Development and Testing of a Low-Cost Inactivation Buffer That Allows for Direct SARS-CoV-2 Detection in Saliva
Abstract
:1. Introduction
2. Materials and Methods
2.1. COVID-19-Positive Saliva Samples
2.2. PKTP Buffer
2.3. RT-qPCR Reaction
2.4. Self-Collection of Saliva Samples
2.5. Total Saliva RNA Integrity Test
2.6. Statistical Analysis
3. Results
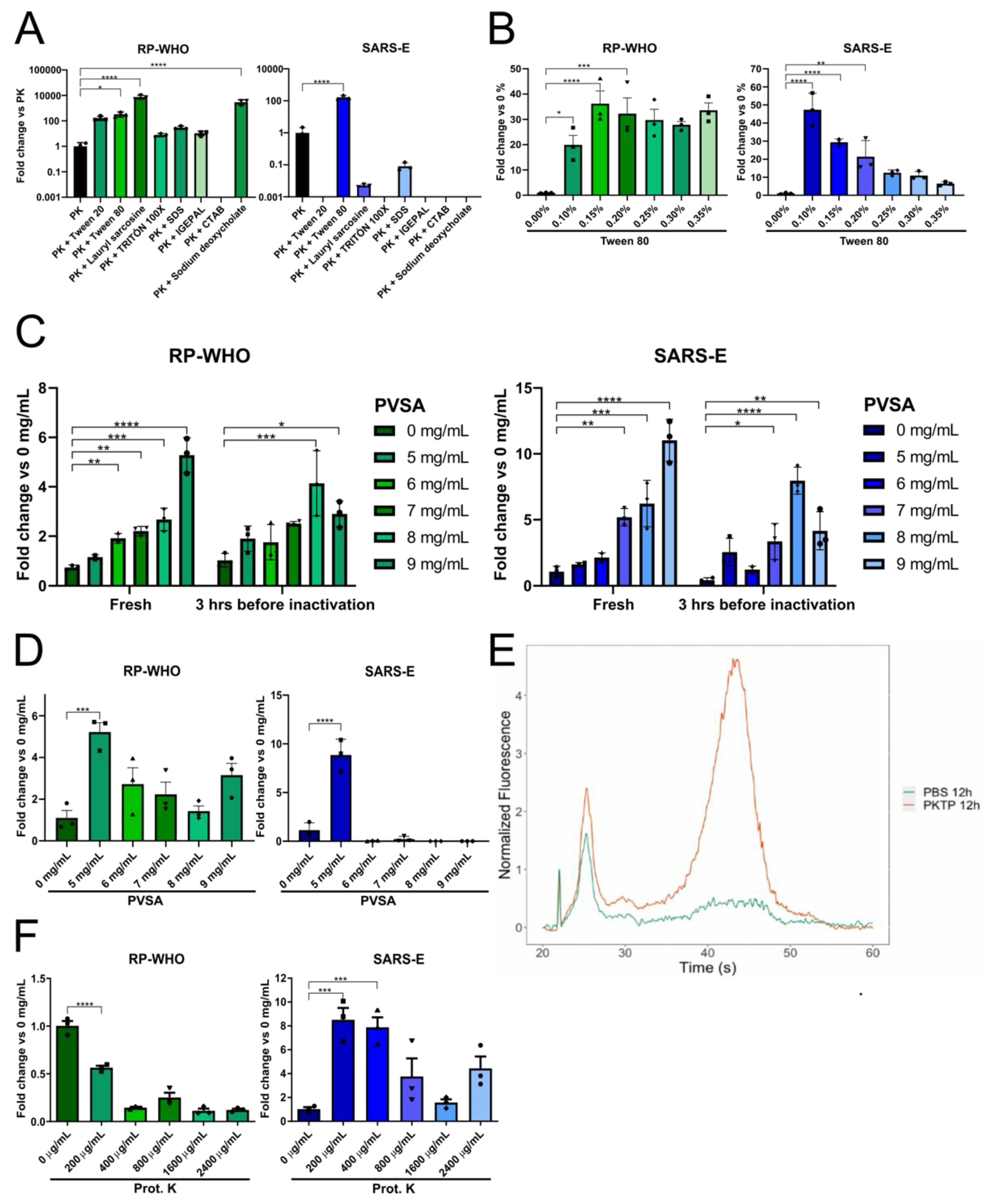
3.1. Detergent Screening for Direct SARS-CoV-2 Detection in Saliva by RT-qPCR
3.2. Addition of an RNase Inhibitor Enhances Viral Genetic Material Detection in Saliva
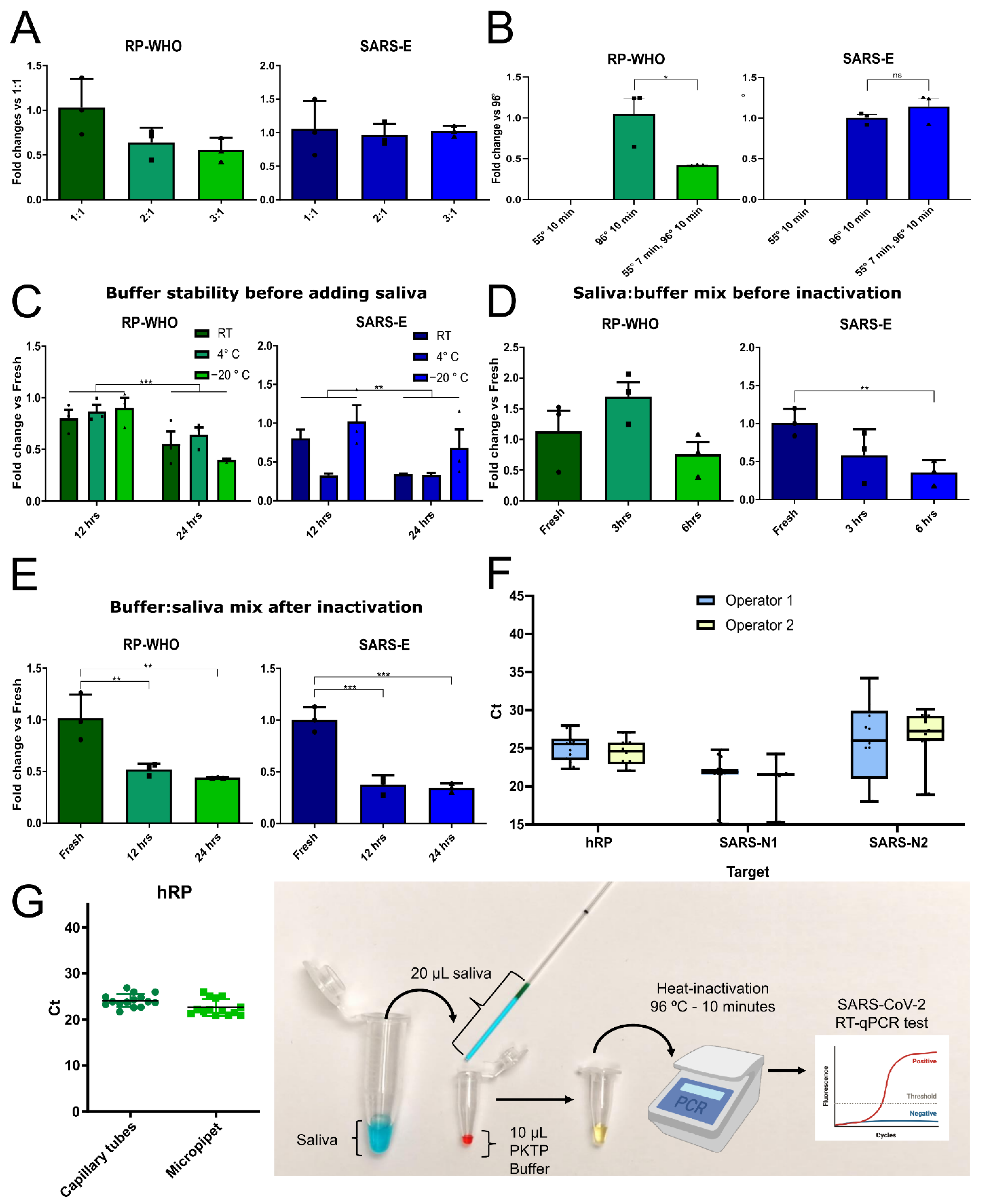
3.3. Optimization of Saliva Lysis Conditions
3.4. Saliva Self-Collection Protocol
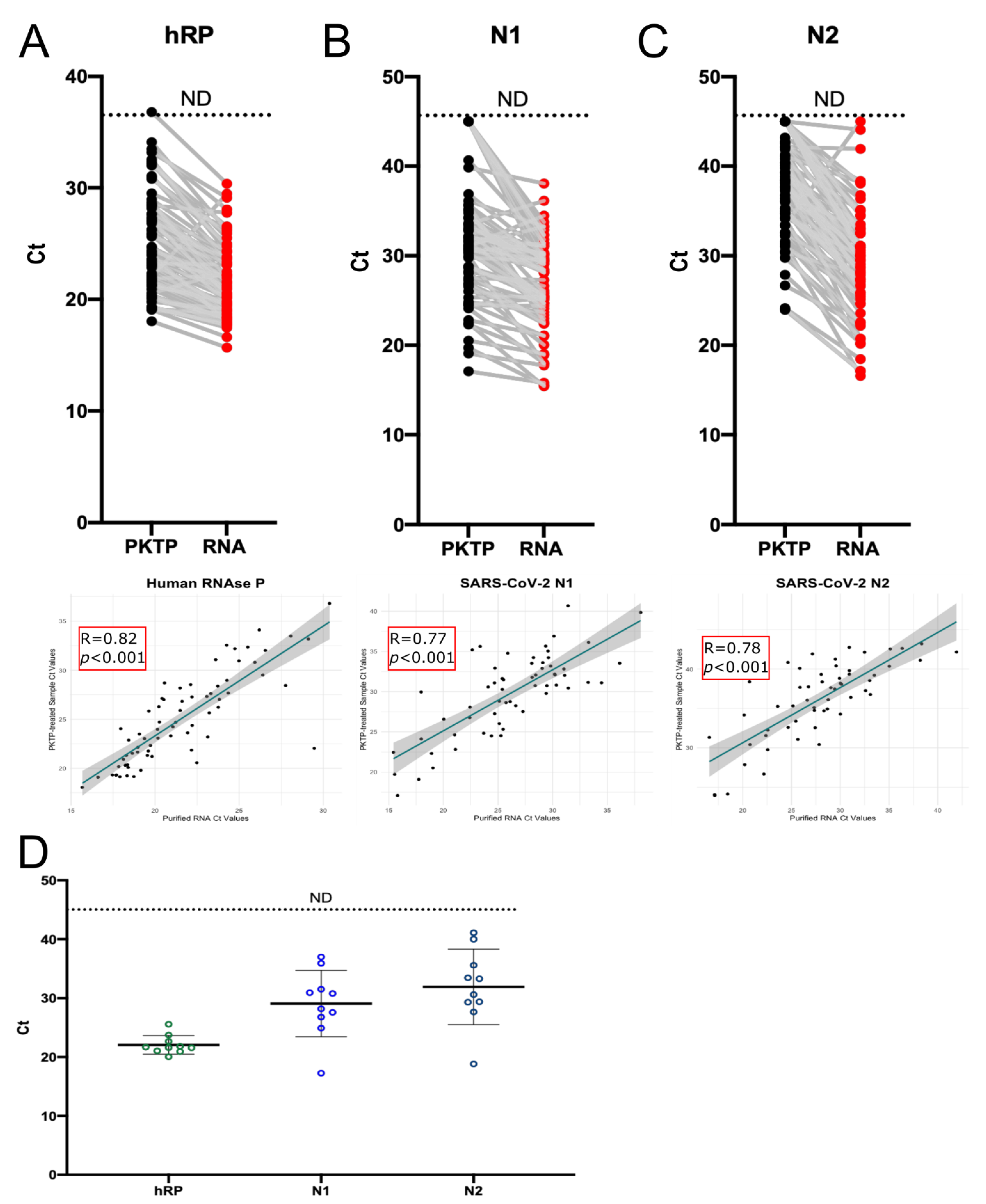
3.5. Use of PKTP Buffer in a Clinical Setting
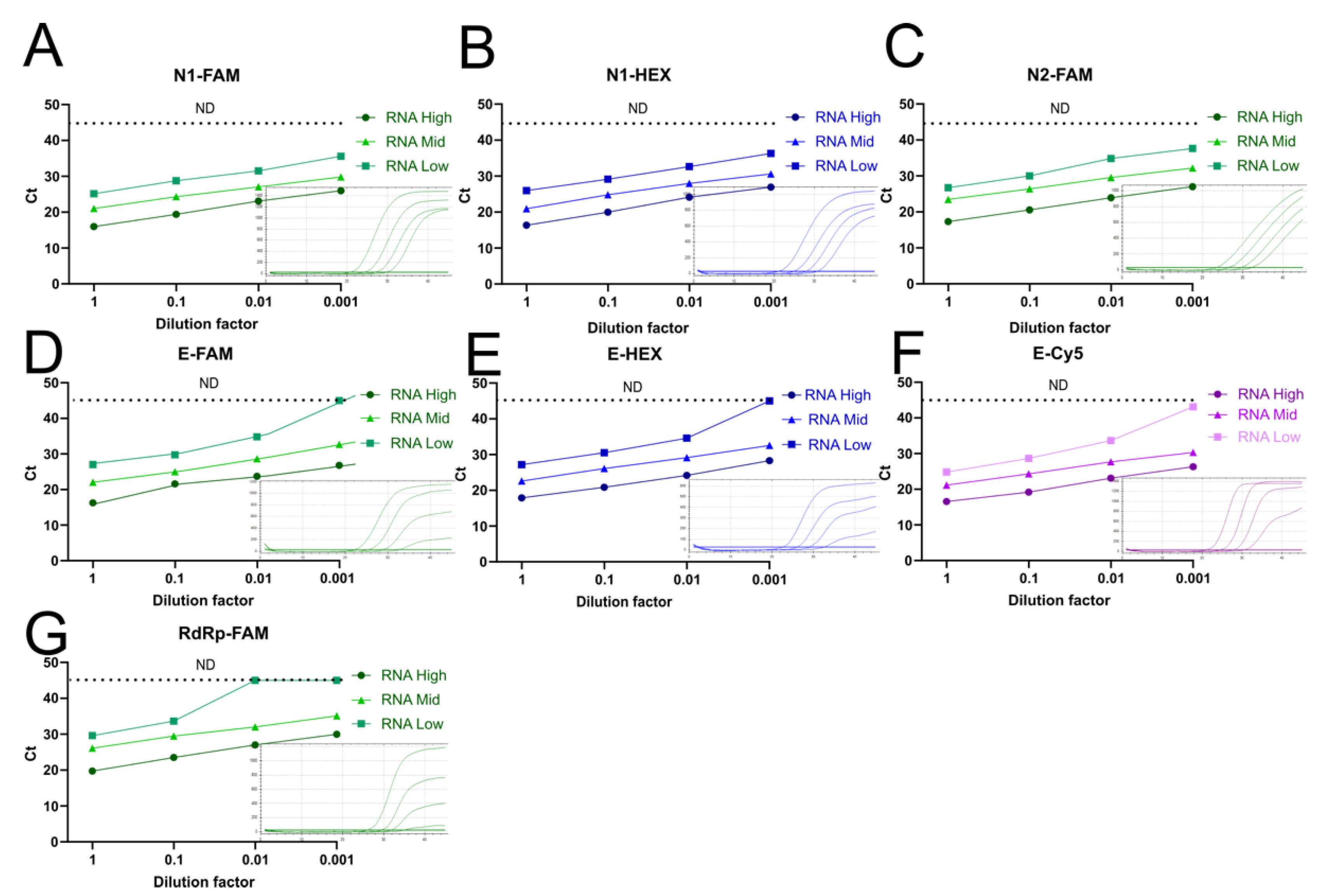
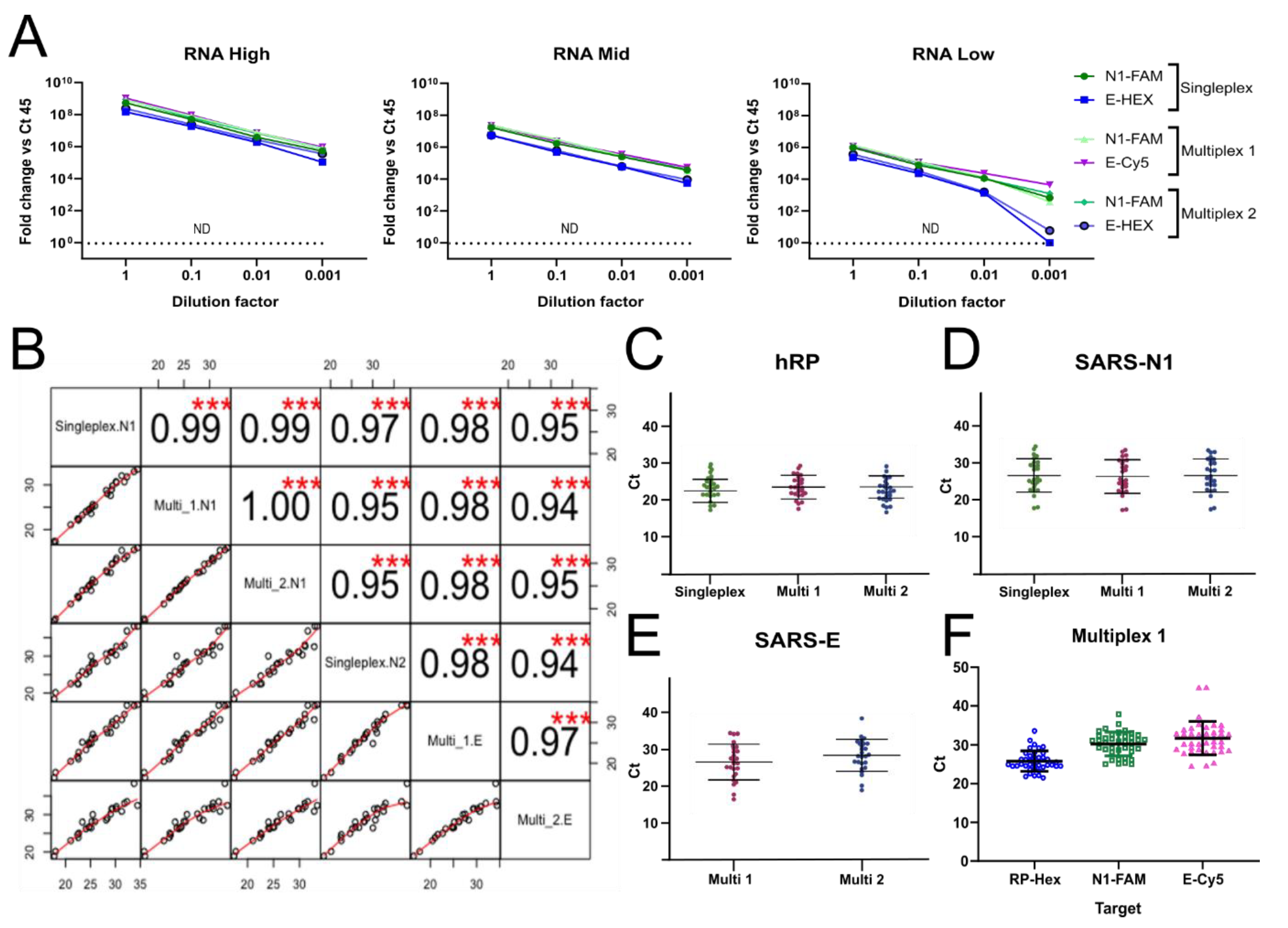
3.6. Reducing the Number of RT-qPCR Reactions by Multiplexing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Callaway, E. Heavily mutated Omicron variant puts scientists on alert. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.M.; Taha, T.Y.; Tabata, T.; Chen, I.P.; Ciling, A.; Khalid, M.M.; Sreekumar, B.; Chen, P.Y.; Hayashi, J.M.; Soczek, K.M.; et al. Rapid assessment of SARS-CoV-2 evolved variants using virus-like particles. Science 2021, 374, eabl6184. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L. Why scientists worldwide are watching UK COVID infections. Nature 2021, 599, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Wadman, M. Israel’s grim warning: Delta can overwhelm shots. Science 2021, 373, 838–839. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Piot, P. The Potential Future of the COVID-19 Pandemic: Will SARS-CoV-2 Become a Recurrent Seasonal Infection? JAMA 2021, 325, 1249–1250. [Google Scholar] [CrossRef]
- Murray, A.F.; Emanuels, A.; Wolf, C.; Franko, N.; Starita, L.; Englund, J.A.; Chu, H.Y. School-Based Surveillance of Respiratory Pathogens on "High-Touch" Surfaces. Front. Pediatr. 2021, 9, 686386. [Google Scholar] [CrossRef]
- Park, S.K.; Lee, C.W.; Park, D.I.; Woo, H.Y.; Cheong, H.S.; Shin, H.C.; Ahn, K.; Kwon, M.J.; Joo, E.J. Detection of SARS-CoV-2 in Fecal Samples from Patients with Asymptomatic and Mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2021, 19, 1387–1394.e2. [Google Scholar] [CrossRef]
