J. Biomed. Inform. 2015, 53, 405–414. [Google Scholar] [CrossRef]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.Y. HPEPDOCK: A web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.; Biswal, S.; Bhatt, R.; Tiwary, B.N. Computational perspectives revealed prospective vaccine candidates from five structural proteins of novel SARS corona virus 2019 (SARS-CoV-2). PeerJ 2020, 8, e9855. [Google Scholar] [CrossRef] [PubMed]
- Farhani, I.; Yamchi, A.; Madanchi, H.; Khazaei, V.; Behrouzikhah, M.; Abbasi, H.; Salehi, M.; Moradi, N.; Sanami, S. Designing a Multi-epitope Vaccine against the SARS-CoV-2 Variant based on an Immunoinformatics Approach. Curr. Comput.-Aided Drug Des. 2024, 20, 274–290. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, X.; Zhou, H.; Zhu, H.; Jiang, S.; Wang, P. Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses. Nat. Rev. Immunol. 2023, 23, 189–199. [Google Scholar] [CrossRef]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, P.; Zhao, Y.; Zhuang, Z.; Wang, Z.; Song, R.; Zhang, J.; Liu, C.; Gao, Q.; Xu, Q.; et al. Single-Cell Sequencing of Peripheral Mononuclear Cells Reveals Distinct Immune Response Landscapes of COVID-19 and Influenza Patients. Immunity 2020, 53, 685–696.e683. [Google Scholar] [CrossRef]
- Gao, F.; Mallajosyula, V.; Arunachalam, P.S.; van der Ploeg, K.; Manohar, M.; Röltgen, K.; Yang, F.; Wirz, O.; Hoh, R.; Haraguchi, E.; et al. Spheromers reveal robust T cell responses to the Pfizer/BioNTech vaccine and attenuated peripheral CD8(+) T cell responses post SARS-CoV-2 infection. Immunity 2023, 56, 864–878.e864. [Google Scholar] [CrossRef]
- Li, Z.; Xiang, T.; Liang, B.; Deng, H.; Wang, H.; Feng, X.; Quan, X.; Wang, X.; Li, S.; Lu, S.; et al. Characterization of SARS-CoV-2-Specific Humoral and Cellular Immune Responses Induced by Inactivated COVID-19 Vaccines in a Real-World Setting. Front. Immunol. 2021, 12, 802858. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Qin, Y.; Tu, K.; Teng, Q.; Feng, D.; Zhao, Y.; Zhang, G. Identification of Novel T-Cell Epitopes on Infectious Bronchitis Virus N Protein and Development of a Multi-epitope Vaccine. J. Virol. 2021, 95, e0066721. [Google Scholar] [CrossRef]
- Rcheulishvili, N.; Mao, J.; Papukashvili, D.; Feng, S.; Liu, C.; Yang, X.; Lin, J.; He, Y.; Wang, P.G. Development of a Multi-Epitope Universal mRNA Vaccine Candidate for Monkeypox, Smallpox, and Vaccinia Viruses: Design and In Silico Analyses. Viruses 2023, 15, 1120. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Zhu, F.; Pan, P.; Wu, A.; Li, C. Development of multi-epitope vaccines against the monkeypox virus based on envelope proteins using immunoinformatics approaches. Front. Immunol. 2023, 14, 1112816. [Google Scholar] [CrossRef] [PubMed]
- Dastar, S.; Gharesouran, J.; Mortazavi, D.; Hosseinzadeh, H.; Kian, S.J.; Taheri, M.; Ghafouri-Fard, S.; Jamali, E.; Rezazadeh, M. COVID-19 pandemic: Insights into genetic susceptibility to SARS-CoV-2 and host genes implications on virus spread, disease severity and outcomes. Hum. Antibodies 2022, 30, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Godri Pollitt, K.J.; Peccia, J.; Ko, A.I.; Kaminski, N.; Dela Cruz, C.S.; Nebert, D.W.; Reichardt, J.K.V.; Thompson, D.C.; Vasiliou, V. COVID-19 vulnerability: The potential impact of genetic susceptibility and airborne transmission. Hum. Genom. 2020, 14, 17. [Google Scholar] [CrossRef]
