Could SARS-CoV-2 Have Bacteriophage Behavior or Induce the Activity of Other Bacteriophages?
Abstract
:1. Introduction
2. Materials and Methods
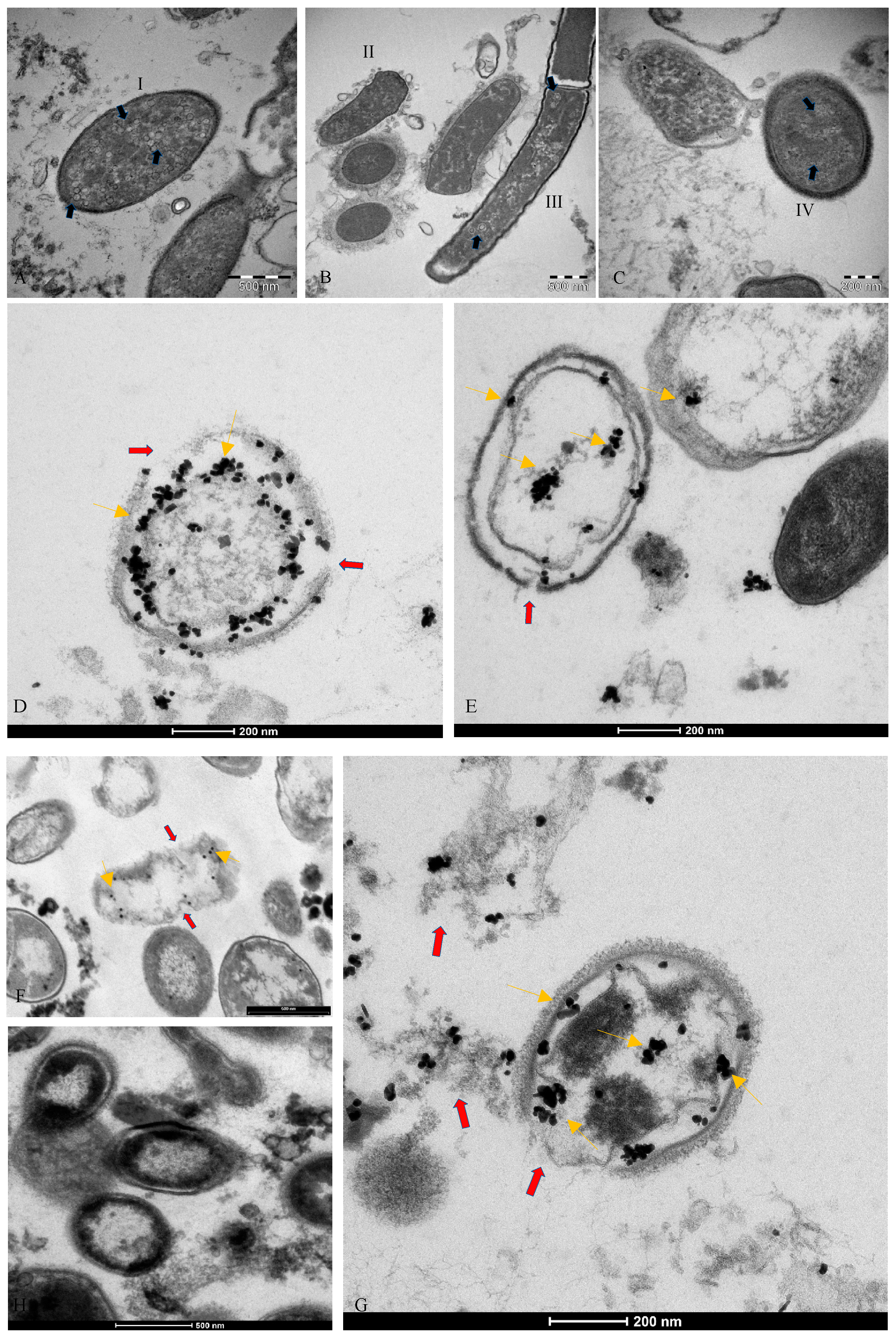
2.1. Electron Microscopy
2.2. Immunogold Labelling Tecnique