- Lee, R.A.; Herigon, J.C.; Benedetti, A.; Pollock, N.R.; Denkinger, C.M. Performance of Saliva, Oropharyngeal Swabs, and Nasal Swabs for SARS-CoV-2 Molecular Detection: A Systematic Review and Meta-analysis. J. Clin. Microbiol. 2021, 59, 1–17. [Google Scholar] [CrossRef]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef]
- Moreno-Contreras, J.; Espinoza, M.A.; Sandoval-Jaime, C.; Cantu-Cuevas, M.A.; Baron-Olivares, H.; Ortiz-Orozco, O.D.; Munoz-Rangel, A.V.; Hernandez-de la Cruz, M.; Eroza-Osorio, C.M.; Arias, C.F.; et al. Saliva Sampling and Its Direct Lysis, an Excellent Option to Increase the Number of SARS-CoV-2 Diagnostic Tests in Settings with Supply Shortages. J. Clin. Microbiol. 2020, 58, e01659-20. [Google Scholar] [CrossRef]
- Herrera, L.A.; Hidalgo-Miranda, A.; Reynoso-Noveron, N.; Meneses-Garcia, A.A.; Mendoza-Vargas, A.; Reyes-Grajeda, J.P.; Vadillo-Ortega, F.; Cedro-Tanda, A.; Penaloza, F.; Frias-Jimenez, E.; et al. Saliva is a reliable and accessible source for the detection of SARS-CoV-2. Int. J. Infect. Dis. 2021, 105, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Matos, P.M.; Maguire, T.J.A.; Poulton, K.; Ortiz-Zapater, E.; Page, R.; Sertkaya, H.; Ortega-Prieto, A.M.; Scourfield, E.; O’Byrne, A.M.; et al. Resilient SARS-CoV-2 diagnostics workflows including viral heat inactivation. PLoS ONE 2021, 16, e0256813. [Google Scholar] [CrossRef] [PubMed]
- Smyrlaki, I.; Ekman, M.; Lentini, A.; Rufino de Sousa, N.; Papanicolaou, N.; Vondracek, M.; Aarum, J.; Safari, H.; Muradrasoli, S.; Rothfuchs, A.G.; et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat. Commun. 2020, 11, 4812. [Google Scholar] [CrossRef] [PubMed]
- Genoud, V.; Stortz, M.; Waisman, A.; Berardino, B.G.; Verneri, P.; Dansey, V.; Salvatori, M.; Remes Lenicov, F.; Levi, V. Extraction-free protocol combining proteinase K and heat inactivation for detection of SARS-CoV-2 by RT-qPCR. PLoS ONE 2021, 16, e0247792. [Google Scholar] [CrossRef]
- Vogels, C.B.F.; Watkins, A.E.; Harden, C.A.; Brackney, D.E.; Shafer, J.; Wang, J.; Caraballo, C.; Kalinich, C.C.; Ott, I.M.; Fauver, J.R.; et al. SalivaDirect: A simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med 2021, 2, 263–280.e266. [Google Scholar] [CrossRef]
