Antimicrobial Activity of Cathelicidin-Derived Peptide from the Iberian Mole Talpa occidentalis
Abstract
1. Introduction
2. Materials and Methods
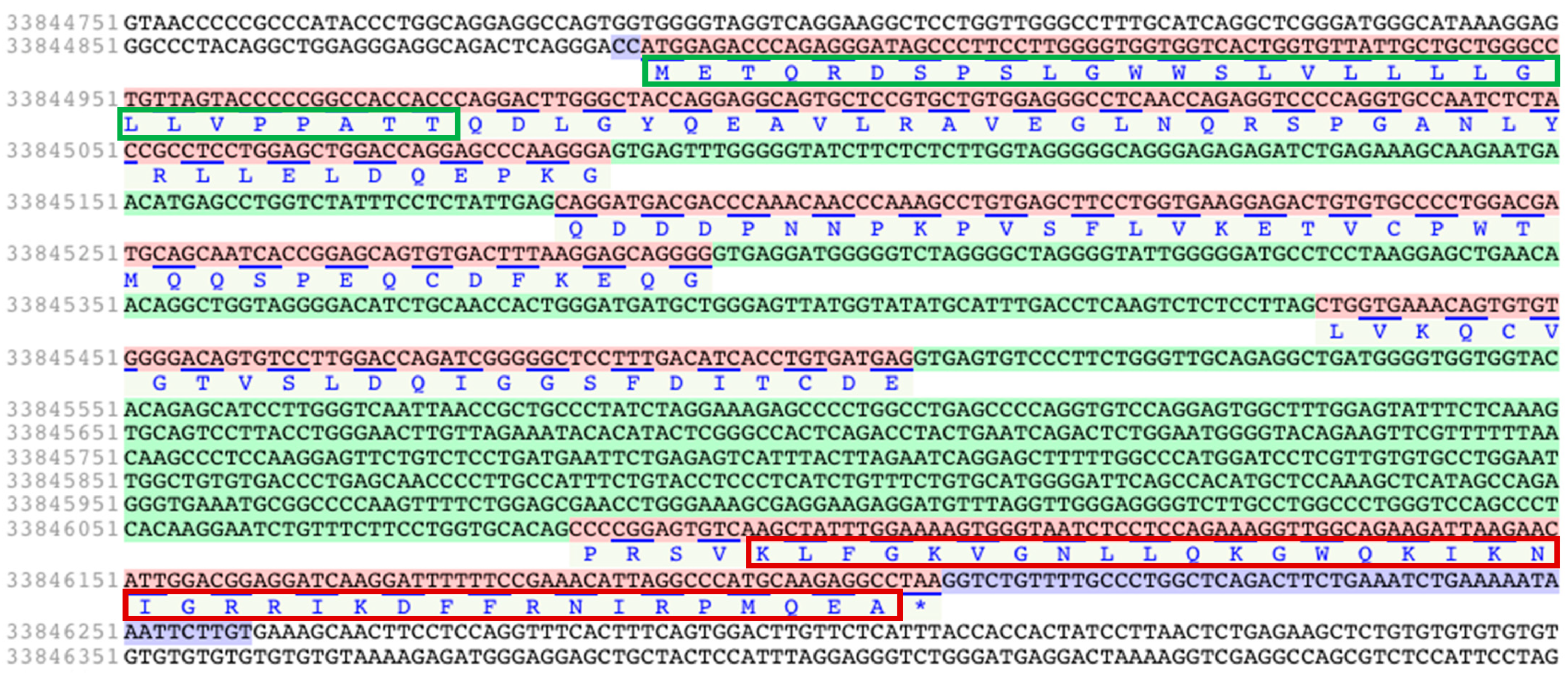
2.1. Retrieval of Iberian Mole Cathelicidin Sequences
2.2. In Silico Analyses of Physicochemical Properties of Peptides
2.3. Multiple Alignment and Phylogenetic Tree
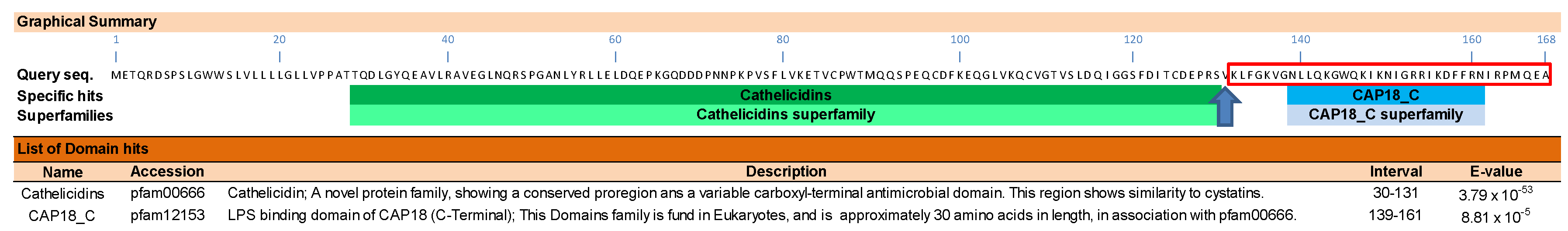
2.4. In Silico Analysis of Biological Activity
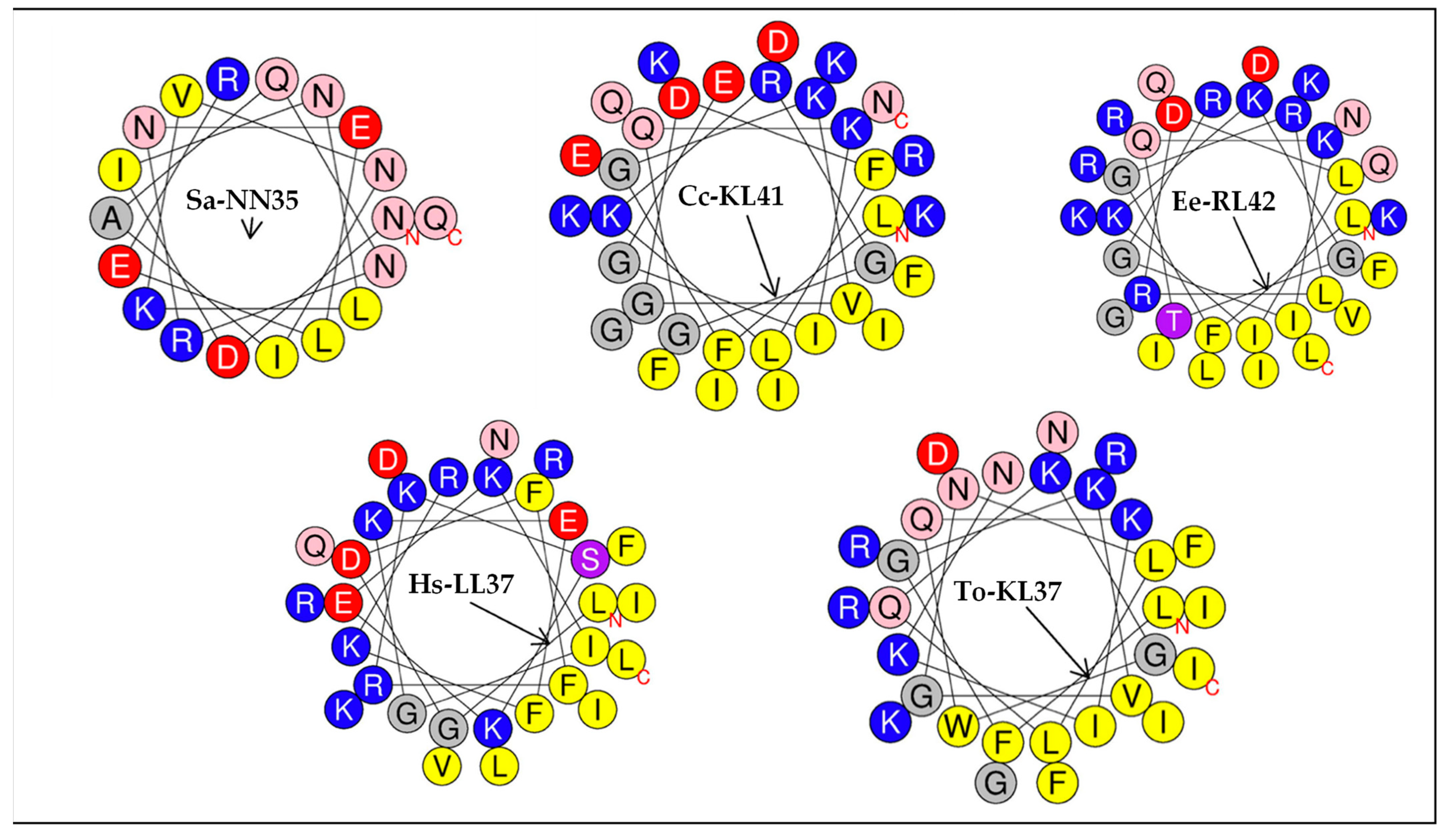
2.5. Structural Analysis of Peptides
2.6. Modeling of Peptides
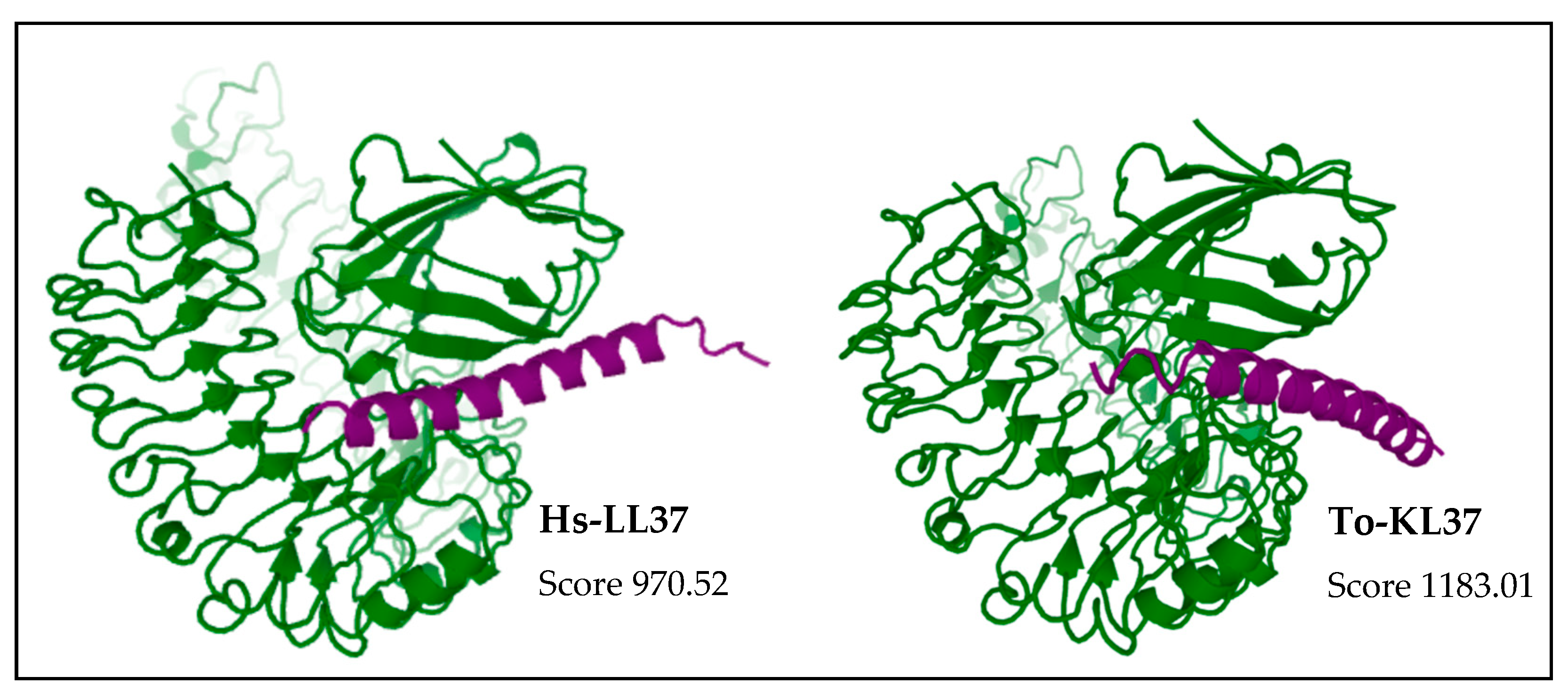
2.7. Molecular Docking Studies with TLR4/MD2 Complex
2.8. Peptide Synthesis
