In Vivo Validation of Novel Synthetic tbp1 Peptide-Based Vaccine Candidates against Haemophilus influenzae Strains in BALB/c Mice
Abstract
:1. Introduction
2. Materials and Methods
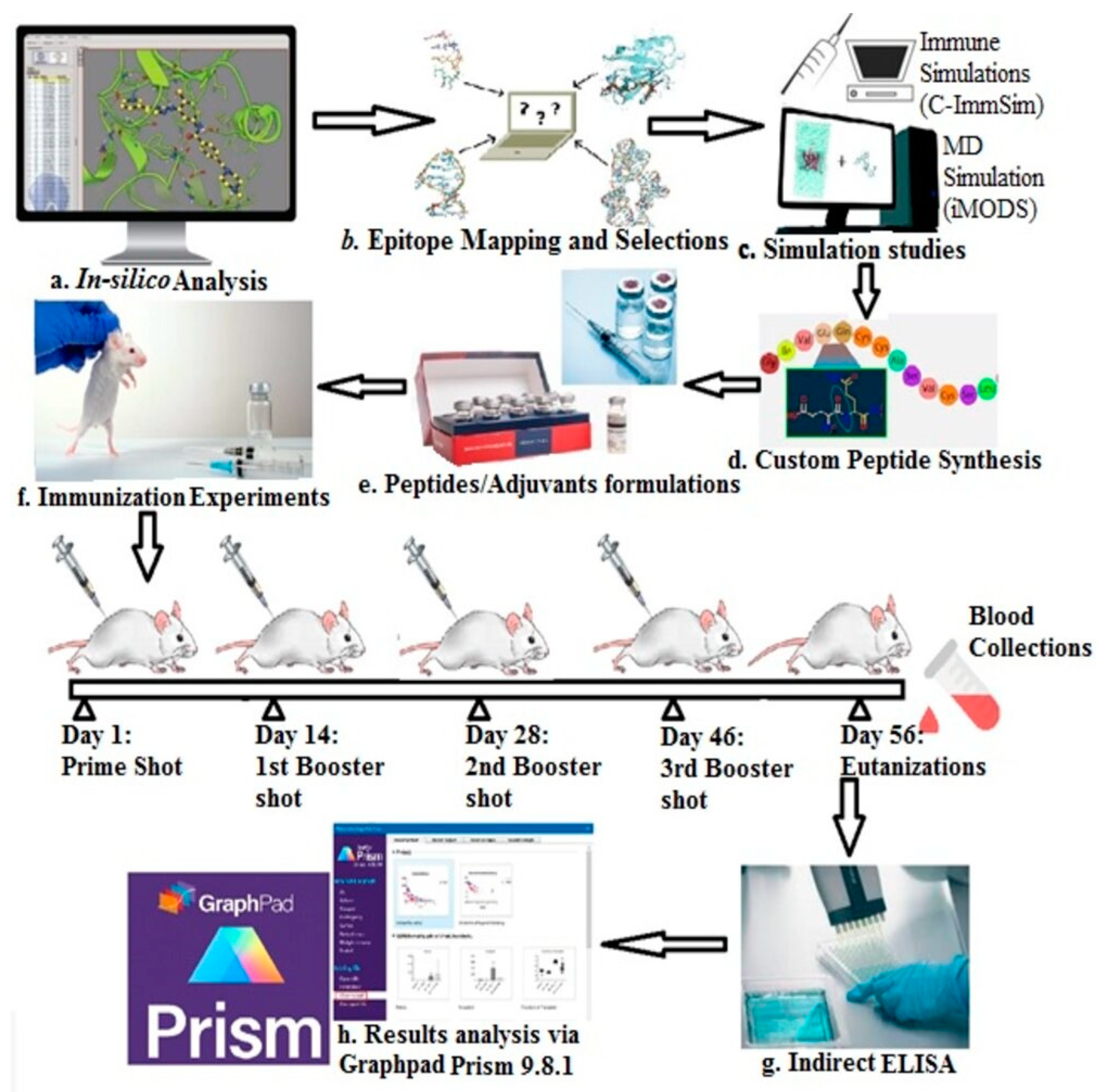
2.1. Scheme of Study
2.2. Prediction and Selection of Overlapping B-Cell and T-Cell Epitopes as Peptide Vaccine Candidates
2.3. Prediction of IFN-γ, IL4, and IL10 Cytokine-Inducing Ability of Peptides
2.4. Molecular Dynamics (MD) Simulations of the Receptor–Ligand Complex
2.5. Immune Simulation of Selected Peptide Antigens
2.6. Molecular Docking: Epitopes and H2-Ad MHC-II Mouse Allele Receptor Binding
2.7. Peptide Design and Synthesis
2.8. Ethics Statement
2.9. Mice Immunizations and Grouping
2.10. Pre-Immunization Procedure
2.11. Detection of Humoral Response with ELISA
2.12. Statistical Analysis
3. Results
3.1. Prediction and Selection of Overlapping B-Cell and T-Cell Epitopes as Putative Candidates for Peptide Vaccine
3.2. Prediction of Cytokine-Inducing Epitopes
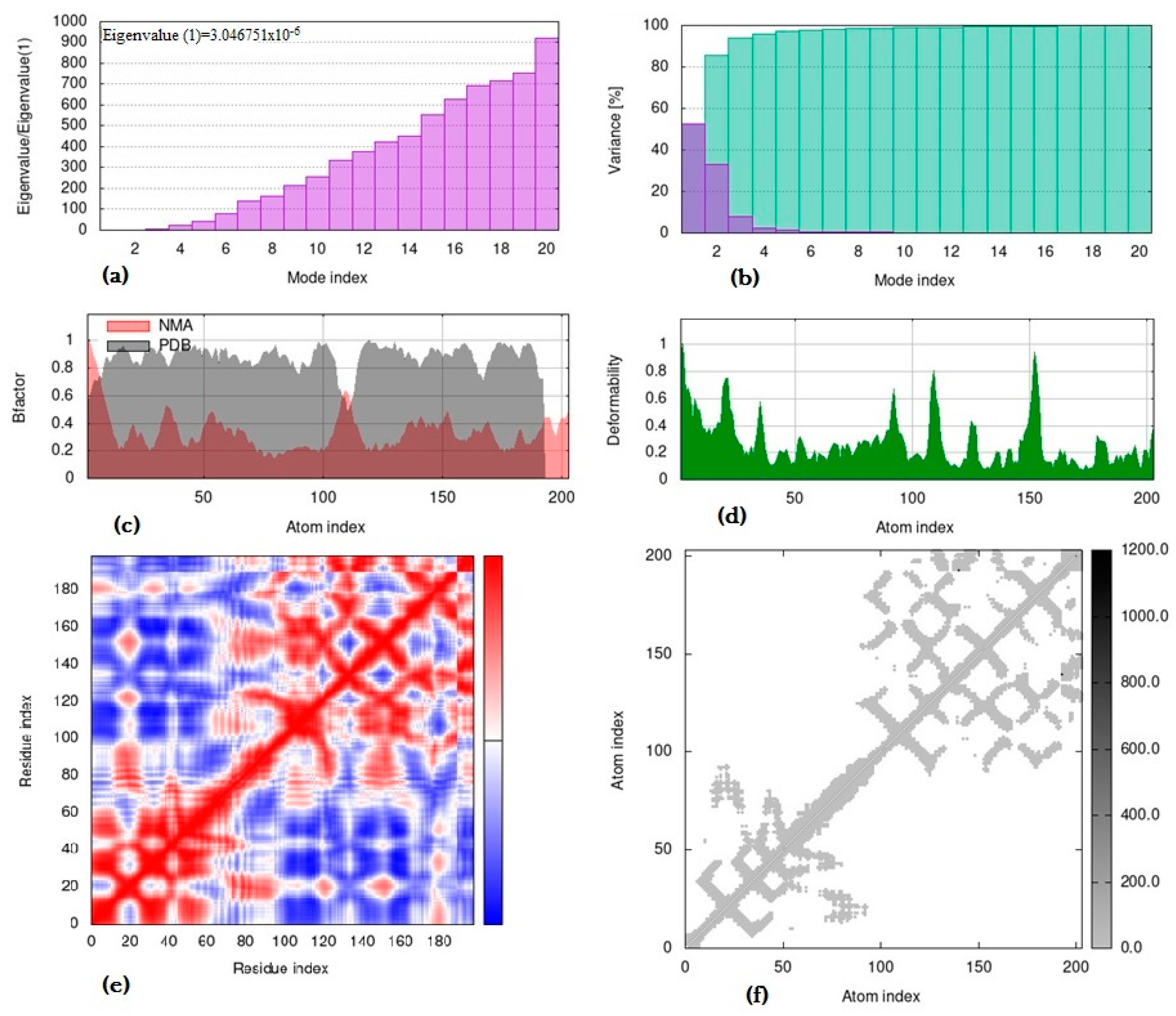
3.3. Molecular Dynamics Simulation
3.4. Immune Simulation Studies
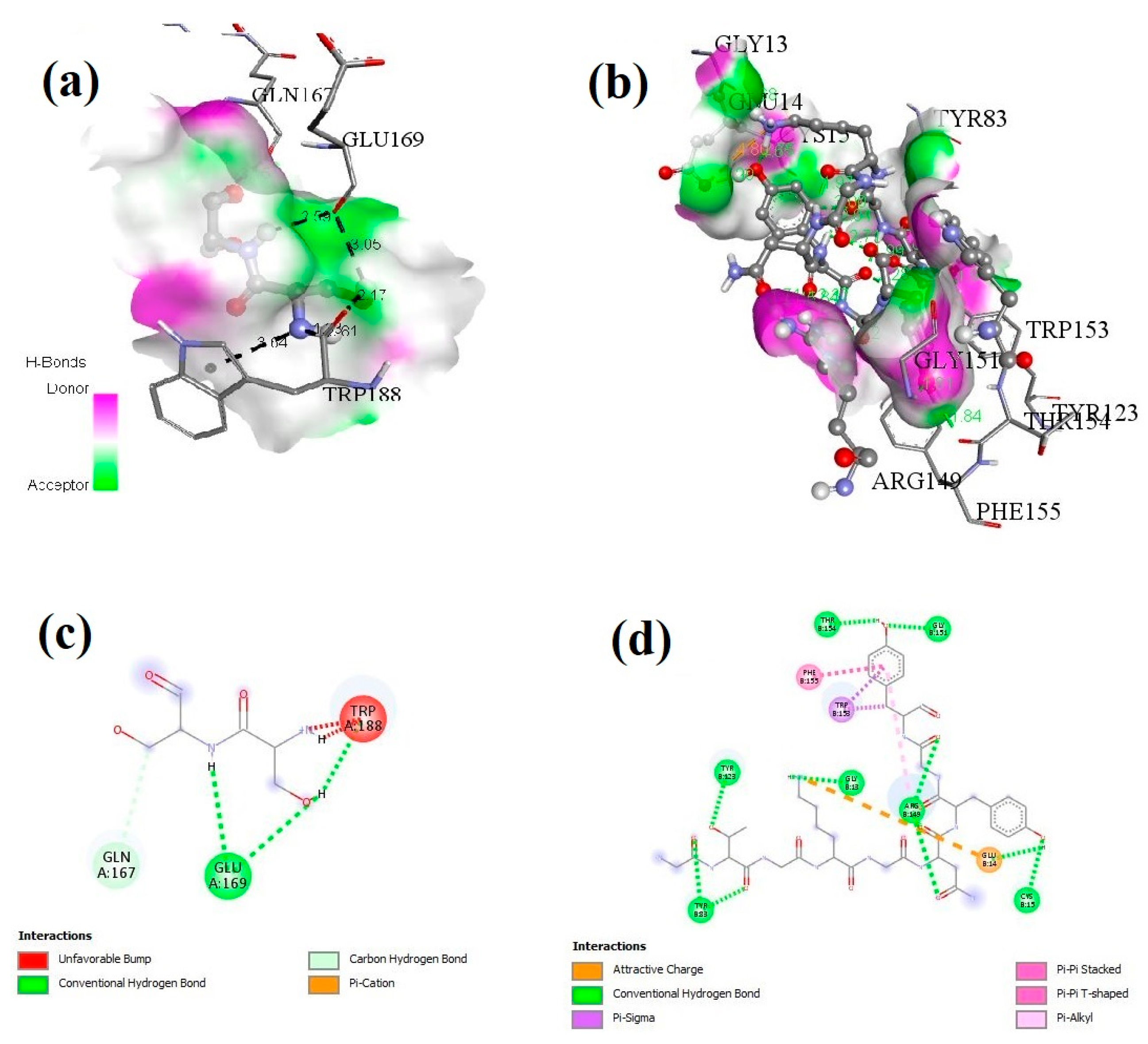
3.5. Docking of H2-Ad Mouse Allele and Selected Epitopes