- Callahan, C.; Ditelberg, S.; Dutta, S.; Littlehale, N.; Cheng, A.; Kupczewski, K.; McVay, D.; Riedel, S.; Kirby, J.E.; Arnaout, R. Saliva is Comparable to Nasopharyngeal Swabs for Molecular Detection of SARS-CoV-2. Microbiol. Spectr. 2021, 9, e0016221. [Google Scholar] [CrossRef]
- Fomsgaard, A.S.; Rosenstierne, M.W. An alternative workflow for molecular detection of SARS-CoV-2—escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill. 2020, 25, 2000398. [Google Scholar] [CrossRef] [Green Version]
- Linke, D. Detergents: An overview. Methods Enzymol. 2009, 463, 603–617. [Google Scholar] [CrossRef]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane proteins, lipids and detergents: Not just a soap opera. Biochim. Biophys. Acta 2004, 1666, 105–117. [Google Scholar] [CrossRef] [Green Version]
- le Maire, M.; Champeil, P.; Moller, J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 2000, 1508, 86–111. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738e713. [Google Scholar] [CrossRef] [PubMed]
- Hardenbrook, N.J.; Zhang, P. A structural view of the SARS-CoV-2 virus and its assembly. Curr. Opin. Virol. 2022, 52, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention, Division of Viral Diseases. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. For Emergency Use Only. Instructions for Use; CDC-006-00019, Revision: 07; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2021. [Google Scholar]
- Lu, X.; Wang, L.; Sakthivel, S.K.; Whitaker, B.; Murray, J.; Kamili, S.; Lynch, B.; Malapati, L.; Burke, S.A.; Harcourt, J.; et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1654–1665. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Peterson, B.G.; Carl, P.; Boudt, K.; Bennett, R.; Ulrich, J.; Zivot, E.; Cornilly, D.; Hung, E.; Lestel, M.; Balkissoon, K.; et al. Performance Analytics: Econometric Tools for Performance and Risk Analysis, R Package Version 2.0.4. 2020. Available online: https://cran.r-project.org/web/packages/PerformanceAnalytics/PerformanceAnalytics.pdf (accessed on 4 August 2021).
- MedCalc Software Ltd. Diagnostic Test Evaluation Calculator. Available online: https://www.medcalc.org/calc/diagnostic_test.php (accessed on 8 September 2021).
