Evaluation of Salmonella Typhimurium Lacking fruR, ssrAB, or hfq as a Prophylactic Vaccine against Salmonella Lethal Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media and Culture Conditions
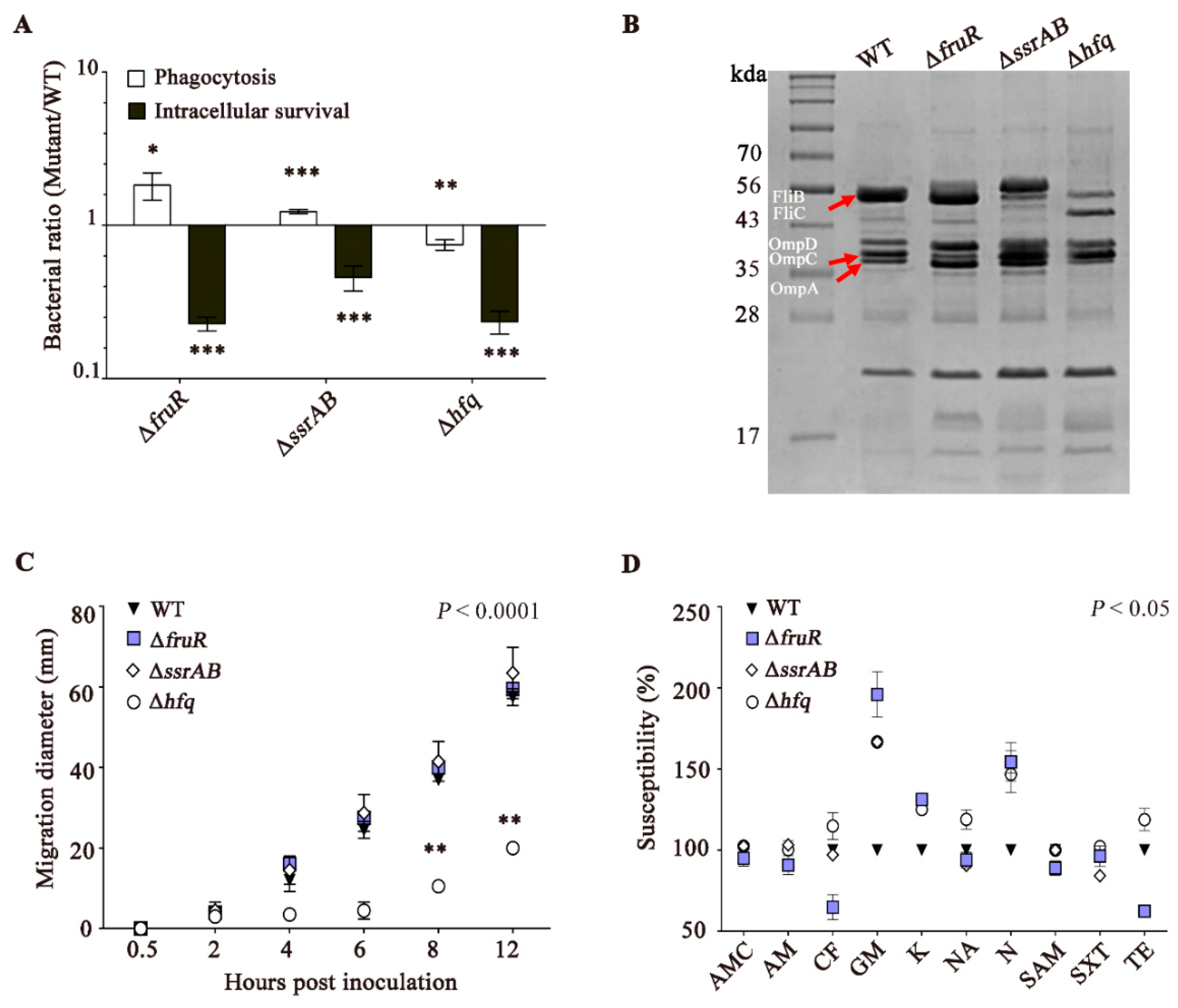
2.2. Intracellular Survival Assay
2.3. Motility Assays
2.4. Analysis of Outer Membrane Proteins (OMPs)
2.5. Antibiotic Susceptibility Test
2.6. Animal Ethics
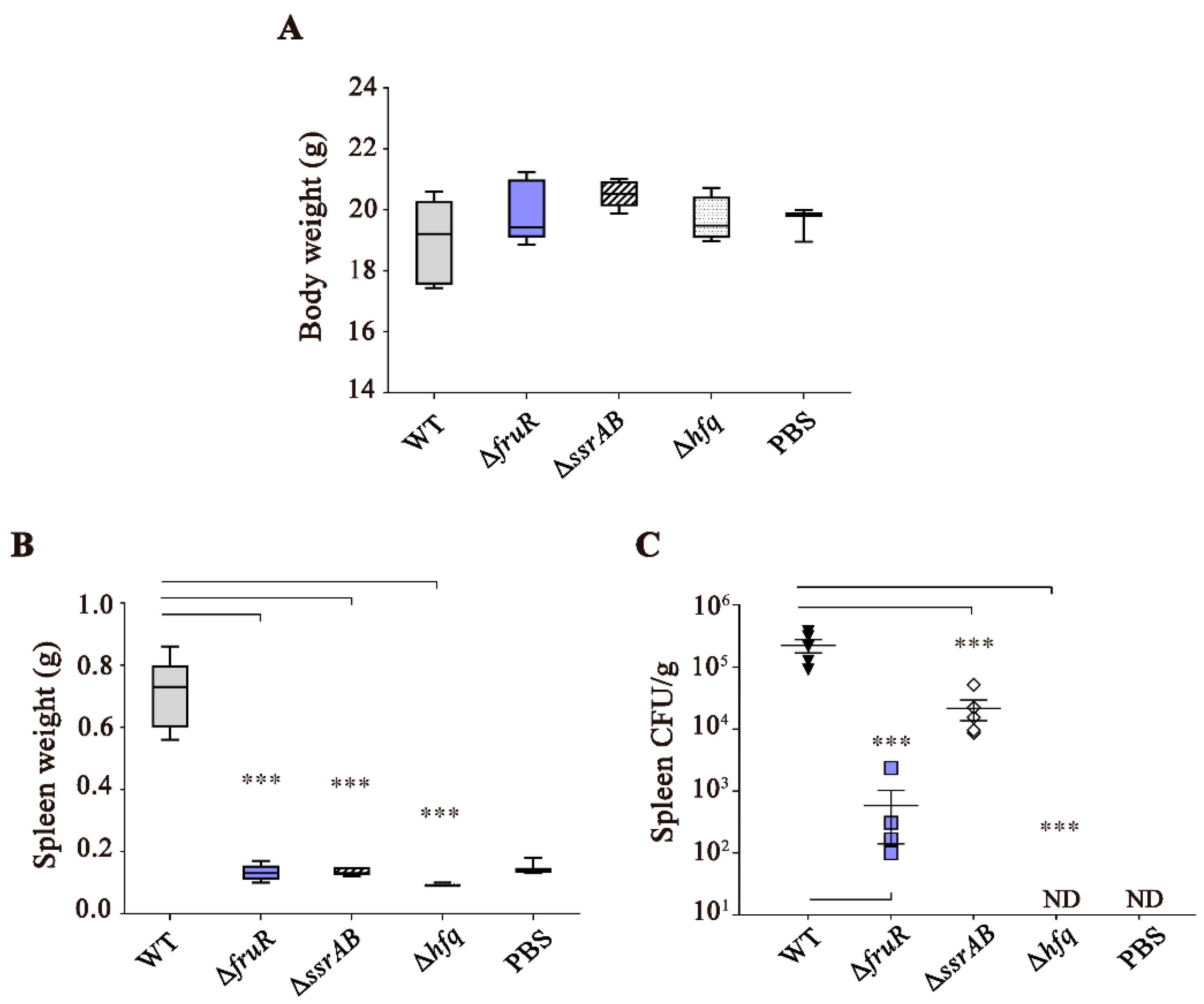
2.7. Determination of Bacterial Virulence in Mice
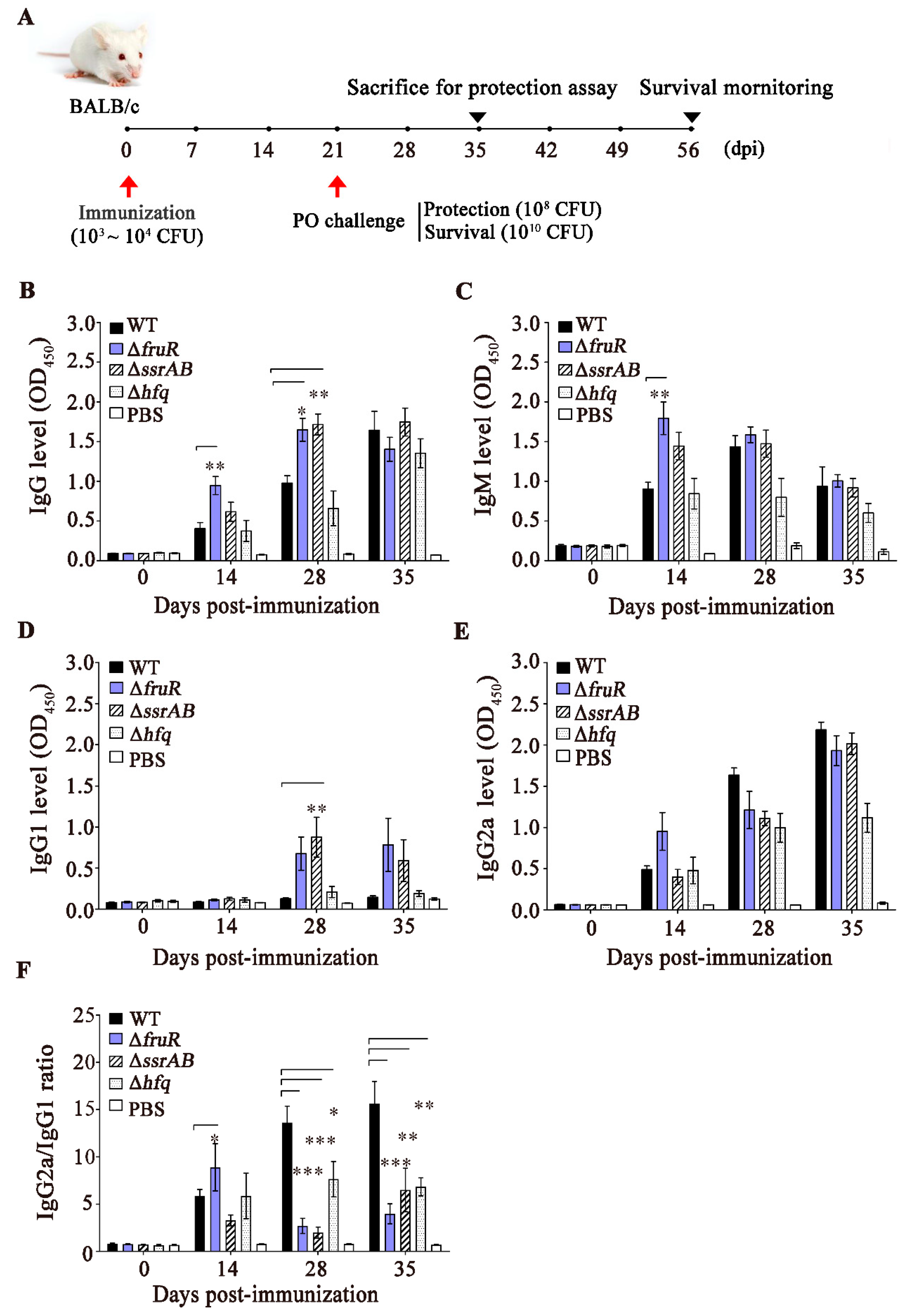
2.8. Mouse Immunization and Challenge Experiments
2.9. Enzyme-Linked Immunosorbent Assay (ELISA) Specific to S. Typhimurium OMPs
2.10. Statistical Analysis
3. Results
3.1. Attenuated Virulence of ΔfruR, ΔssrAB, and Δhfq Mutants in Mice Infection
3.2. The ΔfruR, ΔssrAB, and Δhfq Mutants Are Defective in Survival Inside Macrophages
3.3. Evaluation of the Immune Responses Induced by the ΔfruR, ΔssrAB, and Δhfq Mutants
3.4. Prophylactic Effect of the ΔfruR Mutant against Virulent Salmonella Infection in Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Briggs, A.M. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Angulo, F.J. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Wilairatana, P.; Mala, W.; Klangbud, W.K.; Kotepui, K.U.; Rattaprasert, P.; Kotepui, M. Prevalence, probability, and outcomes of typhoidal/non-typhoidal Salmonella and malaria co-infection among febrile patients: A systematic review and meta-analysis. Sci. Rep. 2021, 11, 21889. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Anderson, C.M.; Mosci, R.E.; Newton, D.W.; Lephart, P.; Salimnia, H.; Manning, S.D. Increasing frequencies of antibiotic resistant non-typhoidal Salmonella infections in Michigan and risk factors for disease. Front. Med. 2019, 6, 250. [Google Scholar] [CrossRef] [PubMed]