3.6. Detection of Humoral Response with ELISA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Behrouzi, A.; Vaziri, F.; Afrough, F.R.-J.P.; Rahbar, M. Vaccine Candidates against Nontypeable Haemophilus influenzae: A Review. Iran. Biomed. J. 2017, 21, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.; Litt, D.; Almond, R.; Findlow, J.; Linley, E.; Ramsay, M.; Borrow, R.; Ladhani, S. Haemophilus influenzae type b (Hib) seroprevalence and current epidemiology in England and Wales. J. Infect. 2018, 76, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.; Tsang, R.S.W.; Morgan, A.; Jamieson, F.B.; Ulanova, M. Invasive disease caused by Haemophilus influenzae type a in Northern Ontario First Nations communities. J. Med. Microbiol. 2011, 60, 384–390. [Google Scholar] [CrossRef]
- Pintoa, M.; González-Díazb, A.; Machadod, M.P.; Duartee, S.; Vieira, L.; Carriçod, J.A.; Martin, S.; Bajanca-Lavado, M.P.; Gomes, J.P. Insights into the population structure and pan-genome of Haemophilus influenzae. Infect. Genet. Evol. 2019, 67, 126–135. [Google Scholar] [CrossRef]
- Soeters, H.M.; Blain, A.; Pondo, T.; Monica, B.D.; Farley, M.; Harrison, L.H.; Lynfield, R.; Miller, L.; Petit, S.; Reingold, A.; et al. Current Epidemiology and Trends in Invasive Haemophilus influenzae Disease—United States, 2009–2015. Clin. Infect. Dis. 2018, 67, 881–889. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Srivastava, S. Association between Haemophilus influenza type B (Hib) vaccination and child anthropometric outcomes in Andhra Pradesh (India): Evidence from the Young Lives Study. J. Public Health 2017, 25, 581–589. [Google Scholar] [CrossRef]
- Zaidi, A.K.M.; Khan, H.; Sherali, A.R.; Lasi, R.; The Sindh Meningitis Study Group. Burden of haemophilus influenzae type b disease in Pakistani children. East. Mediterr. Health J. 2010, 16, 590–594. [Google Scholar] [CrossRef]
- Fischer, N.J. Haemophilus influenzae serotype a septic arthritis in an immunized central Australian indigenous child. Int. J. Infect. Dis. 2014, 21, 15–16. [Google Scholar] [CrossRef]
- Potter, A.A.; Schryvers, A.B.; Ogunnariwo, J.A.; Hutchins, W.A.; Lo, R.Y.C.; Watts, T. Protective capacity of the Pasteurella haemolytica transferrin-binding proteins TbpA and TbpB in cattle. Microb. Pathog. 1999, 27, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Iskander, M.; Hayden, K.; Van Domselaar, G. First Complete Genome Sequence of Haemophilus influenzae Serotype a. Genome Announc. 2017, 5, 1–2. [Google Scholar] [CrossRef]
- Naito, S.; Takeuchi, N.; Ohkusu, M.; Takahashi-Nakaguchi, A.; Takahashi, H.; Imuta, N.; Nishi, J.; Shibayama, K.; Matsuoka, M.; Sasaki, Y.; et al. Clinical and bacteriologic analysis of nontypeable Haemophilus influenzae strains isolated from children with invasive diseases in Japan from 2008 to 2015. J. Clin. Microbiol. 