- Shajahan, A.; Pepi, L.E.; Rouhani, D.S.; Heiss, C.; Azadi, P. Glycosylation of SARS-CoV-2: Structural and functional insights. Anal. Bioanal. Chem. 2021, 413, 7179–7193. [Google Scholar] [CrossRef]
- Nishibata, Y.; Koshimoto, S.; Ogaki, K.; Ishikawa, E.; Wada, K.; Yoshinari, M.; Tamura, Y.; Uozumi, R.; Masuda, S.; Tomaru, U.; et al. RNase in the saliva can affect the detection of severe acute respiratory syndrome coronavirus 2 by real-time one-step polymerase chain reaction using saliva samples. Pathol. Res. Pract. 2021, 220, 153381. [Google Scholar] [CrossRef]
- Earl, C.C.; Smith, M.T.; Lease, R.A.; Bundy, B.C. Polyvinylsulfonic acid: A Low-cost RNase inhibitor for enhanced RNA preservation and cell-free protein translation. Bioengineered 2018, 9, 90–97. [Google Scholar] [CrossRef] [Green Version]
- Singanayagam, A.; Patel, M.; Charlett, A.; Lopez Bernal, J.; Saliba, V.; Ellis, J.; Ladhani, S.; Zambon, M.; Gopal, R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020, 25, 2001483. [Google Scholar] [CrossRef]
- Antonelli, M.; Penfold, R.S.; Merino, J.; Sudre, C.H.; Molteni, E.; Berry, S.; Canas, L.S.; Graham, M.S.; Klaser, K.; Modat, M.; et al. Risk factors and disease profile of post-vaccination SARS-CoV-2 infection in UK users of the COVID Symptom Study app: A prospective, community-based, nested, case-control study. Lancet Infect. Dis. 2021, 22, 43–55. [Google Scholar] [CrossRef]
- Burton, J.; Love, H.; Richards, K.; Burton, C.; Summers, S.; Pitman, J.; Easterbrook, L.; Davies, K.; Spencer, P.; Killip, M.; et al. The effect of heat-treatment on SARS-CoV-2 viability and detection. J. Virol. Methods 2021, 290, 114087. [Google Scholar] [CrossRef]
- Agullo, V.; Fernandez-Gonzalez, M.; Ortiz de la Tabla, V.; Gonzalo-Jimenez, N.; Garcia, J.A.; Masia, M.; Gutierrez, F. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. J. Infect. 2021, 82, 186–230. [Google Scholar] [CrossRef] [PubMed]
- Seitz, T.; Schindler, S.; Winkelmeyer, P.; Zach, B.; Wenisch, C.; Zoufaly, A.; Allerberger, F. Evaluation of rapid antigen tests based on saliva for the detection of SARS-CoV-2. J. Med. Virol. 2021, 93, 4161–4162. [Google Scholar] [CrossRef] [PubMed]
- Asai, N.; Sakanashi, D.; Ohashi, W.; Nakamura, A.; Kawamoto, Y.; Miyazaki, N.; Ohno, T.; Yamada, A.; Chida, S.; Shibata, Y.; et al. Efficacy and validity of automated quantitative chemiluminescent enzyme immunoassay for SARS-CoV-2 antigen test from saliva specimen in the diagnosis of COVID-19. J. Infect. Chemother. 2021, 27, 1039–1042. [Google Scholar] [CrossRef] [PubMed]
- Brummer, L.E.; Katzenschlager, S.; Gaeddert, M.; Erdmann, C.; Schmitz, S.; Bota, M.; Grilli, M.; Larmann, J.; Weigand, M.A.; Pollock, N.R.; et al. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: A living systematic review and meta-analysis. PLoS Med. 2021, 18, e1003735. [Google Scholar] [CrossRef]
- Yoon, J.; Yun, S.G.; Nam, J.; Choi, S.H.; Lim, C.S. The use of saliva specimens for detection of influenza A and B viruses by rapid influenza diagnostic tests. J. Virol. Methods 2017, 243, 15–19. [Google Scholar] [CrossRef]