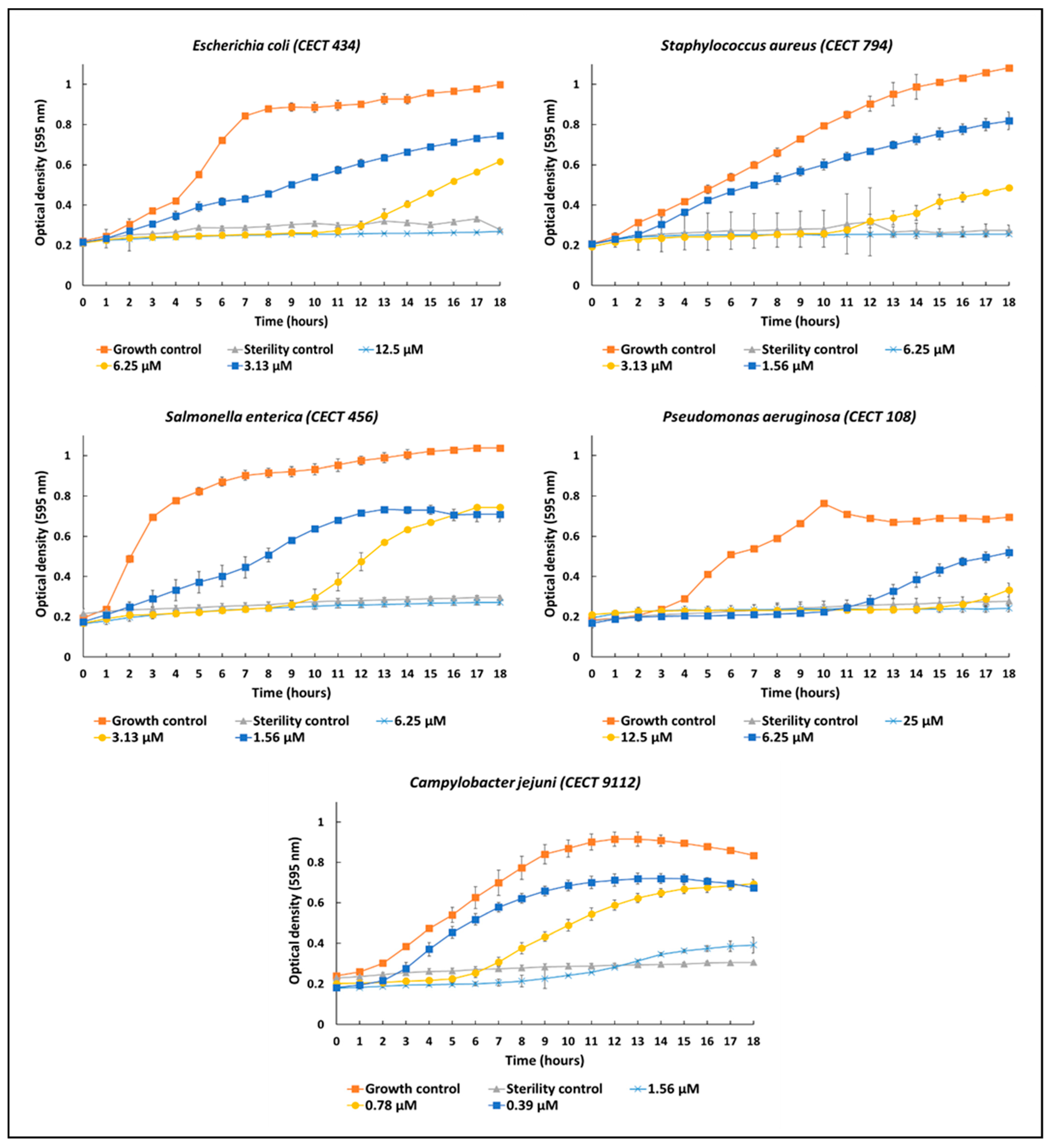
2.9. Antimicrobial Activity In Vitro
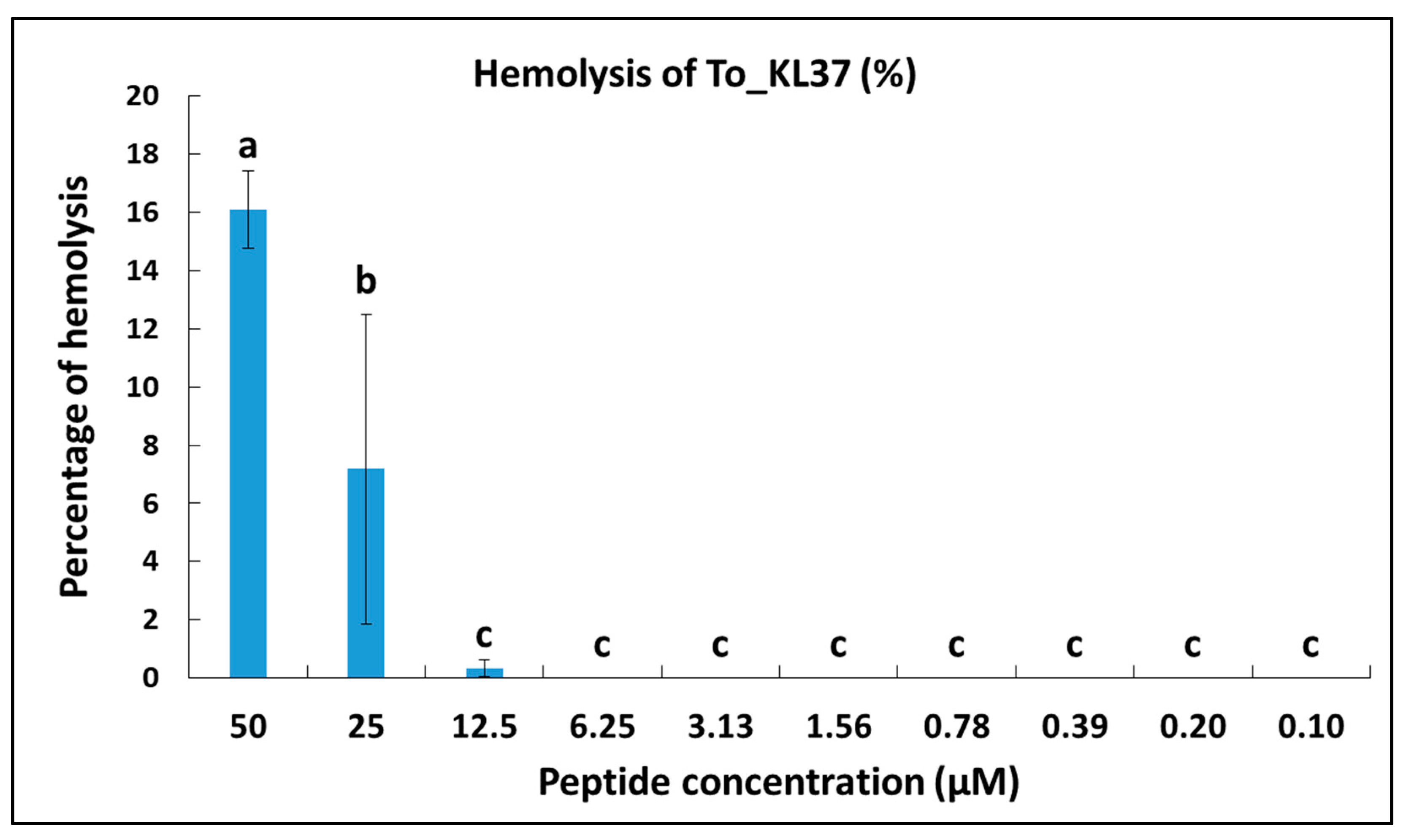
2.10. Hemolytic Activity
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boto, A.; Pérez de la Lastra, J.M.; González, C.C. The road from host-defense peptides to a new generation of antimicrobial drugs. Molecules 2018, 23, 311. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh Moghaddam, M.; Mirnejad, R. Antimicrobial peptides: Features, action, and their resistance mechanisms in bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Bhatia, G.; Sharma, A.; Saxena, S. Host defense peptides: An insight into the antimicrobial world. J. Oral Maxillofac. Pathol. JOMFP 2018, 22, 239. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Vishnepolsky, B.; Grigolava, M.; Managadze, G. Physicochemical Features and Peculiarities of Interaction of AMP with the Membrane. Pharmaceuticals 2021, 14, 471. [Google Scholar] [CrossRef]
- Young-Speirs, M.; Drouin, D.; Cavalcante, P.A.; Barkema, H.W.; Cobo, E.R. Host defense cathelicidins in cattle: Types, production, bioactive functions and potential therapeutic and diagnostic applications. Int. J. Antimicrob. Agents 2018, 51, 813–821. [Google Scholar] [CrossRef]
- Van Harten, R.M.; Van Woudenbergh, E.; Van Dijk, A.; Haagsman, H.P. Cathelicidins: Immunomodulatory antimicrobials. Vaccines 2018, 6, 63. [Google Scholar] [CrossRef]
- Chakraborty, S.; Chatterjee, R.; Chakravortty, D. Evolving and assembling to pierce through: Evolutionary and Structural Aspects of Antimicrobial Peptides. Comput. Struct. Biotechnol. J. 2022, 20, 2247–2258. [Google Scholar] [CrossRef]
- D’Agostino, J. Insectivores (Insectivora, Macroscelidea, Scandentia). In Fowler’s Zoo and Wild Animal Medicine; Elsevier Health Sciences, 2014; Volume 8, pp. 275–281. [Google Scholar] [CrossRef]
- Barrionuevo, F.J.; Zurita, F.; Burgos, M.; Jiménez, R. Developmental Stages and Growth Rate of the Mole Talpa occidentals (Insectivora, Mammalia). J. Mammal. 2004, 85, 120–125. [Google Scholar] [CrossRef]
- Douady, C.; Douzery, E. Hedgehogs, shrews, moles, and solenodons (Eulipotyphla). In Timetree of Life; Hedges, S.B., Kumar, S., Eds.; Oxford University Press: Oxford, UK, 2009; pp. 495–498. [Google Scholar]
- Bowdish, D.; Davidson, D.; Hancock, R. Immunomodulatory properties of defensins and cathelicidins. Antimicrob. Pept. Hum. Dis. 2006, 306, 27–66. [Google Scholar] [CrossRef]
- Souvorov, A.; Kapustin, Y.; Kiryutin, B.; Chetvernin, V.; Tatusova, T.; Lipman, D. Gnomon–NCBI eukaryotic gene prediction tool. Natl. Cent. Biotechnol. Inf. 2010, 1–24. Available online: https://www.ncbi.nlm.nih.gov/genome/annotation_euk/gnomon (accessed on 9 May 2022).
