Design and In Silico Validation of a Novel MZF-1-Based Multi-Epitope Vaccine to Combat Metastatic Triple Negative Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Retrieval of MZF-1 Protein
2.2. Prediction and Prioritization of Immunogenic Epitopes
2.3. Population Coverage Analysis and Multi-Epitope Vaccine Formulation
2.4. Evaluation of Physicochemical Characteristics of the Vaccine
2.5. Modelling and Validation of Secondary and Tertiary Structure
2.6. Analysis of Discontinuous B Cell Epitopes
2.7. Molecular Docking Analysis
2.8. Molecular Dynamics Simulation Analysis
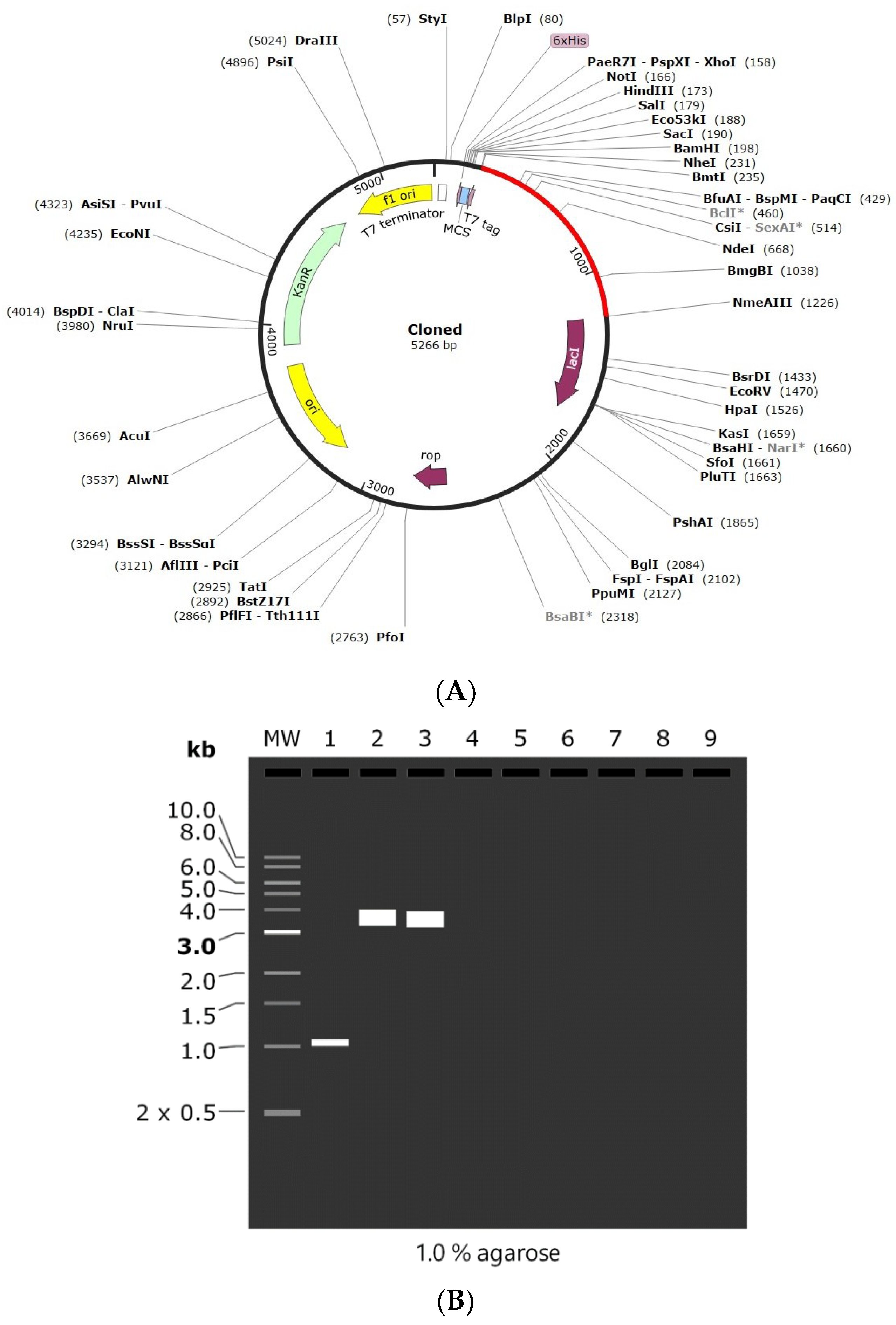
2.9. Codon Adaptation and In Silico Cloning
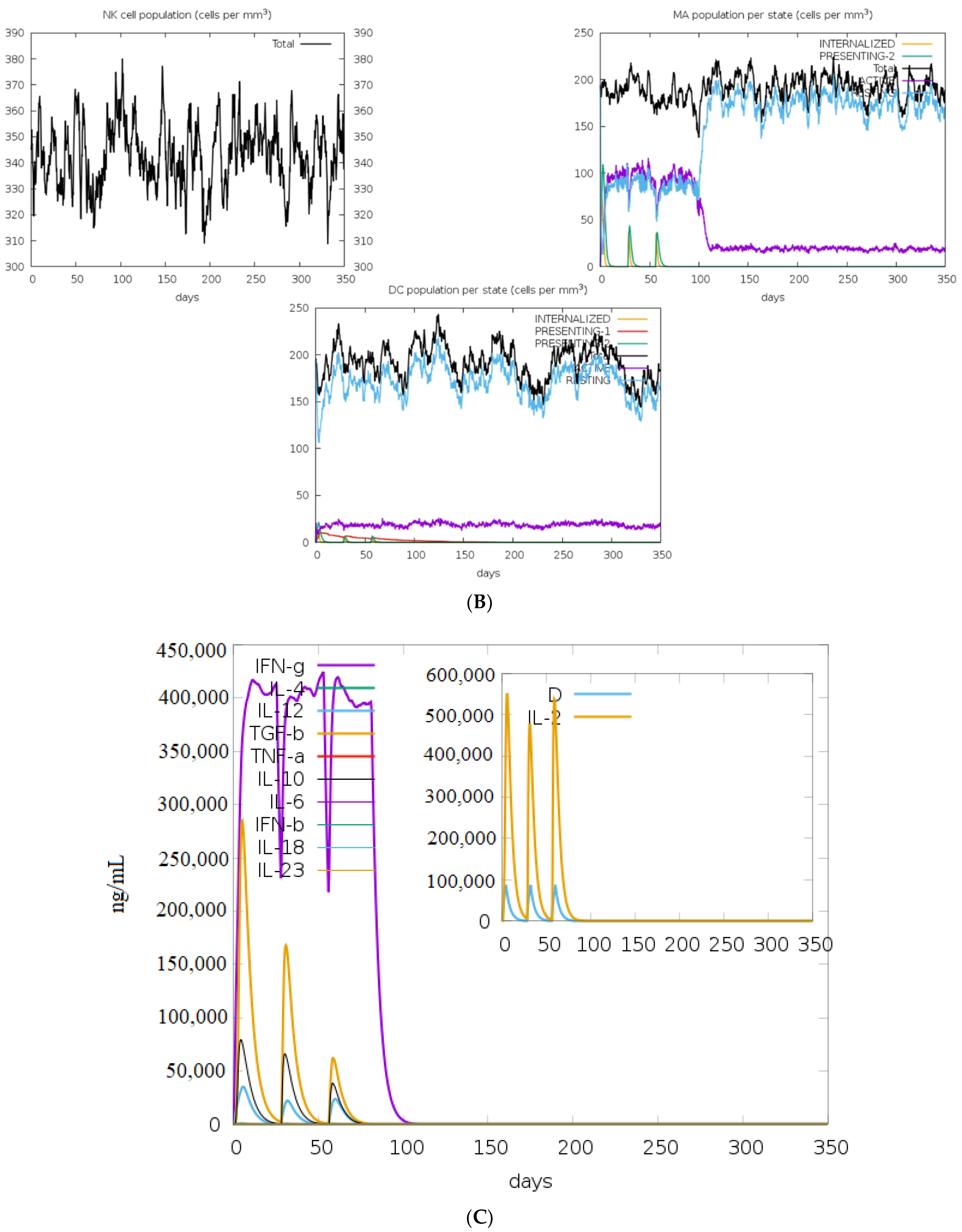
2.10. In Silico Immune Simulation Analysis
3. Results
3.1. Prediction and Prioritization Phase of Potential Peptides
3.2. Multi-Epitope Vaccine Engineering
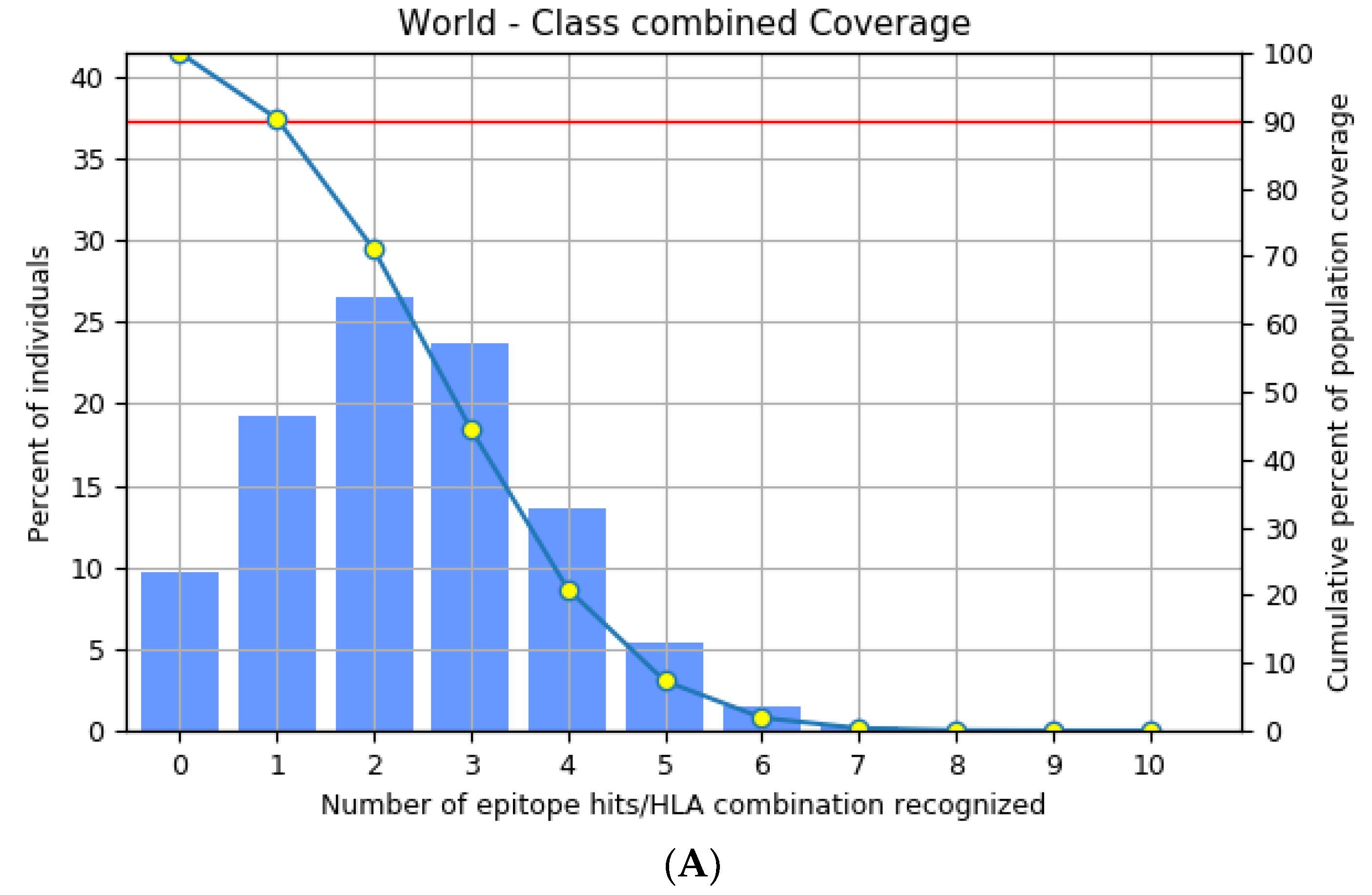
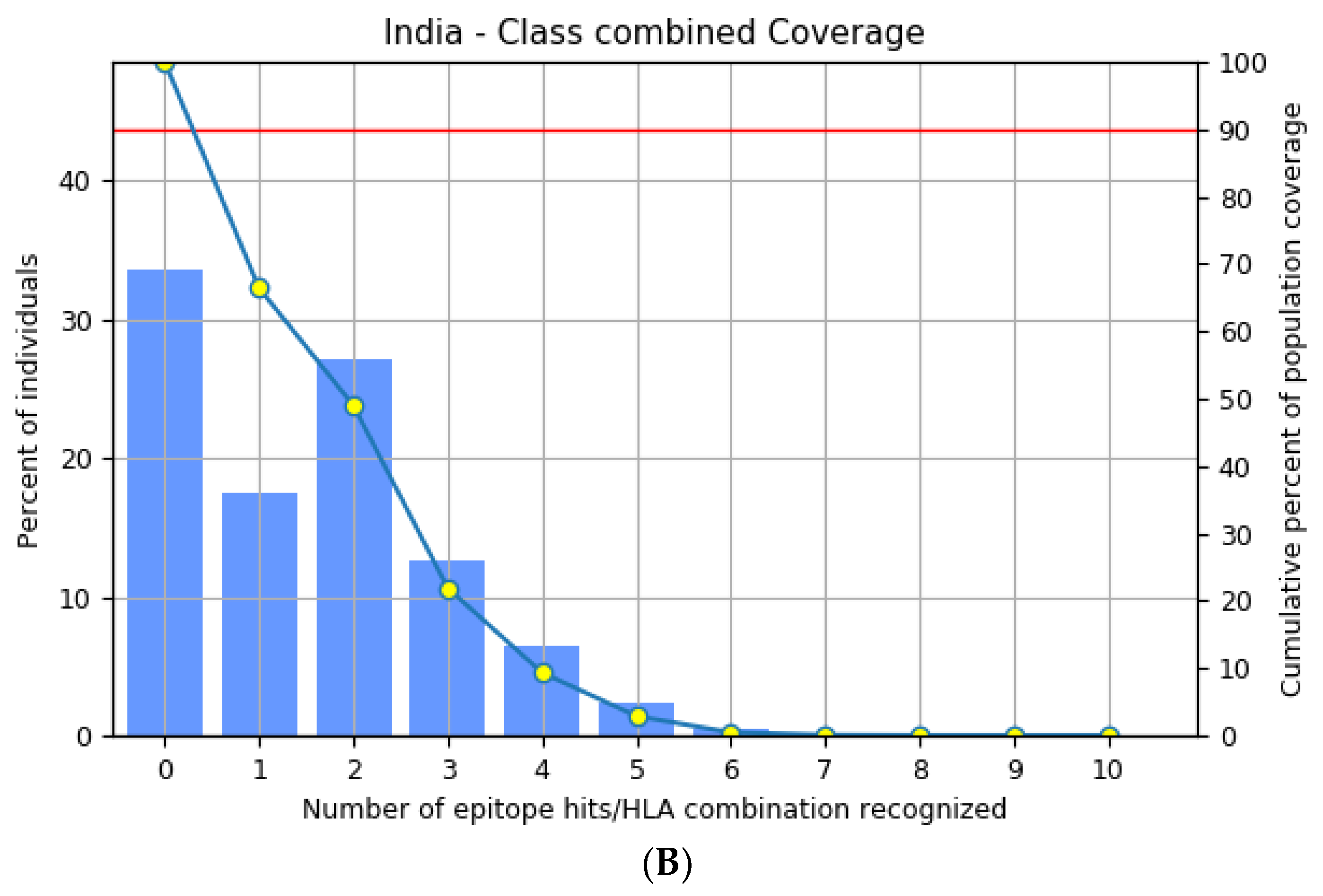
3.3. Population Distribution Analysis of the Estimated Epitopes
3.4. Assessment of Immunogenic and Physicochemical Properties of the Vaccine
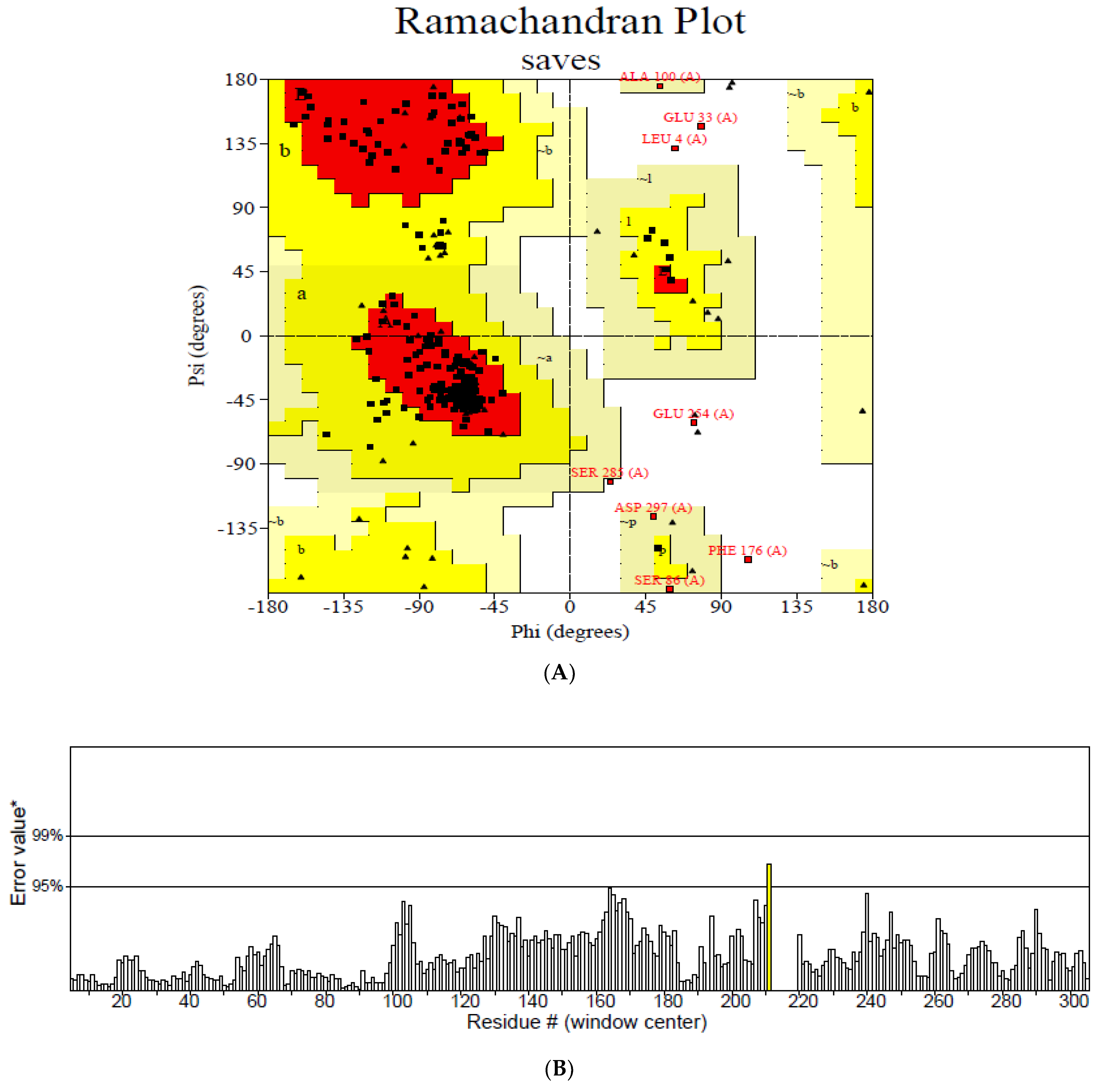
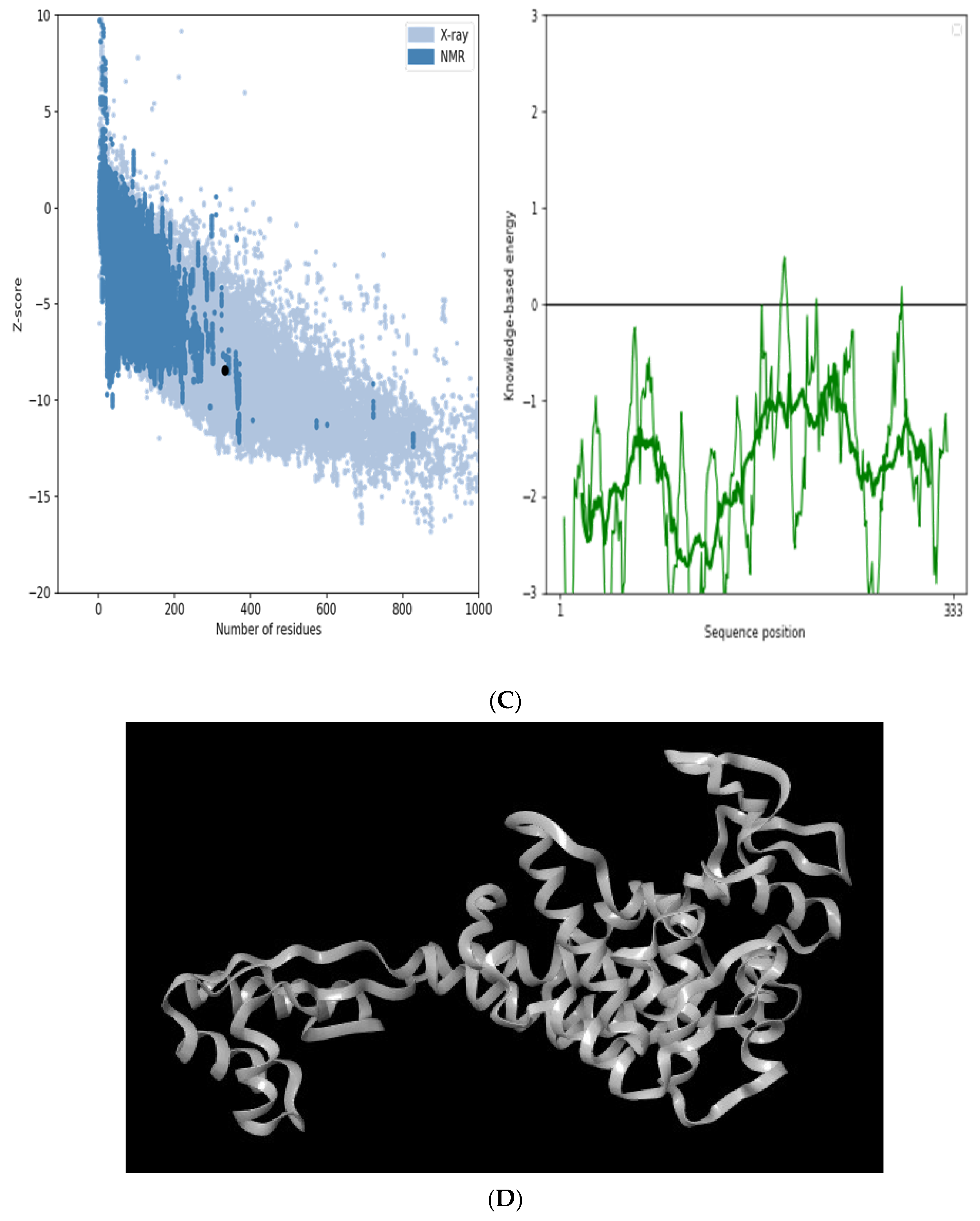
3.5. Modelling and Validation of the Secondary and Tertiary Structure
3.6. Identification of Discontinuous B Cell Epitopes
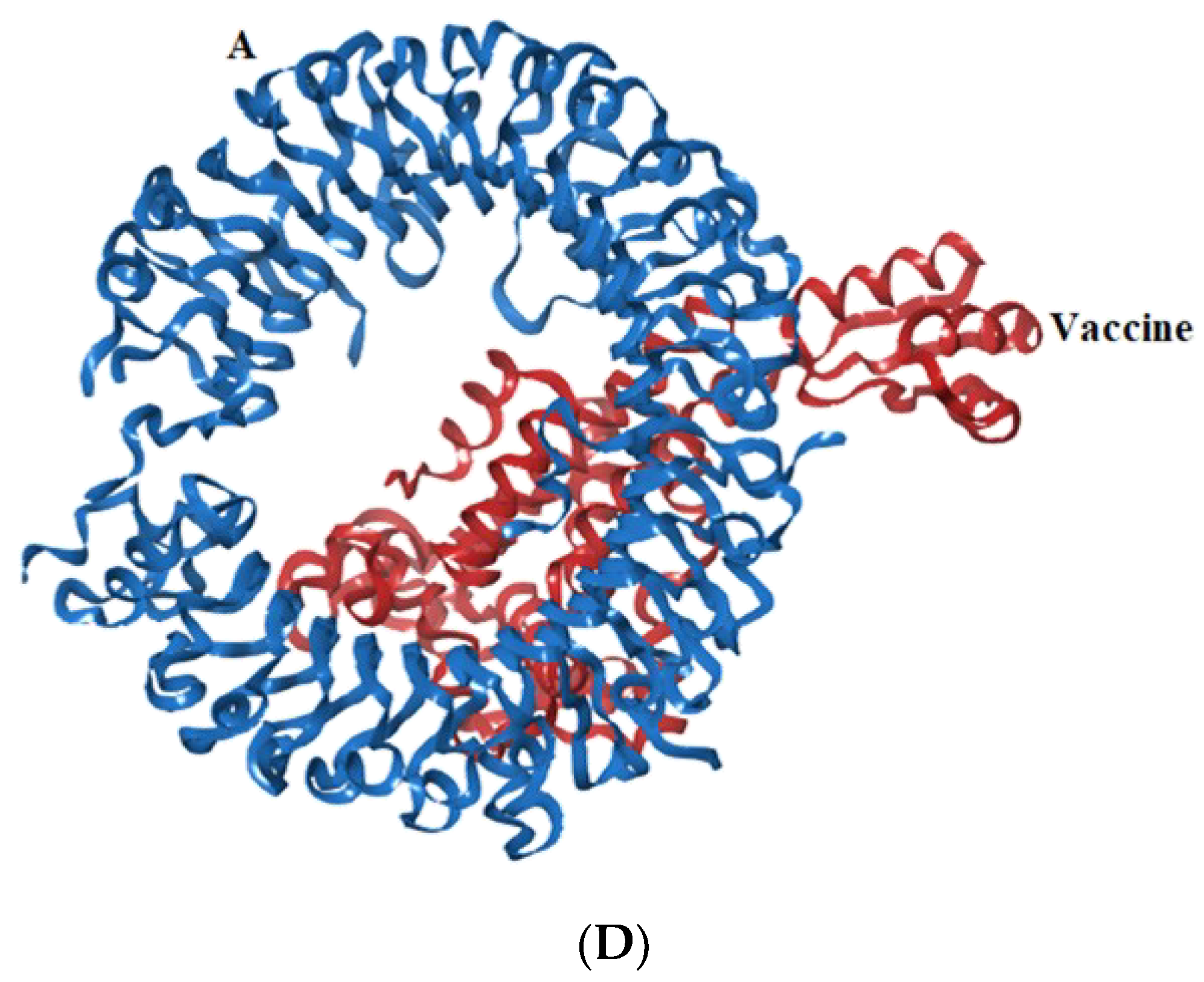
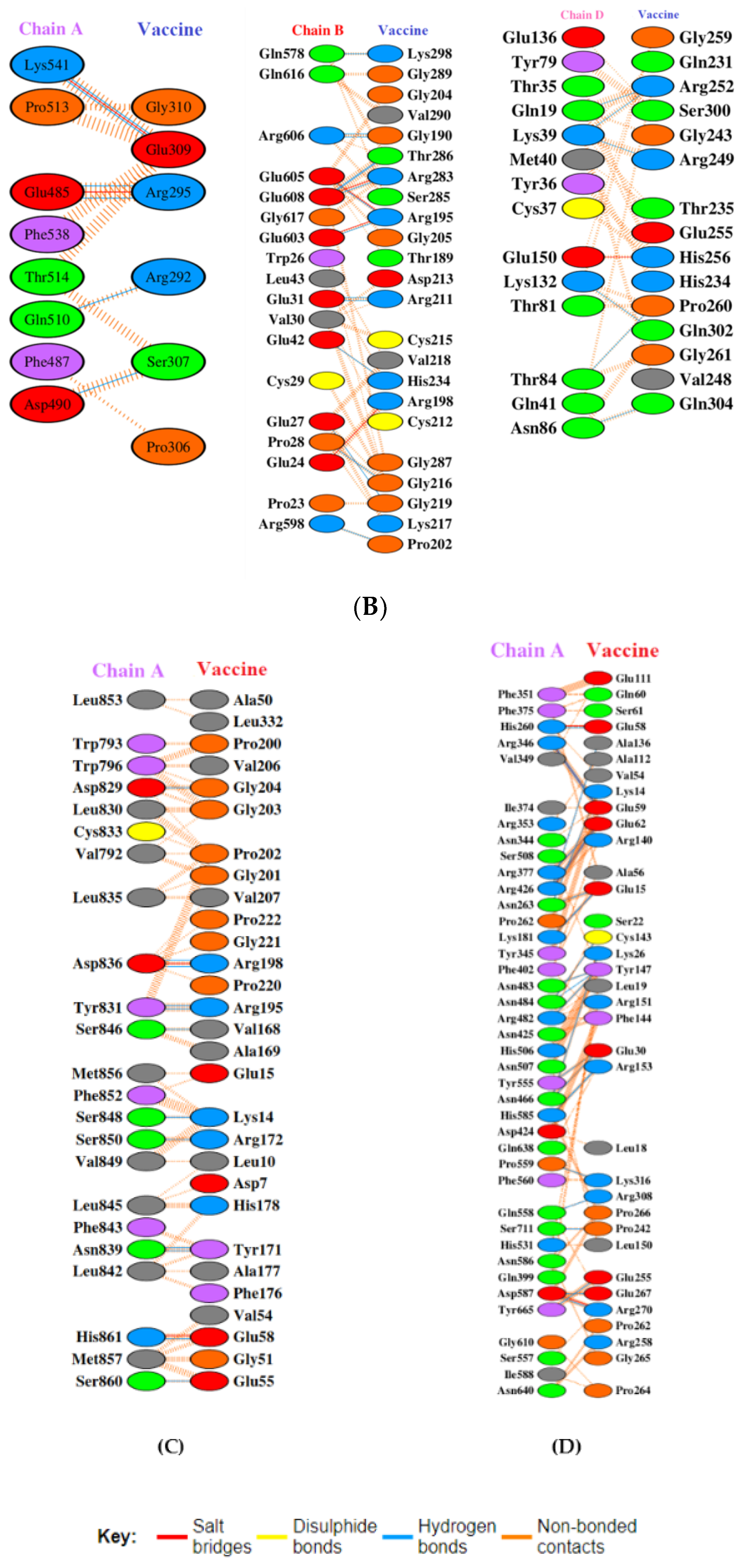
3.7. Docking Studies of Vaccine with TLR-2, 4, 7, and 9
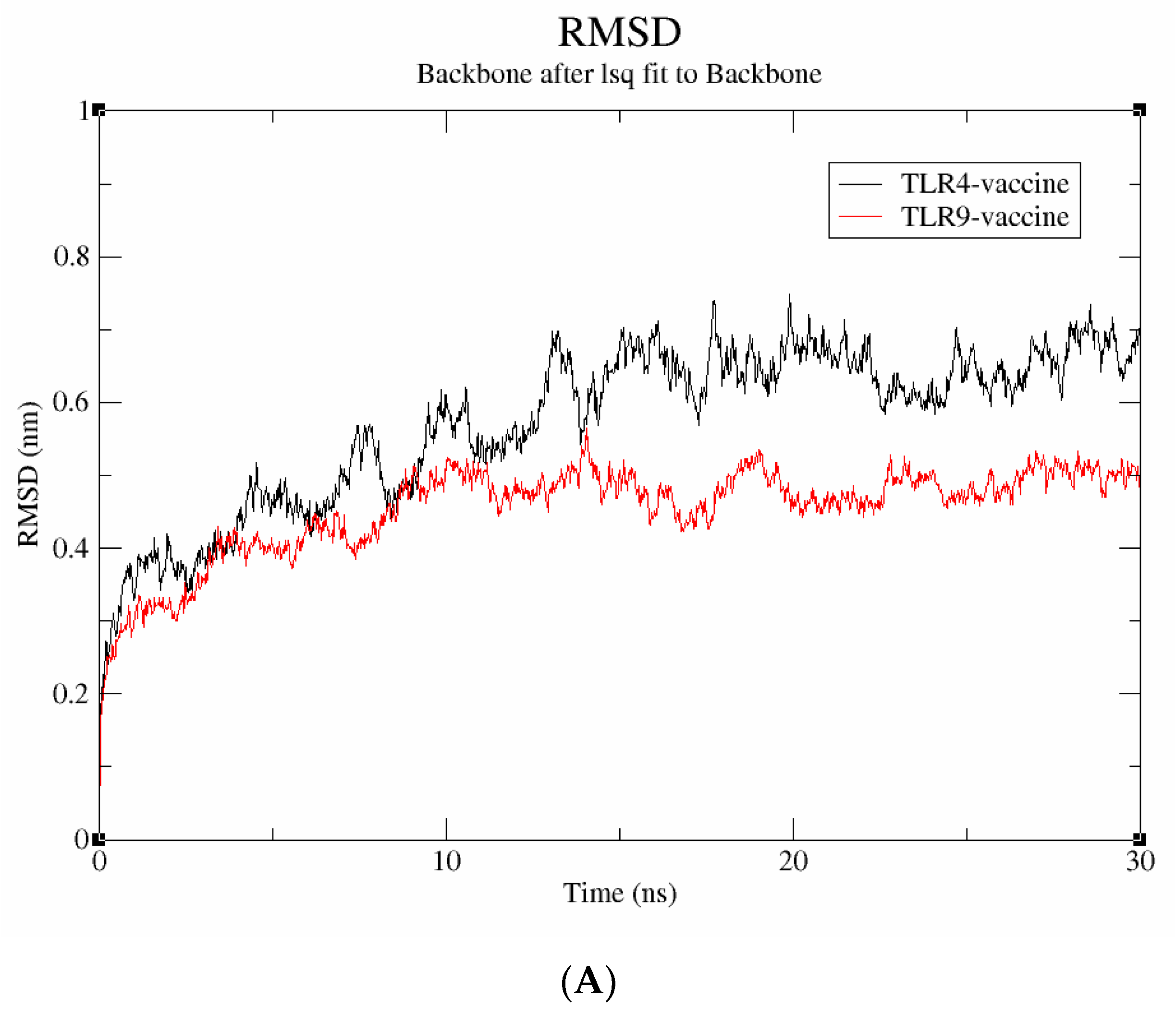
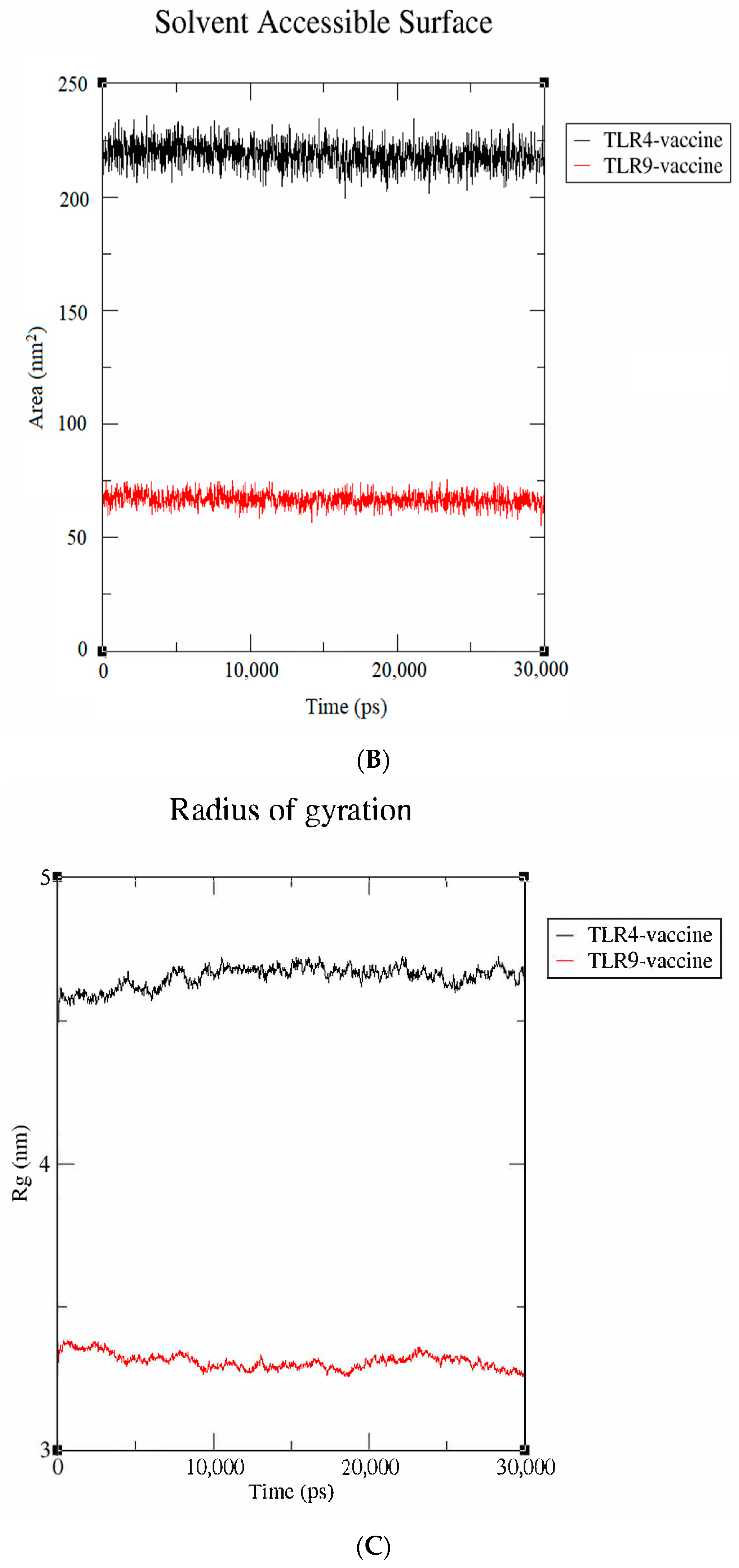
3.8. Stability Assessment by Molecular Dynamics Simulation
3.9. Codon Adaptation and In Silico Cloning of the Vaccine
3.10. Immune Simulation Studies