- Fischer, J.C.; Schmidt, A.G.; Bölke, E.; Uhrberg, M.; Keitel, V.; Feldt, T.; Jensen, B.; Häussinger, D.; Adams, O.; Schneider, E.M.; et al. Association of HLA genotypes, AB0 blood type and chemokine receptor 5 mutant CD195 with the clinical course of COVID-19. Eur. J. Med. Res. 2021, 26, 107. [Google Scholar] [CrossRef]
- Balas, A.; Moreno-Hidalgo, M.; de la Calle-Prieto, F.; Vicario, J.L.; Arsuaga, M.; Trigo, E.; de Miguel-Buckley, R.; Bellón, T.; Díaz-Menéndez, M. Coronavirus-19 disease risk and protective factors associated with HLA/KIR polymorphisms in Ecuadorian patients residing in Madrid. Hum. Immunol. 2023, 84, 571–577. [Google Scholar] [CrossRef]
- Jiang, D.B.; Zhang, J.P.; Cheng, L.F.; Zhang, G.W.; Li, Y.; Li, Z.C.; Lu, Z.H.; Zhang, Z.X.; Lu, Y.C.; Zheng, L.H.; et al. Hantavirus Gc induces long-term immune protection via LAMP-targeting DNA vaccine strategy. Antivir. Res. 2018, 150, 174–182. [Google Scholar] [CrossRef]
- Santos-López, G.; Cortés-Hernández, P.; Vallejo-Ruiz, V.; Reyes-Leyva, J. SARS-CoV-2: Basic concepts, origin and treatment advances. Gac. Medica Mex. 2021, 157, 84–89. [Google Scholar] [CrossRef]
- Singh, D.; Yi, S.V. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547. [Google Scholar] [CrossRef]
- Wolf, J.M.; Wolf, L.M.; Bello, G.L.; Maccari, J.G.; Nasi, L.A. Molecular evolution of SARS-CoV-2 from December 2019 to August 2022. J. Med. Virol. 2023, 95, e28366. [Google Scholar] [CrossRef]
- Chavda, V.P.; Patel, A.B.; Vaghasiya, D.D. SARS-CoV-2 variants and vulnerability at the global level. J. Med. Virol. 2022, 94, 2986–3005. [Google Scholar] [CrossRef] [PubMed]
- McLean, G.; Kamil, J.; Lee, B.; Moore, P.; Schulz, T.F.; Muik, A.; Sahin, U.; Türeci, Ö.; Pather, S. The Impact of Evolving SARS-CoV-2 Mutations and Variants on COVID-19 Vaccines. mBio 2022, 13, e0297921. [Google Scholar] [CrossRef] [PubMed]
- Jimenez Ruiz, J.A.; Lopez Ramirez, C.; Lopez-Campos, J.L. A Comparative Study between Spanish and British SARS-CoV-2 Variants. Curr. Issues Mol. Biol. 2021, 43, 2036–2047. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Strych, U.; Hotez, P.J.; Bottazzi, M.E. The SARS-CoV-2 Vaccine Pipeline: An Overview. Curr. Trop. Med. Rep. 2020, 7, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Wnuk, K.; Sudol, J.; Spilman, P.; Soon-Shiong, P. Peptidome Surveillance Across Evolving SARS-CoV-2 Lineages Reveals HLA Binding Conservation in Nucleocapsid Among Variants With Most Potential for T-Cell Epitope Loss in Spike. Front. Immunol. 2022, 13, 918928. [Google Scholar] [CrossRef]
- Rak, A.; Isakova-Sivak, I.; Rudenko, L. Overview of Nucleocapsid-Targeting Vaccines against COVID-19. Vaccines 2023, 11, 1810. [Google Scholar] [CrossRef]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020, 94, e00510-20. [Google Scholar] [CrossRef]