- Cheng, Y.; Ma, J.; Wang, H.; Wang, X.; Hu, Z.; Li, H.; Zhang, H.; Liu, X. Co-infection of influenza A virus and SARS-CoV-2: A retrospective cohort study. J. Med. Virol. 2021, 93, 2947–2954. [Google Scholar] [CrossRef]
| Primer/Probe | Sequence 5′-3′ | Concentration (nM) | Reference | |
|---|---|---|---|---|
| Singleplex | Multiplex | |||
| WHO-hRP-F | AGATTTGGACCTGCGAGCG | 500 | 200 | [24] |
| WHO-hRP-R | GAGCGGCTGTCTCCACAAGT | 500 | 200 | [24] |
| WHO-hRP_FAM | [FAM]TTCTGACCTGAAGGCTCTGCGCG[BHQ-1] | 125 | - | [24] |
| WHO-hRP_TxR | [TxRd]TTCTGACCTGAAGGCTCTGCGCG | - | 100 | [24] |
| WHO-hRP_Probe P1_HEX | [HEX]TTCTGACCTGAAGGCTCTGCGCG[BHQ1] | - | 100 | [24] |
| 2019-nCoV_N1-F | GACCCCAAAATCAGCGAAAT | 500 | 400 | [24] |
| 2019-nCoV_N1-R | TCTGGTTACTGCCAGTTGAATCTG | 500 | 400 | [24] |
| 2019-nCov_N1-P_FAM | [FAM]ACCCCGCATTACGTTTGGTGGACC[BHQ1] | 125 | 100 | [24] |
| 2019-nCoV_N2-F | TTACAAACATTGGCCGCAAA | 500 | - | [24] |
| 2019-nCoV_N2-R | GCGCGACATTCCGAAGAA | 500 | - | [24] |
| 2019-nCov_N2-P_FAM | [FAM]ACAATTTGCCCCCAGCGCTTC AG[BHQ1] | 125 | - | [24] |
| WHO-SARS E-F1 | ACAGGTACGTTAATAGTTAATAGCGT | 400 | 400 | [25] |
| WHO-SARS E-R1 | ATATTGCAGCAGTACGCACACA | 400 | 400 | [25] |
| WHO-SARS E_HEX | [HEX]ACACTAGCCATCCTTACTGCGCTTCG[BHQ3] | 200 | 200 | [25] |
| WHO-SARS E_CY5 | [Cyanine5]ACACTAGCCATCCTTACTGCGCTTCG[BHQ3] | 200 | 100 | [25] |
| WHO-SARS E_FAM | [FAM]ACACTAGCCATCCTTACTGCGCTTCG[BBQ] | 200 | - | [25] |
| RdRP_SARSr-F | GTGARATGGTCATGTGTGGCGG | 600 | - | [25] |
| RdRP_SARSr-R | CARATGTTAAASACACTATTAGCATA | 800 | - | [25] |
| RdRP_SARSr-P2 | [FAM]CAGGTGGAACCTCATCAGGAGATGC[BBQ] | 200 | - | [25] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bustos-Garcia, B.; Garza-Manero, S.; Cano-Dominguez, N.; Lopez-Sanchez, D.M.; Salgado-Montes de Oca, G.; Salgado-Aguayo, A.; Recillas-Targa, F.; Avila-Rios, S.; Valdes, V.J. Development and Testing of a Low-Cost Inactivation Buffer That Allows for Direct SARS-CoV-2 Detection in Saliva. Vaccines 2022, 10, 730. https://doi.org/10.3390/vaccines10050730
Bustos-Garcia B, Garza-Manero S, Cano-Dominguez N, Lopez-Sanchez DM, Salgado-Montes de Oca G, Salgado-Aguayo A, Recillas-Targa F, Avila-Rios S, Valdes VJ. Development and Testing of a Low-Cost Inactivation Buffer That Allows for Direct SARS-CoV-2 Detection in Saliva. Vaccines. 2022; 10(5):730. https://doi.org/10.3390/vaccines10050730
Chicago/Turabian StyleBustos-Garcia, Brandon, Sylvia Garza-Manero, Nallely Cano-Dominguez, Dulce Maria Lopez-Sanchez, Gonzalo Salgado-Montes de Oca, Alfonso Salgado-Aguayo, Felix Recillas-Targa, Santiago Avila-Rios, and Victor Julian Valdes. 2022. "Development and Testing of a Low-Cost Inactivation Buffer That Allows for Direct SARS-CoV-2 Detection in Saliva" Vaccines 10, no. 5: 730. https://doi.org/10.3390/vaccines10050730
APA StyleBustos-Garcia, B., Garza-Manero, S., Cano-Dominguez, N., Lopez-Sanchez, D. M., Salgado-Montes de Oca, G., Salgado-Aguayo, A., Recillas-Targa, F., Avila-Rios, S., & Valdes, V. J. (2022). Development and Testing of a Low-Cost Inactivation Buffer That Allows for Direct SARS-CoV-2 Detection in Saliva. Vaccines, 10(5), 730. https://doi.org/10.3390/vaccines10050730