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.; Zhang, H.; Merkher, Y.; Chen, L.; Liu, N.; Leonov, S.; Chen, Y. Recent advances in therapeutic strategies for triple-negative breast cancer. J. Hematol. Oncol. 2022, 15, 121. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Immunotherapy for triple-negative breast cancer: A molecular insight into the microenvironment, treatment, and resistance. J. Natl. Cancer Inst. 2021, 1, 75–87. [Google Scholar] [CrossRef]
- Dixon-Douglas, J.; Loibl, S.; Denkert, C.; Telli, M.; Loi, S. Integrating Immunotherapy into the Treatment Landscape for Patients with Triple-Negative Breast Cancer. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dariushnejad, H.; Ghorbanzadeh, V.; Akbari, S.; Hashemzadeh, P. Design of a Novel Recombinant Multi-Epitope Vaccine against Triple-Negative Breast Cancer. Iran. Biomed. J. 2022, 26, 160–174. [Google Scholar] [PubMed]
- Lin, X.; Chen, H.; Xie, Y.; Zhou, X.; Wang, Y.; Zhou, J.; Liu, L. Combination of CTLA-4 blockade with MUC1 mRNA nanovaccine induces enhanced anti-tumor CTL activity by modulating tumor microenvironment of triple negative breast cancer. Transl. Oncol. 2022, 15, 101298. [Google Scholar] [CrossRef]
- Fan, Y.; He, S. The Characteristics of Tumor Microenvironment in Triple Negative Breast Cancer. Cancer Manag. Res. 2022, 14, 1–17. [Google Scholar] [CrossRef]
- Liu, J.; Fu, M.; Wang, M.; Wan, D.; Wei, Y.; Wei, X. Cancer vaccines as promising immuno-therapeutics: Platforms and current progress. J. Hematol. Oncol. 2022, 15, 28. [Google Scholar] [CrossRef]
- Corti, C.; Giachetti, P.P.; Eggermont, A.M.; Delaloge, S.; Curigliano, G. Therapeutic vaccines for breast cancer: Has the time finally come? Eur. J. Cancer 2022, 160, 150–174. [Google Scholar] [CrossRef]
- Qiu, D.; Zhang, G.; Yan, X.; Xiao, X.; Ma, X.; Lin, S.; Ma, M. Prospects of Immunotherapy for Triple-Negative Breast Cancer. Front. Oncol. 2022, 11, 797092. [Google Scholar] [CrossRef]