- Lineburg, K.E.; Grant, E.J.; Swaminathan, S.; Chatzileontiadou, D.S.M.; Szeto, C.; Sloane, H.; Panikkar, A.; Raju, J.; Crooks, P.; Rehan, S.; et al. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity 2021, 54, 1055–1065.e1055. [Google Scholar] [CrossRef]
- Moballegh Naseri, M.; Moballegh Naseri, M.; Maurya, V.K.; Shams, S.; Pitaloka, D.A.E. Epitope-based vaccine design against the membrane and nucleocapsid proteins of SARS-CoV-2. Dent. Med. Probl. 2023, 60, 489–495. [Google Scholar] [CrossRef]
- Ezzemani, W.; Kettani, A.; Sappati, S.; Kondaka, K.; El Ossmani, H.; Tsukiyama-Kohara, K.; Altawalah, H.; Saile, R.; Kohara, M.; Benjelloun, S.; et al. Reverse vaccinology-based prediction of a multi-epitope SARS-CoV-2 vaccine and its tailoring to new coronavirus variants. J. Biomol. Struct. Dyn. 2023, 41, 4917–4938. [Google Scholar] [CrossRef]
- Khan, M.S.; Khan, I.M.; Ahmad, S.U.; Rahman, I.; Khan, M.Z.; Khan, M.S.Z.; Abbas, Z.; Noreen, S.; Liu, Y. Immunoinformatics design of B and T-cell epitope-based SARS-CoV-2 peptide vaccination. Front. Immunol. 2022, 13, 1001430. [Google Scholar] [CrossRef]
| MHC-IHaplotypes | Prediction Tools | NP Epitopes | NP (Short-Listed) |
|---|---|---|---|
| HLA-A1 | IEDB | 38 | 25 |
| NetMHCpan | 25 | ||
| Rankpep | 9 | ||
| SMMPMBEC | 11 | ||
| SYFPEITHI | 0 | ||
| HLA-A2 | IEDB | 25 | 21 |
| NetMHCpan | 23 | ||
| Rankpep | 27 | ||
| SMMPMBEC | 6 | ||
| SYFPEITHI | 8 | ||
| HLA-A3 | IEDB | 57 | 48 |
| NetMHCpan | 49 | ||
| Rankpep | 28 | ||
| SMMPMBEC | 25 | ||
| SYFPEITHI | 16 | ||
| HLA-A24 | IEDB | 18 | 16 |
| NetMHCpan | 14 | ||
| Rankpep | 9 | ||
| SMMPMBEC | 7 | ||
| SYFPEITHI | 8 | ||
| HLA-3201 | IEDB | 14 | 19 |
| NetMHCpan | 19 | ||
| Rankpep | 0 | ||
| SMMPMBEC | 5 | ||
| SYFPEITHI | 0 | ||
| HLA-B7 | IEDB | 42 | 22 |
| NetMHCpan | 33 | ||
| Rankpep | 15 | ||
| SMMPMBEC | 19 | ||
| SYFPEITHI | 32 | ||
| HLA-B8 | IEDB | 18 | 10 |
| NetMHCpan | 13 | ||
| Rankpep | 0 | ||
| SMMPMBEC | 5 | ||
| SYFPEITHI | 0 | ||
| HLA-B15 | IEDB | 23 | 17 |
| NetMHCpan | 15 | ||
| Rankpep | 0 | ||
| SMMPMBEC | 4 | ||
| SYFPEITHI | 8 | ||
| HLA-B44 | IEDB | 23 | 15 |
| NetMHCpan | 15 | ||
| Rankpep | 9 | ||
| SMMPMBEC | 2 | ||
| SYFPEITHI | 16 | ||
| HLA-B58 | IEDB | 17 | 11 |
| NetMHCpan | 11 | ||
| Rankpep | 15 | ||
| SMMPMBEC | 6 | ||
| SYFPEITHI | 8 | ||
| HLA-B46 | IEDB | 21 | 15 |
| NetMHCpan | 15 | ||
| Rankpep | 0 | ||
| SMMPMBEC | 4 | ||
| SYFPEITHI | 0 | ||
| HLA-B62 | IEDB | 0 | |
| NetMHCpan | 23 | ||
| Rankpep | 0 | ||
| SMMPMBEC | 0 | ||
| SYFPEITHI | 0 | ||
| HLA-C0401 | IEDB | 22 | 19 |
| NetMHCpan | 16 | ||
| Rankpep | 0 | ||
| SMMPMBEC | 5 | ||
| SYFPEITHI | 0 |
| MHC-IHaplotypes | Prediction Tools | NP Epitopes | NP (Short-Listed) |
|---|---|---|---|
| H-2 Db | IEDB | 15 | 9 |
| NetMHCpan | 10 | ||
| Rankpep | 9 | ||
| SMMPMBEC | 5 | ||
| SYFPEITHI | 8 | ||
| H-2 Dd | IEDB | 31 | 19 |
| NetMHCpan | 15 | ||
| Rankpep | 9 | ||
| SMMPMBEC | 3 | ||
| SYFPEITHI | 0 | ||
| H-2 Kb | IEDB | 19 | 13 |
| NetMHCpan | 11 | ||
| Rankpep | 9 | ||
| SMMPMBEC | 6 | ||
| SYFPEITHI | 0 | ||
| H-2 Kd | IEDB | 20 | 7 |
| NetMHCpan | 13 | ||
| Rankpep | 9 | ||
| SMMPMBEC | 4 | ||
| SYFPEITHI | 8 | ||
| H-2 Kk | IEDB | 11 | 9 |
| NetMHCpan | 7 | ||
| Rankpep | 9 | ||