2018, 56, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Mousav, S.F.; Fatemi, S.; Siadat, S.D.; Zahraei, S.M.; Nikanpour, E.; Malekan, M.A.; Khabiri, A.R.; Janani, A.R. Development and optimization of a homemade ELISA kit for detection of antibodies against Haemophilus influenzae type b. Jundishapur J. Microbiol. 2016, 9, e30629. [Google Scholar] [CrossRef] [PubMed]
- Van Elder, J.; Slack, M.P.E.; Ladhani, S.; Cripps, A.W. Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect. Dis. 2014, 14, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Hawdon, N.; Nix, E.B.; Tsang, R.S.W.; Ferroni, G.; McCready, W.G.; Ulanova, M. Immune Response to Haemophilus influenzae Type b Vaccination in Patients with Chronic Renal Failure. Clin. Vaccine Immunol. 2012, 19, 967–969. [Google Scholar] [CrossRef]
- Townsend, K.; Ladhani, S.N.; Findlow, H.; Borrow, R. Evaluation and validation of a serum bactericidal antibody assay for Haemophilus influenzae type b and the threshold of protection. Vaccine 2014, 32, 5650–5656. [Google Scholar] [CrossRef]
- CDC Centers for Disease Control. Safety Information for Haemophilus Influenza Type B (Hib) Vaccines _ CDC. Available online: https://www.cdc.gov/vaccinesafety/vaccines/hib-vaccine.html (accessed on 15 October 2023).