- Abdelmoneim, A.H.; Mustafa, M.I.; Abdelmageed, M.I.; Murshed, N.S.; Dawoud, E.D.; Ahmed, E.M.; Makhawi, A.M. Immunoinformatics design of multiepitopes peptide-based universal cancer vaccine using matrix metalloproteinase-9 protein as a target. Immunol. Med. 2021, 44, 35–52. [Google Scholar] [CrossRef]
- Tao, Z.; Wu, X. Targeting Transcription Factors in Cancer: From “Undruggable” to “Druggable”. In Transcription Factor Regulatory Networks, 1st ed.; Song, Q., Tao, Z., Eds.; Humana: New York, NY, USA, 2022; pp. 107–131. [Google Scholar]
- Yue, C.H.; Liu, J.Y.; Chi, C.S.; Hu, C.W.; Tan, K.T.; Huang, F.M.; Lee, C.J. Myeloid Zinc Finger 1 (MZF1) Maintains the Mesenchymal Phenotype by Down-regulating IGF1R/p38 MAPK/ERα Signaling Pathway in High-level MZF1-expressing TNBC cells. Anticancer Res. 2019, 39, 4149–4164. [Google Scholar] [CrossRef] [PubMed]
- Prawiningrum, A.F.; Paramita, R.I.; Panigoro, S.S. Immunoinformatics Approach for Epitope-Based Vaccine Design: Key Steps for Breast Cancer Vaccine. Diagnostics 2022, 12, 2981. [Google Scholar] [CrossRef] [PubMed]
- Khairkhah, N.; Aghasadeghi, M.R.; Namvar, A.; Bolhassani, A. Design of novel multiepitope constructs-based peptide vaccine against the structural S, N and M proteins of human COVID-19 using immunoinformatics analysis. PLoS ONE 2020, 15, e0240577. [Google Scholar] [CrossRef] [PubMed]
- Khairkhah, N.; Bolhassani, A.; Agi, E.; Namvar, A.; Nikyar, A. Immunological investigation of a multiepitope peptide vaccine candidate based on main proteins of SARS-CoV-2 pathogen. PLoS ONE 2022, 17, e0268251. [Google Scholar] [CrossRef] [PubMed]
- Akbari, E.; Kardani, K.; Namvar, A.; Ajdary, S.; Ardakani, E.M.; Khalaj, V.; Bolhassani, A. In silico design and in vitro expression of novel multiepitope DNA constructs based on HIV-1 proteins and Hsp70 T-cell epitopes. Biotechnol. Lett. 2021, 43, 1513–1550. [Google Scholar] [CrossRef] [PubMed]