- Ingle, D.J.; Ambrose, R.L.; Baines, S.L.; Duchene, S.; Gonçalves da Silva, A.; Lee, D.Y.; Williamson, D.A. Evolutionary dynamics of multidrug resistant Salmonella enterica serovar 4,[5], 12:i:-in Australia. Nat. Commun. 2021, 12, 4786. [Google Scholar] [CrossRef]
- Manore, C.; Graham, T.; Carr, A.; Feryn, A.; Jakhar, S.; Mukundan, H.; Highlander, H.C. Modeling and cost benefit analysis to guide deployment of POC diagnostics for non-typhoidal Salmonella infections with antimicrobial resistance. Sci. Rep. 2019, 9, 11245. [Google Scholar] [CrossRef]
- Okpa, B.O.; Gberikon, G.M.; Aguoru, C.U.; Ogbonna, I.O. ESBL production and multidrug resistance of Salmonella serovars isolates in Benue State. Am. J. Mol. Biol. 2020, 10, 200. [Google Scholar] [CrossRef]
- Perera, S.R.; Sokaribo, A.S.; White, A.P. Polysaccharide vaccines: A perspective on non-typhoidal Salmonella. Polysaccharides 2021, 2, 691–714. [Google Scholar] [CrossRef]
- Piccini, G.; Montomoli, E. Pathogenic signature of invasive non-typhoidal Salmonella in Africa: Implications for vaccine development. Hum. Vaccin. Immunother. 2020, 16, 2056–2071. [Google Scholar] [CrossRef]
- Galen, J.E.; Wahid, R.; Buskirk, A.D. Strategies for enhancement of live-attenuated Salmonella-based carrier vaccine immunogenicity. Vaccines 2021, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Hensel, M. Systematic analysis of the SsrAB virulon of Salmonella enterica. Infect. Immun. 2010, 78, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, B.; Mulder, D.T.; Little, D.J.; Elhenawy, W.; Banda, M.M.; Pérez-Morales, D.; Coombes, B.K. Regulatory evolution drives evasion of host inflammasomes by Salmonella Typhimurium. Cell Rep. 2018, 25, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Sittka, A.; Lucchini, S.; Papenfort, K.; Sharma, C.M.; Rolle, K.; Binnewies, T.T.; Vogel, J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008, 4, e1000163. [Google Scholar] [CrossRef] [PubMed]