- Dworkin, M.S.; Park, L.; Borchardt, S.M. The Changing Epidemiology of Invasive Haemophilus influenzae Disease, Especially in Persons >= 65 Years Old. Clin. Infect. Dis. 2007, 44, 810–816. [Google Scholar] [CrossRef]
- Cox, A.D.; Williams, D.; Cairns, C.; Michael, F.S.; Fleming, P.; Vinogradov, E.; Arbour, M.; Masson, L.; Zou, W. Investigating the candidacy of a capsular polysaccharide-based glycoconjugate as a vaccine to combat Haemophilus influenzae type a disease: A solution for an unmet public health need. Vaccine 2017, 35, 6129–6136. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, A.; Verdijk, P.; Kreeftenberg, H. Preclinical evaluation of a Haemophilus influenzae type b conjugate vaccine process intended for technology transfer. Hum. Vaccines Immunother. 2014, 10, 2691–2696. [Google Scholar] [CrossRef]
- Kim, H.S.; Yoo, T.H.; Jang, Y.S. An animal model to evaluate the protective Efficacy of H. influnzae type b Conjugate vaccine. Biotechnol. Bioprocess Eng. 2004, 9, 490–494. [Google Scholar] [CrossRef]
- Mirza, M.U.; Rafique, S.; Ali, A.; Munir, M.; Ikram, N.; Manan, A.; Salo-Ahen, O.M.; Idrees, M. Towards peptide vaccines against Zika virus: Immunoinformatics combined with molecular dynamics simulations to predict antigenic epitopes of Zika viral proteins. Sci. Rep. 2016, 6, 37313. [Google Scholar] [CrossRef]
- Rojas-Caraballo, J.; López-Abán, J.; del Villar, L.P.; Vizcaíno, C.; Vicente, B.; Fernández-Soto, P.; del Olmo, E.; Patarroyo, M.A.; Muro, A. In Vitro and In Vivo Studies for Assessing the Immune Response and Protection-Inducing Ability Conferred by Fasciola hepatica-Derived Synthetic Peptides Containing. PLoS ONE 2014, 9, e105323. [Google Scholar] [CrossRef] [PubMed]
- Azmi, F.; Al, A.; Ahmad, H.; Skwarczynski, M.; Toth, I. Recent progress in adjuvant discovery for peptide-based subunit vaccines. Hum. Vaccines Immunother. 2014, 10, 778–796. [Google Scholar] [CrossRef]
- Akter, T.; Atanelishvili, I.; Noguchi, A.; Silver, R.M.; Bogatkevich, G.S. Establishment of an indirect ELISA for detection of the novel antifibrotic peptide M10. PLoS ONE 2017, 12, e0188588. [Google Scholar] [CrossRef] [PubMed]
- Hos, B.J.; Tondini, E.; Van Kasteren, S.I.; Ossendorp, F. Approaches to Improve Chemically Defined Synthetic Peptide Vaccines. Front. Immunol. 2018, 9, 884. [Google Scholar] [CrossRef]
- Kateregga, J.; Lubega, G.W.; Lindblad, E.B.; Authié, E.; Coetzer, T.H.T.; Boulangé, A.F.V. Effect of adjuvants on the humoral immune response to congopain in mice and cattle. BMC Vet. Res. 2012, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Buckwalter, C.M.; Currie, E.G.; Tsang, R.S.W.; Gray-Owen, S.D. Discordant Effects of Licensed Meningococcal Serogroup B Vaccination on Invasive Disease and Nasal Colonization in a Humanized Mouse Model. J. Infect. Dis. 2017, 215, 1590–1598. [Google Scholar] [CrossRef]