- Jahangirian, E.; Jamal, G.A.; Nouroozi, M.; Mohammadpour, A. A Novel Multiepitope Vaccine Against Bladder Cancer Based on CTL and HTL Epitopes for Induction of Strong Immune Using Immunoinformatics Approaches. Int. J. Pept. Res. Ther. 2022, 28, 71. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sahu, U.; Kumari, P.; Dixit, A.; Khare, P. Designing of multi-epitope chimeric vaccine using immunoinformatic platform by targeting oncogenic strain HPV 16 and 18 against cervical cancer. Sci. Rep. 2022, 12, 9521. [Google Scholar] [CrossRef]
- Dar, M.A.; Kumar, P.; Kumar, P.; Shrivastava, A.; Dar, M.A.; Chauhan, R.; Dhingra, S. Designing of peptide based multi-epitope vaccine construct against gallbladder cancer using immunoinformatics and computational approaches. Vaccines 2022, 10, 1850. [Google Scholar] [CrossRef]
- Rajendran Krishnamoorthy, H.; Karuppasamy, R. Designing a novel SOX9 based multi-epitope vaccine to combat metastatic triple-negative breast cancer using immunoinformatics approach. Mol. Divers. 2022, 1–14. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct. Funct. Genet. 2006, 65, 40–48. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v. 2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Open-Source Drug Discovery Consortium; Raghava, G.P. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmud, S.; Rafi, M.; Paul, G.K.; Promi, M.M.; Shimu, M.; Sultana, S.; Saleh, M. Designing a multi-epitope vaccine candidate to combat MERS-CoV by employing an immunoinformatics approach. Sci. Rep. 2021, 11, 15431. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Vir, P.; Raghava, G.P. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct 2013, 8, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bui, H.H.; Sidney, J.; Dinh, K.; Southwood, S.; Newman, M.J.; Sette, A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 2006, 7, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dey, A.K.; Malyala, P.; Singh, M. Physicochemical and functional characterization of vaccine antigens and adjuvants. Expert Rev. Vaccines 2014, 13, 671–685. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, 1st ed.; Walker, J.M., Ed.; Humana: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef] [Green Version]
- Ponomarenko, J.; Bui, H.H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M. Lim-Wilby, M. Molecular docking. In Molecular Modeling of Proteins, 1st ed.; Kukol, A., Ed.; Humana: Totowa, NJ, USA, 2008; pp. 365–382. [Google Scholar]
- Comeau, S.R.; Gatchell, D.W.; Vajda, S.; Camacho, C.J. ClusPro: An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 2004, 20, 45–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castiglione, F.; Bernaschi, M. C-immsim: Playing with the immune response. In Proceedings of the Sixteenth International Symposium on Mathematical Theory of Networks and Systems (MTNS2004), Leuven, Belgium, 5–9 July 2004. [Google Scholar]
- Nordin, M.L.; Mohamad Norpi, A.S.; Ng, P.Y.; Yusoff, K.; Abu, N.; Lim, K.P.; Azmi, F. HER2/neu-Based Peptide Vaccination-Pulsed with B-Cell Epitope Induced Efficient Prophylactic and Therapeutic Antitumor Activities in TUBO Breast Cancer Mice Model. Cancers 2021, 13, 4958. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, D.D. In silico vaccine design: A tutorial in immunoinformatics. Health Care Anal 2022, 2, 100044. [Google Scholar] [CrossRef]
- Lee, S.J.; Shin, S.J.; Lee, M.H.; Lee, M.G.; Kang, T.H.; Park, W.S.; Park, Y.M. A potential protein adjuvant derived from Mycobacterium tuberculosis Rv0652 enhances dendritic cells-based tumor immunotherapy. PLoS ONE 2014, 9, e104351. [Google Scholar] [CrossRef] [PubMed]