- Ansong, C.; Yoon, H.; Porwollik, S.; Mottaz-Brewer, H.; Petritis, B.O.; Jaitly, N.; Smith, R.D. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: Implications for virulence and global protein translation. PLoS ONE 2009, 4, e4809. [Google Scholar] [CrossRef]
- Saier, M.H.; Ramseier, T.M. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 1996, 178, 3411–3417. [Google Scholar] [CrossRef]
- Kim, D.; Seo, S.W.; Gao, Y.; Nam, H.; Guzman, G.I.; Cho, B.K.; Palsson, B.O. Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP. Nucleic Acids Res. 2018, 46, 2901–2917. [Google Scholar] [CrossRef]
- Tchawa Yimga, M.; Leatham, M.P.; Allen, J.H.; Laux, D.C.; Conway, T.; Cohen, P.S. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 2006, 74, 1130–1140. [Google Scholar] [CrossRef]
- Kim, S.; Kim, E.; Park, S.; Hahn, T.W.; Yoon, H. Genomic approaches for understanding the characteristics of Salmonella enterica subsp. enterica Serovar Typhimurium ST1120, isolated from swine feces in Korea. J. Microbiol. Biotechnol. 2017, 27, 1983–1993. [Google Scholar] [CrossRef]
- Yoon, H.; McDermott, J.E.; Porwollik, S.; McClelland, M.; Heffron, F. Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog. 2009, 5, e1000306. [Google Scholar] [CrossRef]
- Schmieger, H. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 1972, 119, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jung, B.; Kim, E.; Hong, S.T.; Yoon, H.; Hahn, T.W. Salmonella Typhimurium Lacking YjeK as a candidate live attenuated vaccine against invasive Salmonella infection. Front. Immunol. 2020, 11, 1277. [Google Scholar] [CrossRef]
- Rashid, M.H.; Kornberg, A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 4885–4890. [Google Scholar] [CrossRef] [PubMed]