- Montero, D.A.; Del Canto, F.; Salazar, J.C.; Céspedes, S.; Cádiz, L.; Arenas-Salinas, M.; Reyes, J.; Oñate, A.; Vidal, R.M. Immunization of mice with chimeric antigens displaying selected epitopes confers protection against intestinal colonization and renal damage caused by Shiga toxin-producing Escherichia coli. NPJ Vaccines 2020, 5, 20. [Google Scholar] [CrossRef]
- Guizzo, J.A.; Chaudhuri, S.; Prigol, S.R.; Yu, R.-H.; Dazzi, C.C.; Balbinott, N.; Frandoloso, G.P.; Kreutz, L.C.; Frandoloso, R.; Schryvers, A.B. The amino acid selected for generating mutant TbpB antigens defective in binding transferrin can compromise the in vivo protective capacity. Sci. Rep. 2018, 8, 7372. [Google Scholar] [CrossRef]
- Calmettes, C.; Yu, R.H.; Silva, L.P.; Curran, D.; Schriemer, D.C.; Schryvers, A.B.; Moraes, T.F. Structural variations within the transferrin binding site on transferrin-binding protein B, TbpB. J. Biol. Chem. 2011, 286, 12683–12692. [Google Scholar] [CrossRef]
- Fegan, J.E.; Calmettes, C.; Islam, E.A.; Ahn, S.K.; Chaudhuri, S.; Yu, R.H.; Gray-Owen, S.D.; Moraes, T.F.; Schryvers, A.B. Utility of Hybrid Transferrin Binding Protein Antigens for Protection against Pathogenic Neisseria Species. Front. Immunol. 2019, 10, 247. [Google Scholar] [CrossRef]
- Jaiswal, V.; Chanumolu, S.K.; Gupta, A.; Chauhan, R.S.; Rout, C. Jenner-predict server: Prediction of protein vaccine candidates (PVCs) in bacteria based on host-pathogen interactions. BMC Bioinform. 2013, 14, 211. [Google Scholar] [CrossRef]
- Singh, S.; Singh, V.P.; Cheema, P.S.; Sandey, M.; Ranjan, R.; Gupta, S.K.; Sharma, B. Immune response to DNA vaccine expressing transferrin binding protein a gene of Pasteurella multocida. Braz. J. Microbiol. 2011, 42, 750–760. [Google Scholar] [CrossRef]
- Shivachandra, S.B.; Yogisharadhya, R.; Kumar, A.; Mohanty, N.N.; Nagaleekar, V.K. Recombinant transferrin binding protein A (rTbpA) fragments of Pasteurella multocida serogroup B:2 provide variable protection following homologous challenge in mouse model. Res. Vet. Sci. 2015, 98, 1–6. [Google Scholar] [CrossRef]
- Bibi, N.; Zaidi, N.S.S.; Tahir, M.; Babar, M.M. Vaccinomics-driven proteome-wide screening of Haemophilus influenzae for the prediction of common putative vaccine candidates. Can. J. Microbiol. 2021, 67, 799–812. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Gupta, S.; Vir, P.; Raghava, G.P. Prediction of IL4 inducing peptides. Clin. Dev. Immunol. 2013, 2013, 263952. [Google Scholar] [CrossRef] [PubMed]
- Dhanda, S.K.; Vir, P.; Raghava, G.P.S. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct 2013, 8, 30. [Google Scholar] [CrossRef]
- Nagpal, G.; Usmani, S.S.; Dhanda, S.K.; Kaur, H.; Singh, S.; Sharma, M.; Raghava, G.P. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci. Rep. 2017, 7, 42851. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; Kuriyan, J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. USA 2005, 102, 6679–6685. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bogdan, P.; Nazarian, S. An in silico deep learning approach to multi-epitope vaccine design: A SARS-CoV-2 case study. Sci. Rep. 2021, 11, 3238. [Google Scholar] [CrossRef] [PubMed]