| SMMPMBEC | 0 | ||
| SYFPEITHI | 8 | ||
| H-2 Ld | IEDB | 20 | 7 |
| NetMHCpan | 16 | ||
| Rankpep | 9 | ||
| SMMPMBEC | 6 | ||
| SYFPEITHI | 8 |
| MHC-I Haplotypes | Interspecies− Intraspecies− | Interspecies− Intraspecies+ | Interspecies+ Intraspecies− | Interspecies+ Intraspecies+ |
|---|---|---|---|---|
| H-2 Db | 9 | 0 | 0 | 0 |
| H-2 Dd | 14 | 4 | 0 | 1 |
| H-2 Kb | 9 | 2 | 0 | 2 |
| H-2 Kd | 7 | 0 | 0 | 0 |
| H-2 Kk | 8 | 1 | 0 | 0 |
| H-2 Ld | 5 | 1 | 0 | 1 |
| HLA-A1 | 21 | 3 | 0 | 1 |
| HLA-A2 | 20 | 0 | 0 | 0 |
| HLA-A3 | 45 | 2 | 0 | 0 |
| HLA-A24 | 13 | 3 | 0 | 0 |
| HLA-3201 | 10 | 4 | 0 | 0 |
| HLA-B7 | 20 | 1 | 0 | 1 |
| HLA-B8 | 4 | 0 | 0 | 0 |
| HLA-B15 | 8 | 0 | 0 | 0 |
| HLA-B44 | 11 | 4 | 0 | 0 |
| HLA-B58 | 7 | 3 | 0 | 1 |
| HLA-B4601 | 13 | 1 | 0 | 1 |
| HLA-C0401 | 17 | 1 | 0 | 0 |
| Amino Acid | NC_045512.2 | Variants | Dominant in | Dominant in | HLA-Ⅰ |
|---|---|---|---|---|---|
| Number | Variants | NC_045512.2 | Genotype | ||
| YES | NO | HLA-A2402 | |||
| 151–159 | TWLTYTGAI | TWLTYHGAI | YES | NO | HLA-A2301 |
| YES | NO | HLA-A2601 | |||
| 153–161 | LTYTGAIKL | LTYHGAIKL | NO | YES | HLA-B5101 |
| Pepitides | MERCI Score | BLAST Score | Prediction | ML Score | Hybrid Score | Prediction |
|---|---|---|---|---|---|---|
| NTASWFTAL | 0.33 | 0 | Non-Allergen | 0.56 | 0.56 | Non-Toxin |
| LSPRWYFYY | 0.29 | 0 | Non-Allergen | 0.55 | 0.55 | Non-Toxin |
| SPRWYFYYL | 0.29 | 0 | Non-Allergen | 0.55 | 0.55 | Non-Toxin |
| NNAAIVLQL | 0.29 | 0.5 | Allergen | 0.71 | 0.71 | Toxin |
| DAALALLLL | 0.41 | 0 | Allergen | 0.73 | 0.73 | Toxin |
| LALLLLDRL | 0.36 | 0 | Non-Allergen | 0.76 | 0.76 | Toxin |
| KHWPQIAQF | 0.4 | 0 | Non-Allergen | 0.62 | 0.62 | Non-Toxin |
| LTYTGAIKL | 0.32 | 0 | Non-Allergen | 0.7 | 0.7 | Non-Toxin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, D.; Ma, Z.; Zhang, J.; Sun, Y.; Bai, T.; Liu, R.; Wang, Y.; Guan, L.; Fu, S.; Sun, Y.; et al. Immunoreactivity Analysis of MHC-I Epitopes Derived from the Nucleocapsid Protein of SARS-CoV-2 via Computation and Vaccination. Vaccines 2024, 12, 1214. https://doi.org/10.3390/vaccines12111214
Jiang D, Ma Z, Zhang J, Sun Y, Bai T, Liu R, Wang Y, Guan L, Fu S, Sun Y, et al. Immunoreactivity Analysis of MHC-I Epitopes Derived from the Nucleocapsid Protein of SARS-CoV-2 via Computation and Vaccination. Vaccines. 2024; 12(11):1214. https://doi.org/10.3390/vaccines12111214
Chicago/Turabian StyleJiang, Dongbo, Zilu Ma, Junqi Zhang, Yubo Sun, Tianyuan Bai, Ruibo Liu, Yongkai Wang, Liang Guan, Shuaishuai Fu, Yuanjie Sun, and et al. 2024. "Immunoreactivity Analysis of MHC-I Epitopes Derived from the Nucleocapsid Protein of SARS-CoV-2 via Computation and Vaccination" Vaccines 12, no. 11: 1214. https://doi.org/10.3390/vaccines12111214
APA StyleJiang, D., Ma, Z., Zhang, J., Sun, Y., Bai, T., Liu, R., Wang, Y., Guan, L., Fu, S., Sun, Y., Li, Y., Zhou, B., Yang, Y., Yang, S., Chang, Y., Sun, B., & Yang, K. (2024). Immunoreactivity Analysis of MHC-I Epitopes Derived from the Nucleocapsid Protein of SARS-CoV-2 via Computation and Vaccination. Vaccines, 12(11), 1214. https://doi.org/10.3390/vaccines12111214