- Barenkamp, S.J.; Munson, R.S.; Granoff, D.M. Subtyping isolates of Haemophilus influenzae type b by outer-membrane protein profiles. J. Infect. Dis. 1981, 143, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Kebede, A.; Kemal, J.; Alemayehu, H.; Habte Mariam, S. Isolation, identification, and antibiotic susceptibility testing of Salmonella from slaughtered bovines and ovines in Addis Ababa Abattoir Enterprise, Ethiopia: A cross-sectional study. Int. J. Bacteriol. 2016, 2016, 3714785. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Dogra, V.; Verma, S.; Singh, G.; Wani, A.H.; Chahota, R.; Dhar, P.; Sharma, M. Development of OMP based indirect ELISA to gauge the antibody titers in bovines against Pasteurella multocida. Iran. J. Vet. Res. 2015, 16, 350. [Google Scholar]
- Lin, H.; Zhou, H.; Gao, L.; Li, B.; He, K.; Fan, H. Development and application of an indirect ELISA for the detection of antibodies to porcine epidemic diarrhea virus based on a recombinant spike protein. BMC Vet. Res. 2018, 14, 243. [Google Scholar] [CrossRef]
- Inoue, T.; Parida, S.; Paton, D.J.; Linchongsubongkoch, W.; Mackay, D.; Oh, Y.; Saeki, T. Development and evaluation of an indirect enzyme-linked immunosorbent assay for detection of foot-and-mouth disease virus nonstructural protein antibody using a chemically synthesized 2B peptide as antigen. J. Vet. Diagn. Invest. 2006, 18, 545–552. [Google Scholar] [CrossRef]
- Tam, M.A.; Rydström, A.; Sundquist, M.; Wick, M.J. Early cellular responses to Salmonella infection: Dendritic cells, monocytes, and more. Immunol. Rev. 2008, 225, 140–162. [Google Scholar] [PubMed]
- Jackson, A.; Nanton, M.R.; O’Donnell, H.; Akue, A.D.; McSorley, S.J. Innate immune activation during Salmonella infection initiates extramedullary erythropoiesis and splenomegaly. J. Immunol. 2010, 185, 6198–6204. [Google Scholar] [PubMed] [Green Version]