2.3. Validation of Antibody against Nucleocaspid-Protein of SARS-CoV-2 with Fluorescent Microscopy
2.4. Immunofluorescence
2.5. Proteomic Profiling Analysis
2.6. NGS Sequencing Data
3. Results
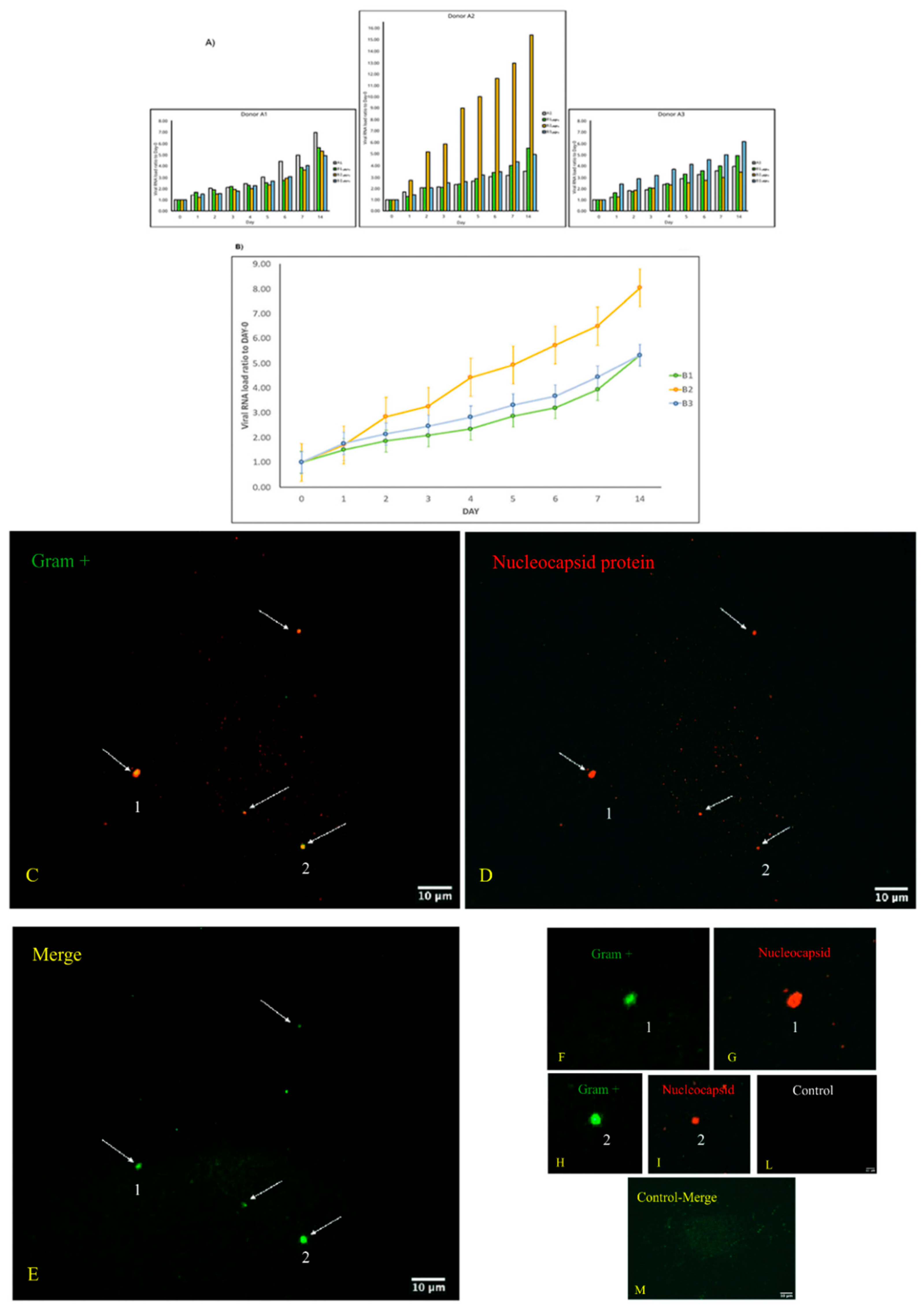