- Sanches, R.C.; Tiwari, S.; Ferreira, L.C.; Oliveira, F.M.; Lopes, M.D.; Passos, M.J.; Lopes, D.O. Immunoinformatics design of multi-epitope peptide-based vaccine against Schistosoma mansoni using transmembrane proteins as a target. Front. Immunol. 2021, 12, 621706. [Google Scholar] [CrossRef]
- Ayyagari, V.S.; Venkateswarulu, T.C.; Srirama, K. Design of a multi-epitope-based vaccine targeting M-protein of SARS-CoV2: An immunoinformatics approach. J. Biomol. Struct. Dyn. 2022, 40, 2963–2977. [Google Scholar] [CrossRef]
- Wieczorek, M.; Abualrous, E.T.; Sticht, J.; Álvaro-Benito, M.; Stolzenberg, S.; Noé, F.; Freund, C. Major histocompatibility complex (MHC) class I and MHC class II proteins: Conformational plasticity in antigen presentation. Front. Immunol. 2017, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.T.; Islam, R.; Jerin, T.J.; Mahmud, A.; Khatun, S.; Kobir, A.; Mondal, S.I. Immunoinformatics and molecular dynamics approaches: Next generation vaccine design against West Nile virus. PLoS ONE 2021, 16, e0253393. [Google Scholar] [CrossRef]
- Kringelum, J.V.; Lundegaard, C.; Lund, O.; Nielsen, M. Reliable B cell epitope predictions: Impacts of method development and improved benchmarking. PLoS Comput. Biol. 2012, 8, e1002829. [Google Scholar] [CrossRef]
- Kumar, S.; Shuaib, M.; Prajapati, K.S.; Singh, A.K.; Choudhary, P.; Singh, S.; Gupta, S. A candidate triple-negative breast cancer vaccine design by targeting clinically relevant cell surface markers: An integrated immuno and bio-informatics approach. 3 Biotech 2022, 12, 72. [Google Scholar] [CrossRef] [PubMed]