- Confer, A.W.; Ayalew, S. The OmpA family of proteins: Roles in bacterial pathogenesis and immunity. Vet. Microbiol. 2013, 163, 207–222. [Google Scholar] [PubMed]
- Choi, U.; Lee, C.R. Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front. Microbiol. 2019, 10, 953. [Google Scholar] [PubMed]
- Mori, N.; Szvalb, A.D.; Adachi, J.A.; Tarrand, J.J.; Mulanovich, V.E. Clinical presentation and outcomes of non-typhoidal Salmonella infections in patients with cancer. BMC Infect. Dis. 2021, 21, 1021. [Google Scholar]
- Tapia, M.D.; Tennant, S.M.; Bornstein, K.; Onwuchekwa, U.; Tamboura, B.; Maiga, A.; Levine, M.M. Invasive nontyphoidal Salmonella infections among children in Mali, 2002–2014: Microbiological and epidemiologic features guide vaccine development. Clin. Infect. Dis. 2015, 61, S332–S338. [Google Scholar]
- Kingsley, R.A.; Msefula, C.L.; Thomson, N.R.; Kariuki, S.; Holt, K.E.; Gordon, M.A.; Dougan, G. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009, 19, 2279–2287. [Google Scholar]
- Mather, A.E.; Phuong, T.L.T.; Gao, Y.; Clare, S.; Mukhopadhyay, S.; Goulding, D.A.; Baker, S. New variant of multidrug-resistant Salmonella enterica serovar Typhimurium associated with invasive disease in immunocompromised patients in Vietnam. MBio 2018, 9, e01056-18. [Google Scholar]
- Stanaway, J.D.; Parisi, A.; Sarkar, K.; Blacker, B.F.; Reiner, R.C.; Hay, S.I.; Crump, J.A. The global burden of non-typhoidal salmonella invasive disease: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 1312–1324. [Google Scholar]
- Mastroeni, P.; Rossi, O. Immunology, epidemiology and mathematical modelling towards a better understanding of invasive non-typhoidal Salmonella disease and rational vaccination approaches. Expert Rev. Vaccines 2016, 15, 1545–1555. [Google Scholar]