- Chukwudozie, O.S.; Gray, C.M.; Fagbayi, T.A.; Chukwuanukwu, R.C.; Oyebanji, V.O.; Bankole, T.T.; Adewole, R.A.; Daniel, E.M. Immuno-informatics design of a multimeric epitope peptide based vaccine targeting SARS-CoV-2 spike glycoprotein. PLoS ONE 2021, 16, e0248061. [Google Scholar] [CrossRef] [PubMed]
- Sanches, R.C.O.; Tiwari, S.; Ferreira, L.C.G.; Oliveira, F.M.; Lopes, M.D.; Passos, M.J.F.; Maia, E.H.B.; Taranto, A.G.; Kato, R.; Azevedo, V.A.C.; et al. Immunoinformatics Design of Multi-Epitope Peptide-Based Vaccine against Schistosoma mansoni Using Transmembrane Proteins as a Target. Front. Immunol. 2021, 12, 621706. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.; Di, J.; Zurakowski, D. A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 1998, 221, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Tsang, R.S.W.; Proulx, J.F.; Hayden, K.; Shuel, M.; Lefebvre, B.; Boisvert, A.A.; Moore, D. Characteristics of invasive Haemophilus influenzae serotype a (Hia) from Nunavik, Canada and comparison with Hia strains in other North American Arctic regions. Int. J. Infect. Dis. 2017, 57, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Babu, L.; Uppalapati, S.R.; Sripathy, M.H.; Reddy, P.N. Evaluation of Recombinant Multi-Epitope Outer Membrane Protein-Based Klebsiella pneumoniae Subunit Vaccine in Mouse Model. Front. Microbiol. 2017, 8, 1805. [Google Scholar] [CrossRef]
- Dawood, R.M.; Moustafa, R.I.; Abdelhafez, T.H.; El-Shenawy, R.; El-Abd, Y.; El Din, N.G.B.; Dubuisson, J.; El Awady, M.K. A multiepitope peptide vaccine against HCV stimulates neutralizing humoral and persistent cellular responses in mice. BMC Infect. Dis. 2019, 19, 932. [Google Scholar] [CrossRef]
- Herrera-Rodriguez, J.; Meijerhof, T.; Niesters, H.G.; Stjernholm, G.; Hovden, A.-O.; Sørensen, B.; Ökvist, M.; Sommerfelt, M.A.; Huckriede, A. A novel peptide-based vaccine candidate with protective efficacy against influenza A in a mouse model. Virology 2018, 515, 21–28. [Google Scholar] [CrossRef]
- Islam, S.I.; Mahfuj, S.; Alam, A.; Ara, Y.; Sanjida, S.; Mou, M.J. Immunoinformatic Approaches to Identify Immune Epitopes and Design an Epitope-Based Subunit Vaccine against Emerging Tilapia Lake Virus (TiLV). Aquac. J. 2022, 2, 186–202. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Usmani, S.S.; Agrawal, P.; Nagpal, G.; Gautam, A.; Raghava, G.P.S. Novel in silico tools for designing peptide-based subunit vaccines and immunotherapeutics. Brief. Bioinform. 2017, 18, 467–478. [Google Scholar]
- Littleton, L.C.; Gruenke, J.A. A comparison of the effect of three adjuvants on the antibody response to ovalbumin in mice. BIOS 2013, 84, 142–147. [Google Scholar] [CrossRef]