- Marchler-Bauer, A.; Zheng, C.; Chitsaz, F.; Derbyshire, M.K.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Lanczycki, C.J. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2012, 41, D348–D352. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607 . [Google Scholar] [CrossRef]
- Nielsen, H.; Tsirigos, K.D.; Brunak, S.; von Heijne, G. A Brief History of Protein Sorting Prediction. Protein J. 2019, 38, 200–216. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Desper, R.; Gascuel, O. Theoretical foundation of the balanced minimum evolution method of phylogenetic inference and its relationship to weighted least-squares tree fitting. Mol. Biol. Evol. 2004, 21, 587–598. [Google Scholar] [CrossRef]
- Grishin, N.V. Estimation of the number of amino acid substitutions per site when the substitution rate varies among sites. J. Mol. Evol. 1995, 41, 675–679. [Google Scholar] [CrossRef][Green Version]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef]
- Waghu, F.H.; Idicula-Thomas, S. Collection of antimicrobial peptides database and its derivatives: Applications and beyond. Protein Sci. 2020, 29, 36–42. [Google Scholar] [CrossRef]
- Thakur, N.; Qureshi, A.; Kumar, M. AVPpred: Collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. 2012, 40, W199–W204. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Chiangjong, W.; Lee, V.S.; Nantasenamat, C.; Hasan, M.; Shoombuatong, W. Improved prediction and characterization of anticancer activities of peptides using a novel flexible scoring card method. Sci. Rep. 2021, 11, 3017. [Google Scholar]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; Von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the 6th International Conference on Intelligent Systems for Molecular Biology, Montreal, QC, Canada, 28 June–1 July 1998; pp. 175–182. [Google Scholar]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold-Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef]
- Lomize, A.L.; Todd, S.C.; Pogozheva, I.D. Spatial arrangement of proteins in planar and curved membranes by PPM 3.0. Protein Sci. 2022, 31, 209–220. [Google Scholar] [CrossRef]
- Park, T.; Baek, M.; Lee, H.; Seok, C. GalaxyTongDock: Symmetric and asymmetric ab initio protein–protein docking web server with improved energy parameters. J. Comput. Chem. 2019, 40, 2413–2417. [Google Scholar] [CrossRef]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The minimum inhibitory concentration of antibiotics: Methods, interpretation, clinical relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef]
- Zanetti, M. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 2004, 75, 39–48. [Google Scholar] [CrossRef]
- Tomasinsig, L.; Zanetti, M. The cathelicidins-structure, function and evolution. Curr. Protein Pept. Sci. 2005, 6, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Pérez de la Lastra, J.M.; Asensio-Calavia, P.; González-Acosta, S.; Baca-González, V.; Morales-delaNuez, A. Bioinformatic Analysis of Genome-Predicted Bat Cathelicidins. Molecules 2021, 26, 1811. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Pirri, G.; Rinaldi, A.C. Antimicrobial peptides: The LPS connection. In Antimicrobial Peptides; Springer: Cham, Switzerland, 2010; pp. 137–154. [Google Scholar] [CrossRef]
- Scheenstra, M.R.; Van Harten, R.M.; Veldhuizen, E.J.; Haagsman, H.P.; Coorens, M. Cathelicidins modulate TLR-activation and inflammation. Front. Immunol. 2020, 11, 1137. [Google Scholar] [CrossRef] [PubMed]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A new era of antibiotics: The clinical potential of antimicrobial peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Sahl, H.-G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef]
- Ageitos, J.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef]
- Toke, O. Antimicrobial peptides: New candidates in the fight against bacterial infections. Pept. Sci. Orig. Res. Biomol. 2005, 80, 717–735. [Google Scholar] [CrossRef]
- Bedford, J.M.; Mock, O.B.; Goodman, S.M. Novelties of conception in insectivorous mammals (Lipotyphla), particularly shrews. Biol. Rev. 2004, 79, 891–909. [Google Scholar] [CrossRef]
