Increased Receptor Affinity and Reduced Recognition by Specific Antibodies Contribute to Immune Escape of SARS-CoV-2 Variant Omicron
Abstract
:1. Introduction
- (1)
- the transmissibility of the variant,
- (2)
- how well vaccinated and infected people are protected against Omicron,
- (3)
- the virulence of Omicron, and finally
- (4)
- the awareness of the population to take protective measures [2]. In light of the importance of these key parameters, our study aimed at revealing the mechanisms underlying the overserved increased transmissibility and reduced protection by a vaccine- or infection-induced antibodies. Our data demonstrate an increased affinity of Omicron for its receptor ACE2 and reduced recognition by serum antibodies. These factors cause reduced viral neutralization. From a mechanistic point of view, our data demonstrate that Omicron avoids viral neutralization by both classical change of antibody specificity and affinity escape due to the high RBD-ACE2 affinity that outcompetes antibody binding.
2. Materials and Methods
2.1. Human Sera
2.2. RBD-ACE2 Binding Kinetics
2.3. RBD Proteins
2.4. Anti-RBD Titers
2.5. BLI-Based Competitive Assay
2.6. Neutralization Assay (Cytopathic Effect-Based Neutralization Assay)
2.7. Statistics
3. Results
3.1. Omicron VOC Accumulated Many More Mutations than Delta Variant
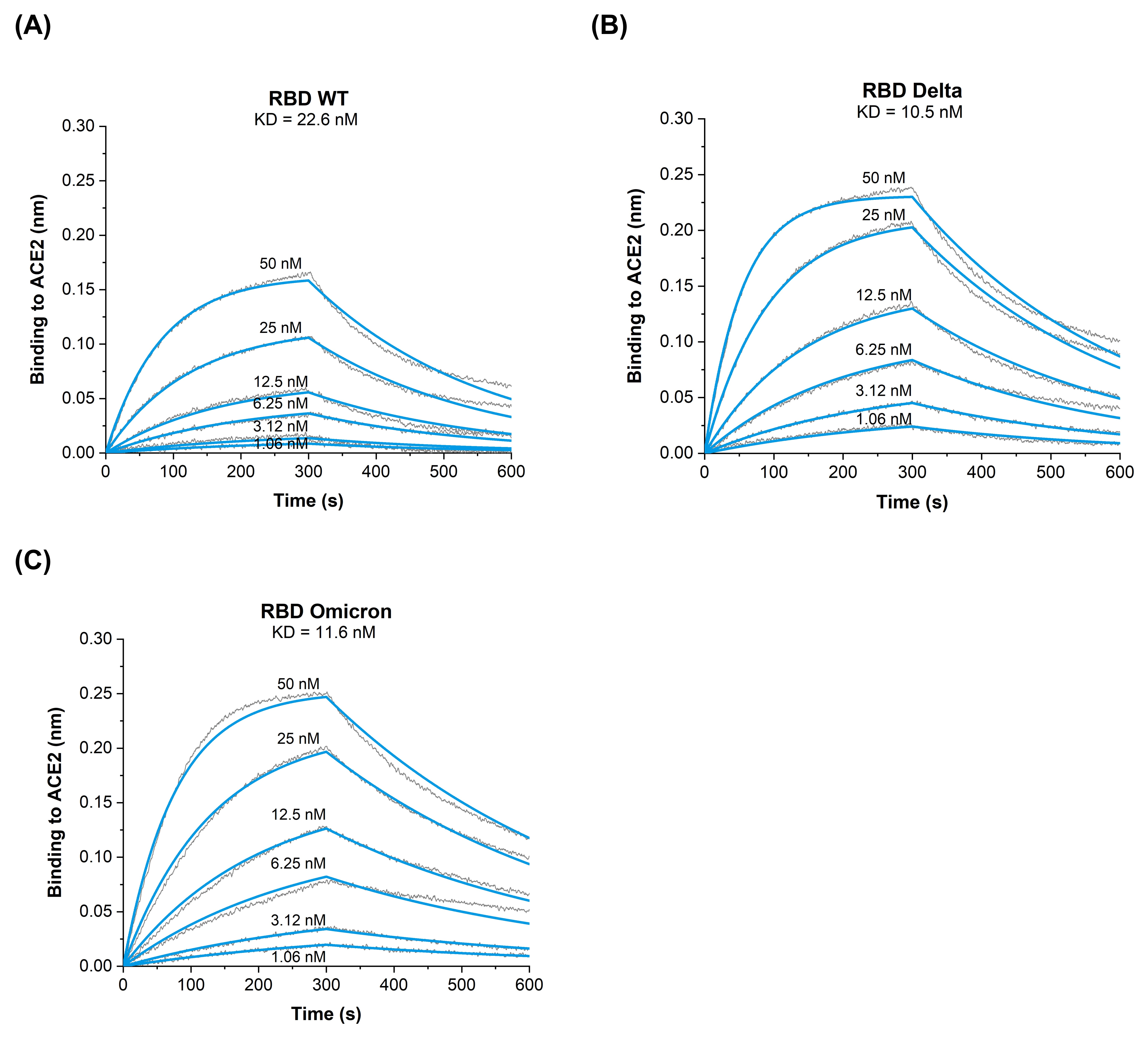
3.2. Binding Kinetics of RBD Omicron (B.1.1.529) to ACE2 Reveal Similar Affinities as RBD Delta (B.1.617.2) VOC