- Licciardi, P.V.; Balloch, A.; Russell, F.M.; Burton, R.L.; Lin, J.; Nahm, M.H.; Mulholland, E.K.; Tang, M.L. Pneumococcal polysaccharide vaccine at 12 months of age produces functional immune responses. J. Allergy Clin. Immunol. 2012, 129, 794–800.e2. [Google Scholar] [CrossRef]
- Ebensen, T.; Paukner, S.; Link, C.; Kudela, P.; de Domenico, C.; Lubitz, W.; Guzmán, C.A. Bacterial Ghosts Are an Efficient Delivery System for DNA Vaccines. J. Immunol. 2004, 172, 6858–6865. [Google Scholar] [CrossRef] [PubMed]
- Mayr, U.B.; Haller, C.; Haidinger, W.; Atrasheuskaya, A.; Bukin, E.; Lubitz, W.; Ignatyev, G. Bacterial Ghosts as an Oral Vaccine: A Single Dose of Escherichia coli O157: H7 Bacterial Ghosts Protects Mice against Lethal Challenge. Infect. Immun. 2005, 73, 4810–4817. [Google Scholar] [CrossRef] [PubMed]
- Hajam, I.A.; Dar, P.A.; Won, G.; Lee, J.H. Bacterial ghosts as adjuvants: Mechanisms and potential. Vet. Res. 2017, 48, 37. [Google Scholar] [CrossRef] [PubMed]


| Group | Subgroup | Day 1 Prime Shot | Day 14 1st Booster Shot | Day 28 2nd Booster Shot | Day 42 |
|---|---|---|---|---|---|
| A | A1 (n = 8) | (tbp1-E1)+CFA | (tbp1-E1)+IFA | (tbp1-E1)+IFA | Euthanization and blood sampling |
| A2 (n = 8) | (tbp1-E1)+BG | (tbp1-E1)+BG | (tbp1-E1)+BG | ||
| B | B1 (n = 8) | (tbp1-E2)+CFA | (tbp1-E2)+IFA | (tbp1-E2)+IFA | |
| B2 (n = 8) | (tbp1-E2)+BG | (tbp1-E2)+BG | (tbp1-E2)+BG | ||
| C | C1 (n = 8) | (tbp1-E1+E2)+CFA | (tbp1-E1+E2)+IFA | (tbp1-E1+E2)+IFA | |
| C2 (n = 8) | (tbp1-E1+E2)+BG | (tbp1-E1+E2)+BG | (tbp1-E1+E2)+BG | ||
| D | D1 (n = 3) | CFA | IFA | IFA | |
| D2 (n = 3) | BGs | BGs | BGs | ||
| D3 (n = 3) | PBS | PBS | PBS |
| S. No. | Peptide/Epitope | IFN-γ | IL4 | IL10 |
|---|---|---|---|---|
| 1. | tbp1-E1 | Positive | Inducer | Inducer |
| 2. | tbp1-E2 | Positive | Inducer | Inducer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bibi, N.; Wajeeha, A.W.; Mukhtar, M.; Tahir, M.; Zaidi, N.u.S.S. In Vivo Validation of Novel Synthetic tbp1 Peptide-Based Vaccine Candidates against Haemophilus influenzae Strains in BALB/c Mice. Vaccines 2023, 11, 1651. https://doi.org/10.3390/vaccines11111651
Bibi N, Wajeeha AW, Mukhtar M, Tahir M, Zaidi NuSS. In Vivo Validation of Novel Synthetic tbp1 Peptide-Based Vaccine Candidates against Haemophilus influenzae Strains in BALB/c Mice. Vaccines. 2023; 11(11):1651. https://doi.org/10.3390/vaccines11111651
Chicago/Turabian StyleBibi, Naseeha, Amtul Wadood Wajeeha, Mamuna Mukhtar, Muhammad Tahir, and Najam us Sahar Sadaf Zaidi. 2023. "In Vivo Validation of Novel Synthetic tbp1 Peptide-Based Vaccine Candidates against Haemophilus influenzae Strains in BALB/c Mice" Vaccines 11, no. 11: 1651. https://doi.org/10.3390/vaccines11111651
APA StyleBibi, N., Wajeeha, A. W., Mukhtar, M., Tahir, M., & Zaidi, N. u. S. S. (2023). In Vivo Validation of Novel Synthetic tbp1 Peptide-Based Vaccine Candidates against Haemophilus influenzae Strains in BALB/c Mice. Vaccines, 11(11), 1651. https://doi.org/10.3390/vaccines11111651