- Furio, M.; Ostende, L.V.D.H.; Agusti, J.; Minwer-Barakat, R. Evolution of the insectivore assemblages (Eulipotyphla, Mammalia) in Spain and their relation with Neogene and Quaternary climatic changes. Ecosistemas 2018, 27, 38–51. [Google Scholar]
- Lukyanova, L.; Ukhova, N.; Ukhova, O.; Gorodilova, Y.V. Common shrew (Sorex araneus, Eulipotyphla) population and the food supply of its habitats in ecologically contrasting environments. Russ. J. Ecol. 2021, 52, 316–328. [Google Scholar] [CrossRef]
- Matei, I.A.; D’Amico, G.; Ionică, A.M.; Kalmár, Z.; Corduneanu, A.; Sándor, A.D.; Fiţ, N.; Bogdan, L.; Gherman, C.M.; Mihalca, A.D. New records for Anaplasma phagocytophilum infection in small mammal species. Parasites Vectors 2018, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M. The role of cathelicidins in the innate host defenses of mammals. Curr. Issues Mol. Biol. 2005, 7, 179–196. [Google Scholar] [PubMed]
- Cho, H.-s.; Soundrarajan, N.; Le Van Chanh, Q.; Jeon, H.; Cha, S.-Y.; Kang, M.; Ahn, B.; Hong, K.; Song, H.; Kim, J.-H. The novel cathelicidin of naked mole rats, Hg-CATH, showed potent antimicrobial activity and low cytotoxicity. Gene 2018, 676, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Narayana, J.L.; Mishra, B.; Zhang, Y.; Wang, F.; Wang, C.; Zarena, D.; Lushnikova, T.; Wang, X. Design of antimicrobial peptides: Progress made with human cathelicidin LL-37. Antimicrob. Pept. 2019, 215–240. [Google Scholar]
- Nagaoka, I.; Tamura, H.; Reich, J. Therapeutic potential of cathelicidin peptide LL-37, an antimicrobial agent, in a murine sepsis model. Int. J. Mol. Sci. 2020, 21, 5973. [Google Scholar] [CrossRef]
- Oliveira, N.G.; Cardoso, M.H.; Velikova, N.; Giesbers, M.; Wells, J.M.; Rezende, T.; de Vries, R.; Franco, O.L. Physicochemical-guided design of cathelicidin-derived peptides generates membrane active variants with therapeutic potential. Sci. Rep. 2020, 10, 9127. [Google Scholar] [CrossRef]
- Shen, J.; Xiao, Z. Cathelicidin in Gastrointestinal Disorders. In Antimicrobial Peptides in Gastrointestinal Diseases; Elsevier: Amsterdam, The Netherlands, 2018; pp. 61–76. [Google Scholar]
- Pérez de Lastra, J.M.; Garrido-Orduña, C.; Borges, L.; Tejera, D.; Marino, M.; González-Guerra, E.; Borges, A.A. Antimicrobial activity of cathelicidins of mammals from avian, aquatic and terrestrial environments. In Antimicrobial Research: Novel Bioknowledge and Educational Programs; Mendez-Vilas, A., Ed.; Formatex: Badajoz, Spain, 2017; pp. 9–18. [Google Scholar]
- Turner, J.; Cho, Y.; Dinh, N.-N.; Waring, A.J.; Lehrer, R.I. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 1998, 42, 2206–2214. [Google Scholar] [CrossRef]
- Rico-Mata, R.; De Leon-Rodriguez, L.M.; Avila, E.E. Effect of antimicrobial peptides derived from human cathelicidin LL-37 on Entamoeba histolytica trophozoites. Exp. Parasitol. 2013, 133, 300–306. [Google Scholar] [CrossRef]
- Luo, X.-L.; Li, J.-X.; Huang, H.-R.; Duan, J.-L.; Dai, R.-X.; Tao, R.-J.; Yang, L.; Hou, J.-y.; Jia, X.-M.; Xu, J.-F. LL37 inhibits Aspergillus fumigatus infection via directly binding to the fungus and preventing excessive inflammation. Front. Immunol. 2019, 10, 283. [Google Scholar] [CrossRef]
- Stepanova, I.; Andreychev, A.; Kulakhmetov, R.; Lobachev, E. Commensals of underground mammals: European mole (Talpa europaea, Eulipotyphla, Talpidae) and the greater mole-rat (Spalax microphthalmus, Rodentia, Spalacidae). Biodiversitas J. Biol. Divers. 2021, 22. [Google Scholar] [CrossRef]
- Humphrey, T.; O’Brien, S.; Madsen, M. Campylobacters as zoonotic pathogens: A food production perspective. Int. J. Food Microbiol. 2007, 117, 237–257. [Google Scholar] [CrossRef] [PubMed]
- Kerkhof, P.-J.; Peruzy, M.F.; Murru, N.; Houf, K. Wild boars as reservoir for Campylobacter and Arcobacter. Vet. Microbiol. 2022, 270, 109462. [Google Scholar] [CrossRef] [PubMed]
- Coorens, M.; Scheenstra, M.R.; Veldhuizen, E.J.; Haagsman, H.P. Interspecies cathelicidin comparison reveals divergence in antimicrobial activity, TLR modulation, chemokine induction and regulation of phagocytosis. Sci. Rep. 2017, 7, 40874. [Google Scholar] [CrossRef] [PubMed]