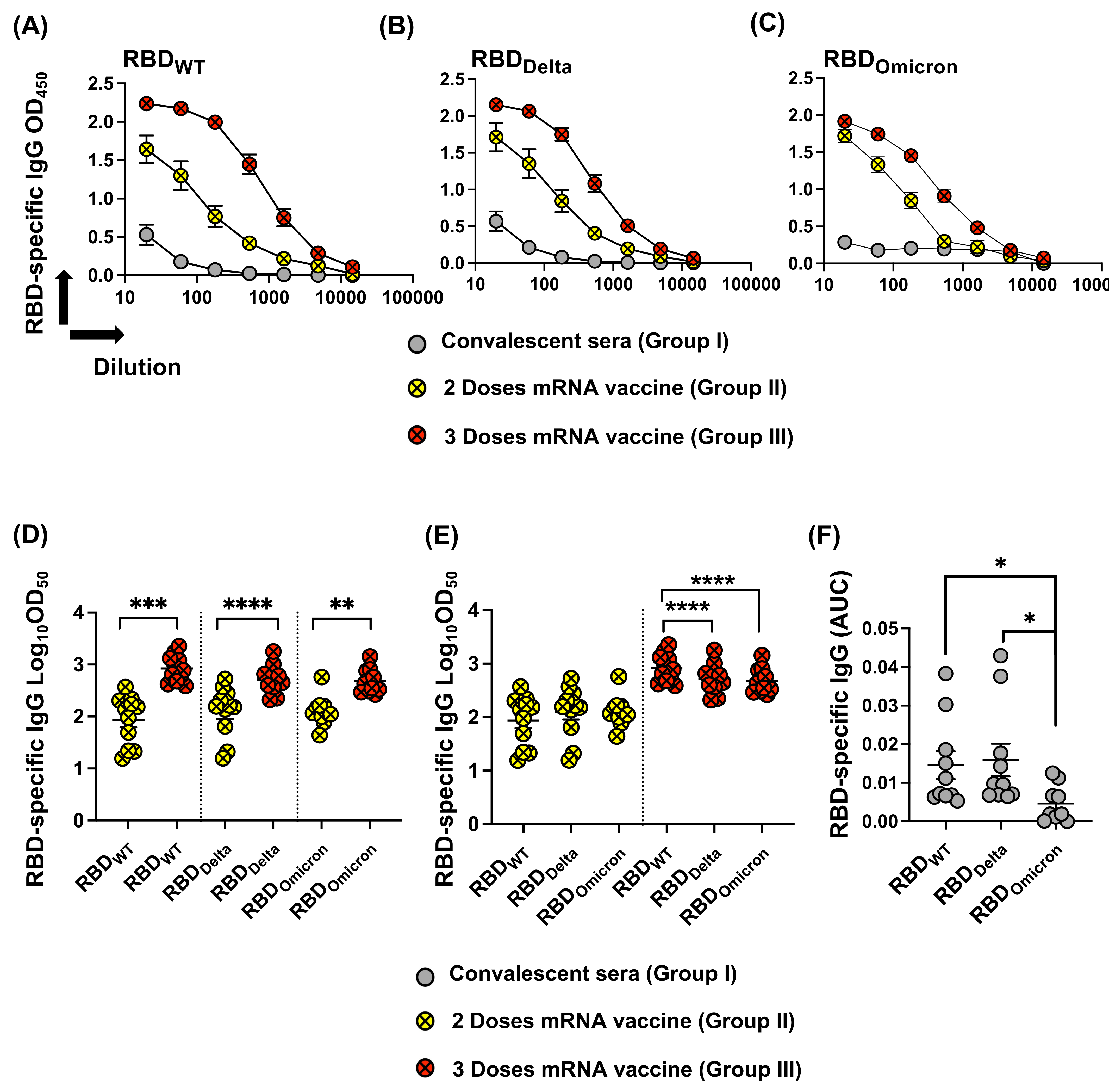
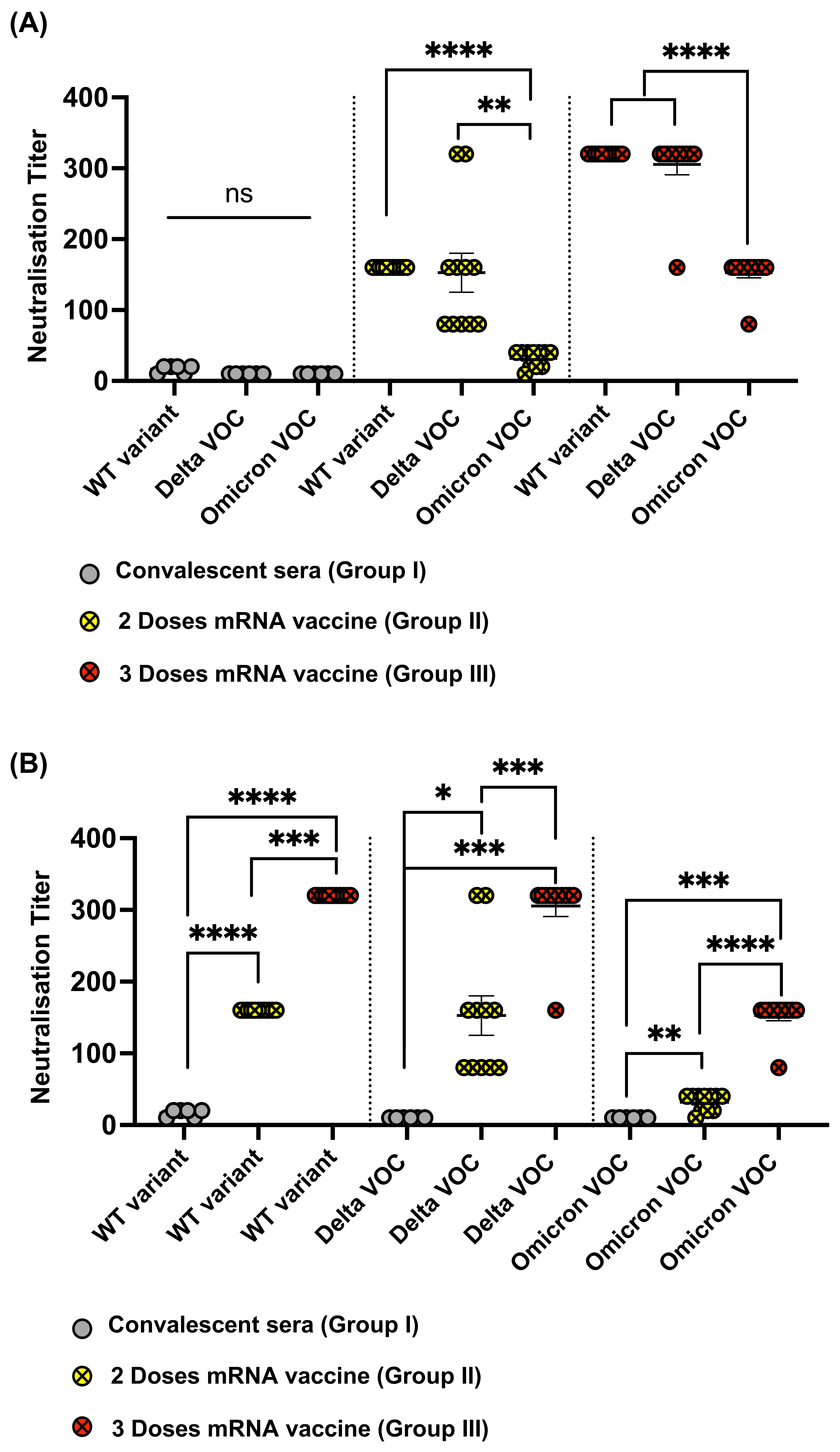
3.3. Reduced Recognition of Omicron Mutant RBDs by Sera from Convalescent Patients and mRNA Vaccinated Individuals
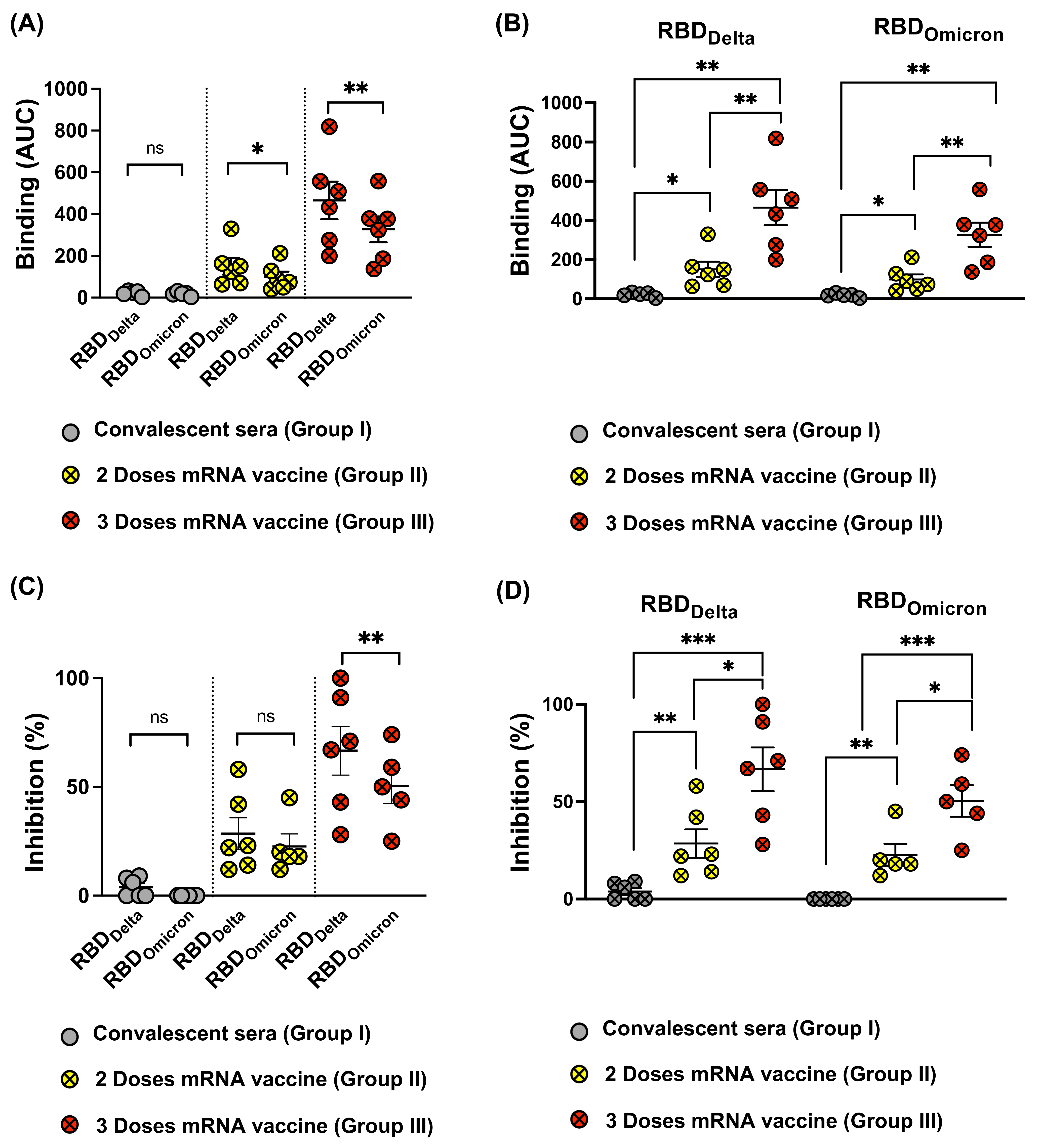
3.4. Omicron RBD Resists Neutralization of Receptor Binding by Immune Sera
3.5. Omicron Resists Viral Neutralization by Immune Sera
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 30 December 2021).