- Luchner, M.; Reinke, S.; Milicic, A. TLR agonists as vaccine adjuvants targeting cancer and infectious diseases. Pharmaceutics 2021, 13, 142. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Khan, S.; Saleem, S.; Nizam-Uddin, N.; Mohammad, A.; Khan, T.; Wei, D.Q. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput. Biol. Med 2021, 133, 104420. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhu, Y.; Niu, C.; Xie, X.; Haimiti, G.; Guo, W.; Zhang, F. Design of a multi-epitope vaccine candidate against Brucella melitensis. Sci. Rep. 2022, 12, 10146. [Google Scholar] [CrossRef] [PubMed]
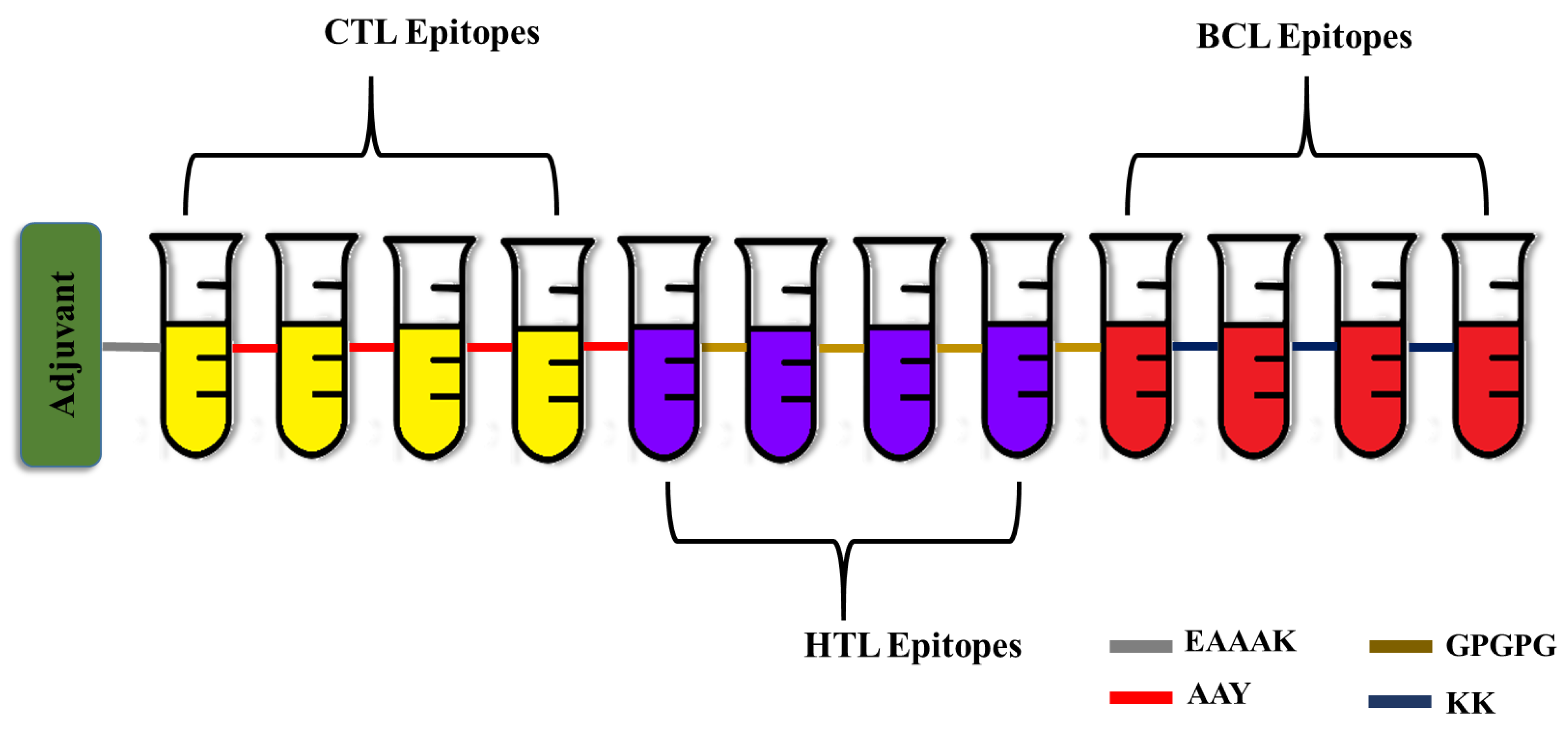





| S. No | Peptides | MHC I Alleles | Vaxijen Score | Allergenicity | Toxicity |
|---|---|---|---|---|---|
| 1 | AARLRFRCF | HLA-B*08:01 | 1.2291 | Non-allergen | Non-toxic |
| 2 | ARLRFRCFR | HLA-B*27:05 | 1.3903 | Non-allergen | Non-toxic |
| 3 | ARLEEHRRV | HLA-B*27:05 | 1.0612 | Non-allergen | Non-toxic |
| 4 | REGGFAHAL | HLA-B*40:01 | 1.3054 | Non-allergen | Non-toxic |
| S. No | Peptides | MHC II Alleles | Vaxijen Score | Allergenicity | Toxicity | IFN-γ Inducers |
|---|---|---|---|---|---|---|
| 1 | RGRPSTGGGVVRGGR | HLA-DQA10401, HLA-DQB10301, HLA-DQA10501, HLA-DQA10505 | 1.1247 | Non-allergen | Non-toxic | Inducer |
| 2 | GGVVRGGRCDVCGKV | HLA-DQB10301, HLA-DQA10501, HLA-DQA10505 | 1.416 | Non-allergen | Non-toxic | Inducer |
| 3 | RAVLLEHQAVHTGDK | HLA-DRB1*0103 | 0.8254 | Non-allergen | Non-toxic | Inducer |
| 4 | GQGFVRSARLEEHRR | HLA-DRB1*1401, HLA-DRB1*1402, HLA-DRB1*1454, HLA-DRB1*0801, HLA-DRB1*0803, HLA-DRB1*1101, HLA-DRB1*1302, HLA-DRB1*1303 | 0.5788 | Non-allergen | Non-toxic | Inducer |
| S. No | Peptides | Vaxijen Score | Allergenicity | Toxicity |
|---|---|---|---|---|
| 1 | PGPEAARLRFRCFRYE | 1.2090 | Non-allergen | Non-toxic |
| 2 | GRPSTGGGVVRGGRCD | 1.4314 | Non-allergen | Non-toxic |
| 3 | SGQIQSPSREGGFAHA | 1.1019 | Non-allergen | Non-toxic |