| Specie | Accession | Name | Predicted Peptide Sequence |
|---|---|---|---|
| Sorex araneus | XP_004615204 | Sa-NN35 | NNDINLKVNIAQNRNELERQDPNICRRKRHKTFPM |
| Erinaceus europaeus | XP_007533667 | Ee-RL42 | RLFGRLRDLIKKGTQKIGRKLRKVGQQIKDFIRNLRPREEDS |
| Condylura cristata | XP_012590677 | Cc-KL41 | KLFGKVGDFLKRGGQKIGEKIEKIGKRIKDFFQNLKPREEA |
| Talpa occidentalis | XP_037384309 | To-KL37 | KLFGKVGNLLQKGWQKIKNIGRRIKDFFRNIRPMQEA |
| Homo sapiens | PDB: 5NNT_A | Hs-LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES |
| Peptide | Hydrophobicity <H> | Net Charge z | Hydrophobic Moment <µH> | Hydrophobic Face |
|---|---|---|---|---|
| Ee-RL42 | 0.205 | 9 | 0.668 | LVILIIFL |
| Sa-NN35 | 0.002 | 0 | 0.143 | LLI |
| Cc-KL41 | 0.187 | 5 | 0.627 | VIILIFI |
| To-KL37 | 0.338 | 7 | 0.683 | LIGIVIILFFGW |
| Hs-LL37 | 0.174 | 6 | 0.693 | LIILFIF |
| Peptide | Position of Embedded Residues | Tilt Angle | Image in Membrane |
|---|---|---|---|
| Ee-RL42 | 1–7,9–10,14,17,21,28,31–32,35,37 | 86 |  |
| Sa-NN35 | 33–35 | 67 |  |
| Cc-KL41 | 1–7,9–10,17,21,32,38,40–41 | 84 | 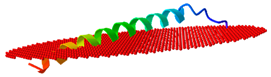 |
| To-KL37 | 2–3,6,9–10,14,17,20,24,27–28,31–32,34–35,37 | 89 | 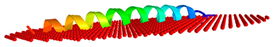 |
| Hs-LL37 | 1–7,9–10,13,17,20–21,24,27–28,31–33 | 85 | 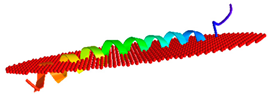 |
| Peptide | Antimicrobial | Antifungal | Antiviral | Anticancer |
|---|---|---|---|---|
| Ee-RL42 | 0.90 | No | 73.58% | No |
| Sa-NN35 | 0.54 | No | No | No |
| Cc-KL41 | 0.80 | No | 76.77% | 0.98 |
| To-KL37 | 0.95 | No | 80.66% | 0.89 |
| Hs-LL37 | 0.76 | No | 78.13% | 0.99 |
| Peptide | Escherichia coli ATCC 25922 | Pseudomonas aeruginosa ATCC 27853 | Klebsiella pneumoniae * | Staphylococcus aureus ATCC 25923 | Bacilus subtilis * | Hemolytic |
|---|---|---|---|---|---|---|
| Ee-RL42 | Active | Active | Not Active | Not Active | Not Active | Not Active |
| Sa-NN35 | Not active | Not active | Not active | Not active | Not active | Not active |
| Cc-KL41 | Active | Active | Not Active | Not Active | Not Active | Active |
| To-KL37 | Active | Active | Active | Active | Active | Not Active |
| Hs-LL37 | Active | Active | Not Active | Not Active | Not Active | Not Active |
| Microorganisms | MIC Ranges (µM) |
|---|---|
| Staphylococcus aureus (CECT 794) | 6.25–3.13 |
| Escherichia coli (CECT 434) | 12.5–6.25 |
| Salmonella enterica (CECT 456) | 6.25–3.13 |
| Pseudomonas aeruginosa (CECT 108) | 25–12.5 |
| Campylobacter jejuni (CECT 9112) | 1.56–0.78 |
| Microorganism | E. coli | S. aureus | S. enterica | P. aeruginosa | C. jejuni |
|---|---|---|---|---|---|
| Growth after 24 h | Negative | Negative | Negative | Positive | Positive |
| Peptide activity | Bactericide | Bactericide | Bactericide | Bacteriostatic | Bacteriostatic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otazo-Pérez, A.; Asensio-Calavia, P.; González-Acosta, S.; Baca-González, V.; López, M.R.; Morales-delaNuez, A.; Pérez de la Lastra, J.M. Antimicrobial Activity of Cathelicidin-Derived Peptide from the Iberian Mole Talpa occidentalis. Vaccines 2022, 10, 1105. https://doi.org/10.3390/vaccines10071105
Otazo-Pérez A, Asensio-Calavia P, González-Acosta S, Baca-González V, López MR, Morales-delaNuez A, Pérez de la Lastra JM. Antimicrobial Activity of Cathelicidin-Derived Peptide from the Iberian Mole Talpa occidentalis. Vaccines. 2022; 10(7):1105. https://doi.org/10.3390/vaccines10071105
Chicago/Turabian StyleOtazo-Pérez, Andrea, Patricia Asensio-Calavia, Sergio González-Acosta, Victoria Baca-González, Manuel R. López, Antonio Morales-delaNuez, and José Manuel Pérez de la Lastra. 2022. "Antimicrobial Activity of Cathelicidin-Derived Peptide from the Iberian Mole Talpa occidentalis" Vaccines 10, no. 7: 1105. https://doi.org/10.3390/vaccines10071105
APA StyleOtazo-Pérez, A., Asensio-Calavia, P., González-Acosta, S., Baca-González, V., López, M. R., Morales-delaNuez, A., & Pérez de la Lastra, J. M. (2022). Antimicrobial Activity of Cathelicidin-Derived Peptide from the Iberian Mole Talpa occidentalis. Vaccines, 10(7), 1105. https://doi.org/10.3390/vaccines10071105