- World Health Organization. Enhancing Response to Omicron SARS-CoV-2 Variant. 2022. Available online: https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states (accessed on 30 March 2022).
- Gu, H.; Krishnan, P.; Ng, D.Y.M.; Chang, L.D.J.; Liu, G.Y.Z.; Cheng, S.S.M.; Hui, M.M.Y.; Fan, M.C.Y.; Wan, J.H.L.; Lau, L.H.K.; et al. Probable Transmission of SARS-CoV-2 Omicron Variant in Quarantine Hotel, Hong Kong, China, November 2021. Emerg. Infect. Dis. 2021, 28, 460–462. [Google Scholar] [CrossRef]
- Clery, D.; Pennisi, E. Omicron leads to fresh wave of meeting cancellations. Science 2022, 375, 132. [Google Scholar] [CrossRef] [PubMed]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, 375, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Gao, Q.; Gao, F.; Wang, Q.; He, Q.; Wu, X.; Mao, Q.; Xu, M.; Liang, Z. Impact of the Delta variant on vaccine efficacy and response strategies. Expert Rev. Vaccines 2021, 20, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.R.; Spratt, A.N.; Cohen, A.R.; Naqvi, S.H.; Chand, H.S.; Quinn, T.P.; Lorson, C.L.; Byrareddy, S.N.; Singh, K. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J. Autoimmun. 2021, 124, 102715. [Google Scholar] [CrossRef] [PubMed]
- Goher, S.S.; Ali, F.; Amin, M. The Delta Variant Mutations in the Receptor Binding Domain of SARS-CoV-2 Show Enhanced Electrostatic Interactions with the ACE2. Med. Drug Discov. 2021, 100114. [Google Scholar] [CrossRef] [PubMed]
- Augusto, G.; Mohsen, M.O.; Zinkhan, S.; Liu, X.; Vogel, M.; Bachmann, M.F. In vitro data suggest that Indian delta variant B.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy 2022, 77, 111–117. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Balke, I.; Zinkhan, S.; Zeltina, V.; Liu, X.; Chang, X.; Krenger, P.S.; Plattner, K.; Gharailoo, Z.; Vogt, A.S.; et al. A scalable and highly immunogenic virus-like particle-based vaccine against SARS-CoV-2. Allergy 2022, 77, 243–257. [Google Scholar] [CrossRef]
- Vogel, M.; Augusto, G.; Chang, X.; Liu, X.; Speiser, D.; Mohsen, M.O.; Bachmann, M.F. Molecular definition of severe acute respiratory syndrome coronavirus 2 receptor-binding domain mutations: Receptor affinity versus neutralization of receptor interaction. Allergy 2022, 77, 143–149. [Google Scholar] [CrossRef]
- Wu, L.Y.; Zhou, L.P.; Mo, M.X.; Liu, T.T.; Wu, C.K.; Gong, C.Y.; Lu, K.; Gong, L.K.; Zhu, W.L.; Xu, Z.J. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Tar. 2022, 7, 8. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Colson, P.; Chahinian, H.; La Scola, B.; Raoult, D. The puzzling mutational landscape of the SARS-2-variant Omicron. J Med Virol 2022, 94, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Mohsen, M.O.; Zha, L.; Vogel, M.; Speiser, D.E. SARS-CoV-2 structural features may explain limited neutralizing-antibody responses. NPJ Vaccines 2021, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Nature. Heavily Mutated Omicron Variant Puts Scientists on Alert. 2022. Available online: https://www.nature.com/articles/d41586-021-03552-w (accessed on 5 January 2022).