- Van Immerseel, F.; Methner, U.; Rychlik, I.; Nagy, B.; Velge, P.; Martin, G.; Barrow, P.A. Vaccination and early protection against non-host-specific Salmonella serotypes in poultry: Exploitation of innate immunity and microbial activity. Epidemiol. Infect. 2005, 133, 959–978. [Google Scholar] [PubMed]
- Hindle, Z.; Chatfield, S.N.; Phillimore, J.; Bentley, M.; Johnson, J.; Cosgrove, C.A.; Lewis, D.J. Characterization of Salmonella enterica derivatives harboring defined aroC and Salmonella pathogenicity island 2 type III secretion system (ssaV) mutations by immunization of healthy volunteers. Infect. Immun. 2002, 70, 3457–3467. [Google Scholar] [PubMed]
- Galán, J.E.; Curtiss, R., III. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb. Pathog. 1989, 6, 433–443. [Google Scholar] [PubMed]
- Na, H.S.; Kim, H.J.; Lee, H.-C.; Hong, Y.; Rhee, J.H.; Choy, H.E. Immune response induced by Salmonella typhimurium defective in ppGpp synthesis. Vaccine 2006, 24, 2027–2034. [Google Scholar] [PubMed]
- Laughlin, R.C.; Knodler, L.A.; Barhoumi, R.; Payne, H.R.; Wu, J.; Gomez, G.; Adams, L.G. Spatial segregation of virulence gene expression during acute enteric infection with Salmonella enterica serovar Typhimurium. MBio 2014, 5, e00946-13. [Google Scholar] [PubMed] [Green Version]
- Gore, A.L.; Payne, S.M. CsrA and Cra influence Shigella flexneri pathogenesis. Infect. Immun. 2010, 78, 4674–4682. [Google Scholar]
- Kuo, C.J.; Wang, S.T.; Lin, C.M.; Chiu, H.C.; Huang, C.R.; Lee, D.Y.; Chen, C.S. A multi-omic analysis reveals the role of fumarate in regulating the virulence of enterohemorrhagic Escherichia coli. Cell Death Dis. 2018, 9, 381. [Google Scholar]
- Hughes, E.A.; Galán, J.E. Immune response to Salmonella: Location, location, location? Immunity 2002, 16, 325–328. [Google Scholar]