| 4 | QVKEESEVTEDSDFLE | 1.2340 | Non-allergen | Non-toxic |
| S. No | Parameters | Score |
|---|---|---|
| 1 | Immunogenicity | 3.72024 |
| 2 | Antigenicity | 0.7833 |
| 3 | Molecular weight | 350 kDa |
| 4 | Theoretical pI | 9.37 |
| 5 | No. of amino acids | 333 |
| 6 | Instability index | 32.80 |
| 7 | Aliphatic index | 71.62 |
| 8 | GRAVY | −0.376 |
| S. No | Residues | Number of Residues | Score |
|---|---|---|---|
| 1 | ILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGAPKPLLVAKEAADEAKAKLEAAGATVTV | 62 | 0.789 |
| 2 | RRGPGPGPGPEA | 12 | 0.721 |
| 3 | GPSTGGGRGGRCDKKSGQQSPSREGGFAH | 31 | 0.673 |
| 4 | MAKLSEPSTGGGVVRGGRGPGPGGGVVRGGRCDVCGKVGPGPGHAVHTGDKGPGPGGQG | 59 | 0.664 |
| 5 | DKKET | 5 | 0.641 |
| 6 | EMTLE | 5 | 0.598 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnamoorthy, H.R.; Karuppasamy, R. Design and In Silico Validation of a Novel MZF-1-Based Multi-Epitope Vaccine to Combat Metastatic Triple Negative Breast Cancer. Vaccines 2023, 11, 577. https://doi.org/10.3390/vaccines11030577
Krishnamoorthy HR, Karuppasamy R. Design and In Silico Validation of a Novel MZF-1-Based Multi-Epitope Vaccine to Combat Metastatic Triple Negative Breast Cancer. Vaccines. 2023; 11(3):577. https://doi.org/10.3390/vaccines11030577
Chicago/Turabian StyleKrishnamoorthy, HemaNandini Rajendran, and Ramanathan Karuppasamy. 2023. "Design and In Silico Validation of a Novel MZF-1-Based Multi-Epitope Vaccine to Combat Metastatic Triple Negative Breast Cancer" Vaccines 11, no. 3: 577. https://doi.org/10.3390/vaccines11030577
APA StyleKrishnamoorthy, H. R., & Karuppasamy, R. (2023). Design and In Silico Validation of a Novel MZF-1-Based Multi-Epitope Vaccine to Combat Metastatic Triple Negative Breast Cancer. Vaccines, 11(3), 577. https://doi.org/10.3390/vaccines11030577