- Chang, X.; Augusto, G.S.; Liu, X.; Kundig, T.M.; Vogel, M.; Mohsen, M.O.; Bachmann, M.F. BNT162b2 mRNA COVID-19 vaccine induces antibodies of broader cross-reactivity than natural infection, but recognition of mutant viruses is up to 10-fold reduced. Allergy 2021, 76, 2895–2998. [Google Scholar] [CrossRef] [PubMed]
- Zha, L.; Chang, X.; Zhao, H.; Mohsen, M.O.; Hong, L.; Zhou, Y.; Chen, H.; Liu, X.; Zhang, J.; Li, D.; et al. Development of a Vaccine against SARS-CoV-2 Based on the Receptor-Binding Domain Displayed on Virus-Like Particles. Vaccines 2021, 9, 395. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Mohsen, M.O.; Speiser, D.E. To the editor: The Future of SARS-CoV-2 Vaccination. N. Engl. J. Med. 2022, 386, 899. [Google Scholar]
- Liu, X.; Chang, X.; Rothen, D.; Derveni, M.; Krenger, P.; Roongta, S.; Wright, E.; Vogel, M.; Tars, K.; Mohsen, M.O.; et al. AP205 VLPs Based on Dimerized Capsid Proteins Accommodate RBM Domain of SARS-CoV-2 and Serve as an Attractive Vaccine Candidate. Vaccines 2021, 9, 403. [Google Scholar] [CrossRef]
- Ren, S.Y.; Wang, W.B.; Gao, R.D.; Zhou, A.M. Omicron variant (B.1.1.529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases 2022, 10, 1–11. [Google Scholar] [CrossRef]
- Tchesnokova, V.; Kulakesara, H.; Larson, L.; Bowers, V.; Rechkina, E.; Kisiela, D.; Sledneva, Y.; Choudhury, D.; Maslova, I.; Deng, K.; et al. Acquisition of the L452R mutation in the ACE2-binding interface of Spike protein triggers recent massive expansion of SARS-Cov-2 variants. bioRxiv 2021. [Google Scholar] [CrossRef]
- Di Giacomo, S.; Mercatelli, D.; Rakhimov, A.; Giorgi, F.M. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J. Med. Virol. 2021, 93, 5638–5643. [Google Scholar] [CrossRef]
- Lupala, C.S.; Ye, Y.; Chen, H.; Su, X.D.; Liu, H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun. 2022, 590, 34–41. [Google Scholar] [CrossRef]
- Plotkin, S.A. Updates on immunologic correlates of vaccine-induced protection. Vaccine 2020, 38, 2250–2257. [Google Scholar] [CrossRef] [PubMed]
- Sevajol, M.; Subissi, L.; Decroly, E.; Canard, B.; Imbert, I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014, 194, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Collier, D.A.; De Marco, A.; Ferreira, I.; Meng, B.; Datir, R.P.; Walls, A.C.; Kemp, S.A.; Bassi, J.; Pinto, D.; Silacci-Fregni, C.; et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021, 593, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Victora, G.D.; Nussenzweig, M.C. Germinal centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; St Denis, K.J.; Hoelzemer, A.; Lam, E.C.; Nitido, A.D.; Sheehan, M.L.; Berrios, C.; Ofoman, O.; Chang, C.C.; Hauser, B.M.; et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. medRxiv 2021. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradnik, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef]
- Muik, A.; Lui, B.G.; Wallisch, A.K.; Bacher, M.; Muhl, J.; Reinholz, J.; Ozhelvaci, O.; Beckmann, N.; Guimil Garcia, R.C.; Poran, A.; et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science 2022, 375, 678–680. [Google Scholar] [CrossRef]
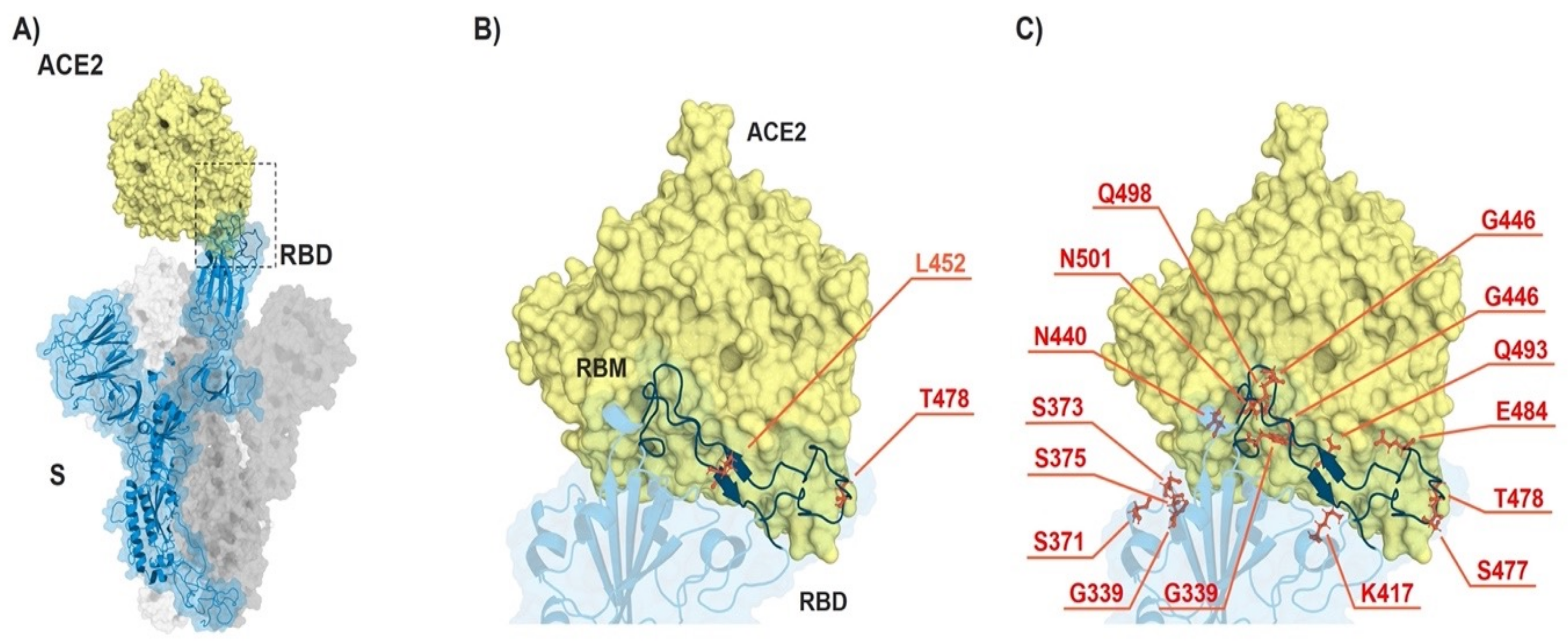
| RBDDelta (B.1.617.2) | RBDOmicron (B.1.1.529) |
|---|---|
| L452R and T478K | K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H |
| Analyte | KD [M] | kon [M−1s−1] | koff [s−1] |
|---|---|---|---|
| RBDWT | 22.6 × 10−9 | 1.7 × 105 | 3.9 × 10−3 |
| RBDDelta | 10.5 × 10−9 | 3.1 × 105 | 2.3 × 10−3 |
| RBDOmicron | 11.6 × 10−9 | 2.1 × 105 | 2.5 × 10−3 |
| RBDL452R/E484Q [9] | 4.6 × 10−9 | 7.2 × 105 | 3.3 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vogt, A.-C.S.; Augusto, G.; Martina, B.; Chang, X.; Nasrallah, G.; Speiser, D.E.; Vogel, M.; Bachmann, M.F.; Mohsen, M.O. Increased Receptor Affinity and Reduced Recognition by Specific Antibodies Contribute to Immune Escape of SARS-CoV-2 Variant Omicron. Vaccines 2022, 10, 743. https://doi.org/10.3390/vaccines10050743
Vogt A-CS, Augusto G, Martina B, Chang X, Nasrallah G, Speiser DE, Vogel M, Bachmann MF, Mohsen MO. Increased Receptor Affinity and Reduced Recognition by Specific Antibodies Contribute to Immune Escape of SARS-CoV-2 Variant Omicron. Vaccines. 2022; 10(5):743. https://doi.org/10.3390/vaccines10050743
Chicago/Turabian StyleVogt, Anne-Cathrine S., Gilles Augusto, Byron Martina, Xinyue Chang, Gheyath Nasrallah, Daniel E. Speiser, Monique Vogel, Martin F. Bachmann, and Mona O. Mohsen. 2022. "Increased Receptor Affinity and Reduced Recognition by Specific Antibodies Contribute to Immune Escape of SARS-CoV-2 Variant Omicron" Vaccines 10, no. 5: 743. https://doi.org/10.3390/vaccines10050743
APA StyleVogt, A.-C. S., Augusto, G., Martina, B., Chang, X., Nasrallah, G., Speiser, D. E., Vogel, M., Bachmann, M. F., & Mohsen, M. O. (2022). Increased Receptor Affinity and Reduced Recognition by Specific Antibodies Contribute to Immune Escape of SARS-CoV-2 Variant Omicron. Vaccines, 10(5), 743. https://doi.org/10.3390/vaccines10050743