- Truong, Q.L.; Cho, Y.; Park, S.; Park, B.K.; Hahn, T.W. Brucella abortus mutants lacking ATP-binding cassette transporter proteins are highly attenuated in virulence and confer protective immunity against virulent B. abortus challenge in BALB/c mice. Microb. Pathog. 2016, 95, 175–185. [Google Scholar]

| Strains | Infection Dose (CFU/Mouse) | Number of Dead Mice/ Total Mice Number | LD50 (CFU/Dose) |
|---|---|---|---|
| Wild-type ST1120 | 105 | 3/4 | 103.1 |
| 104 | 3/4 | ||
| 103 | 0/4 | ||
| 102 | 0/4 | ||
| ΔfruR | 107 | 4/4 | 105.3 |
| 106 | 4/4 | ||
| 105 | 0/4 | ||
| 104 | 0/4 | ||
| ΔssrAB | 107 | 3/4 | 106.5 |
| 106 | 1/4 | ||
| 105 | 0/4 | ||
| 104 | 0/4 | ||
| Δhfq | 107 | 4/4 | 106.5 |
| 106 | 0/4 | ||
| 105 | 0/4 | ||
| 104 | 0/4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Jung, B.; Kim, E.; Yoon, H.; Hahn, T.-W. Evaluation of Salmonella Typhimurium Lacking fruR, ssrAB, or hfq as a Prophylactic Vaccine against Salmonella Lethal Infection. Vaccines 2022, 10, 1413. https://doi.org/10.3390/vaccines10091413
Park S, Jung B, Kim E, Yoon H, Hahn T-W. Evaluation of Salmonella Typhimurium Lacking fruR, ssrAB, or hfq as a Prophylactic Vaccine against Salmonella Lethal Infection. Vaccines. 2022; 10(9):1413. https://doi.org/10.3390/vaccines10091413
Chicago/Turabian StylePark, Soyeon, Bogyo Jung, Eunsuk Kim, Hyunjin Yoon, and Tae-Wook Hahn. 2022. "Evaluation of Salmonella Typhimurium Lacking fruR, ssrAB, or hfq as a Prophylactic Vaccine against Salmonella Lethal Infection" Vaccines 10, no. 9: 1413. https://doi.org/10.3390/vaccines10091413
APA StylePark, S., Jung, B., Kim, E., Yoon, H., & Hahn, T.-W. (2022). Evaluation of Salmonella Typhimurium Lacking fruR, ssrAB, or hfq as a Prophylactic Vaccine against Salmonella Lethal Infection. Vaccines, 10(9), 1413. https://doi.org/10.3390/vaccines10091413