3.1. Replication and Viral Load Increase of SARS-CoV-2 in Bacterial Cultures
3.2. Microscopic Analysis of the Interaction between SARS-CoV-2 and the Human Microbiome
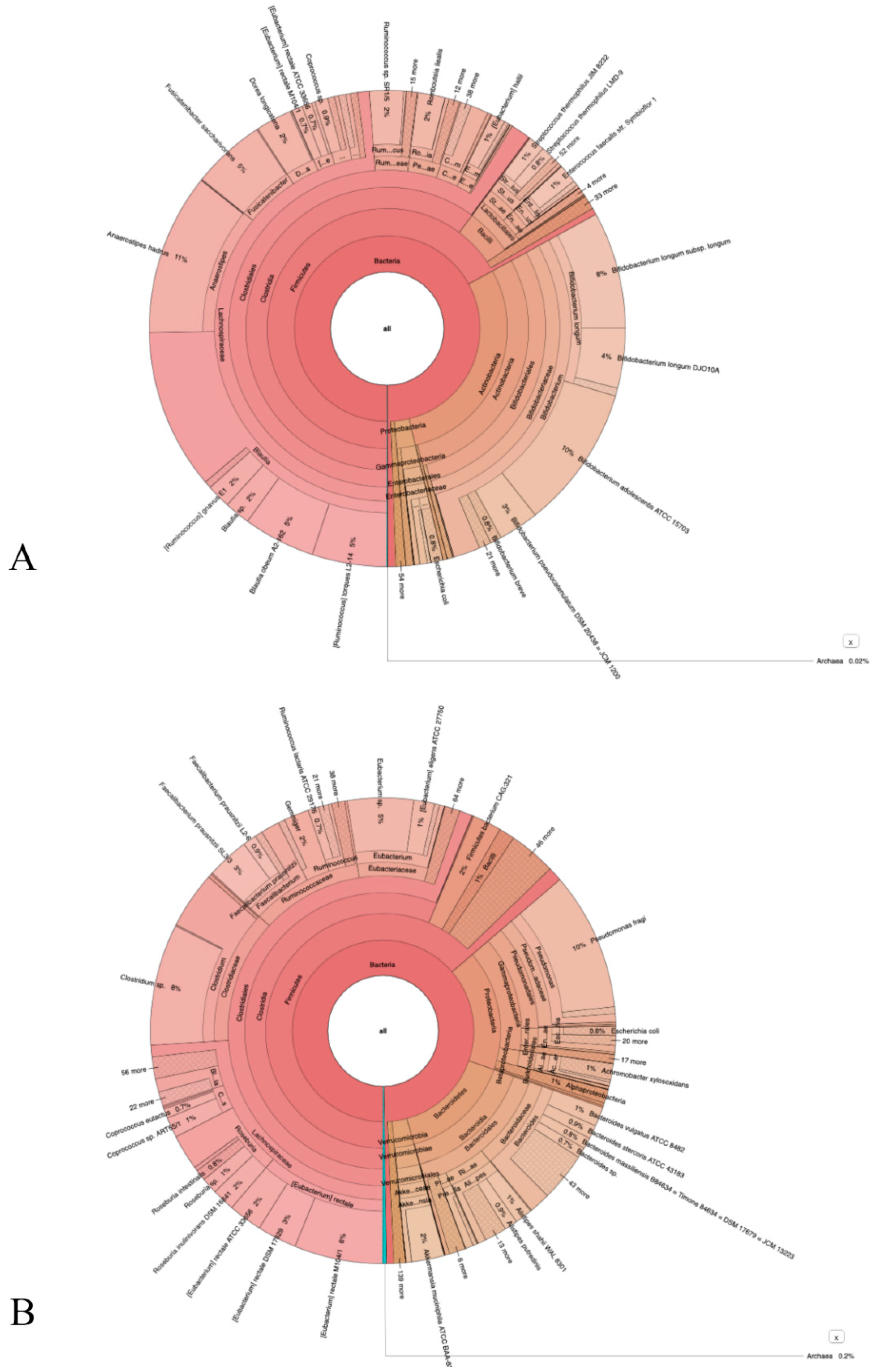
3.3. NGS Sequencing Data
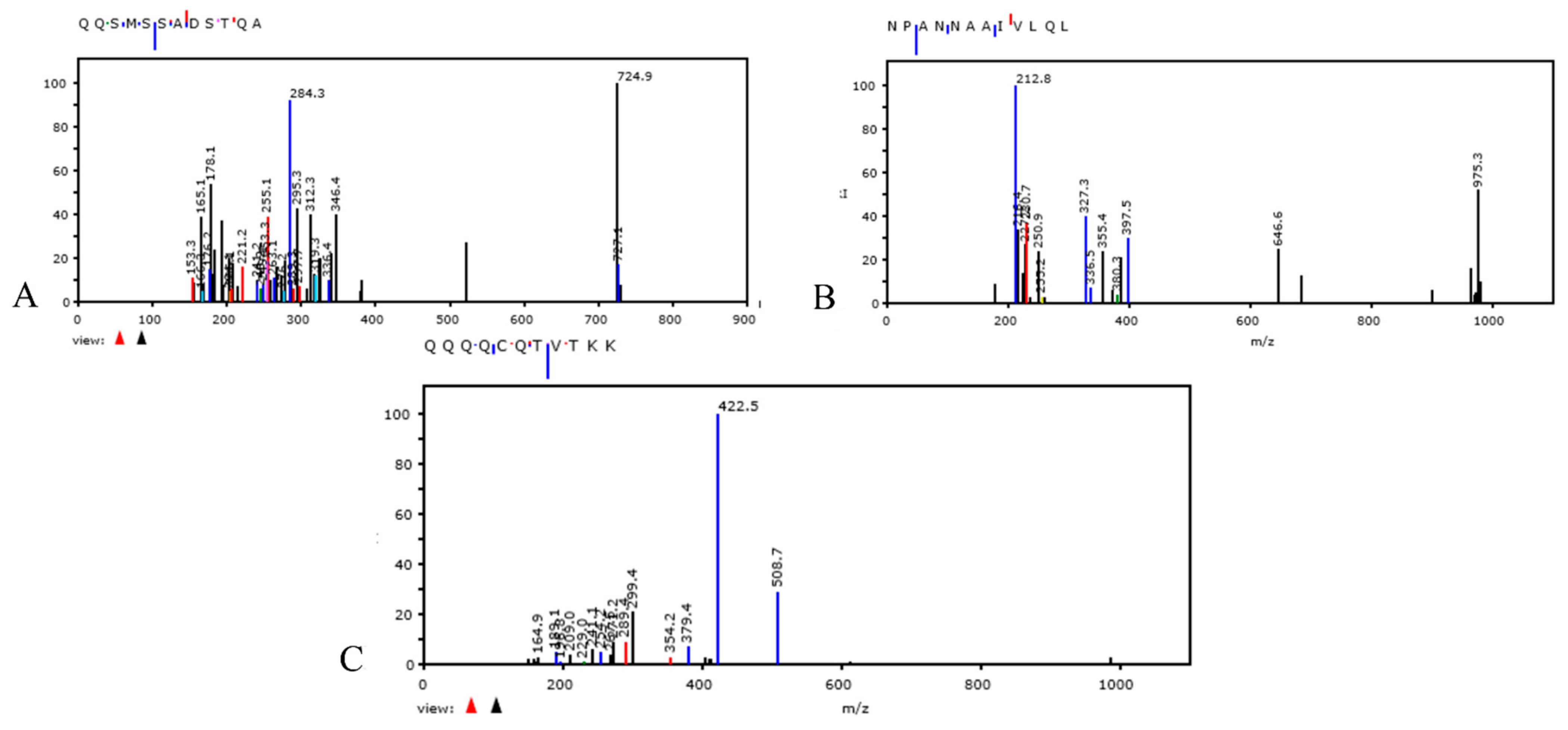
3.4. Nitrogen Isotope 15N Assay in Bacterial Culture Medium. Proteomic Profiling Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://www.statista.com/statistics/1194934/number-of-covid-vaccine-doses-administered-by-county-worldwide (accessed on 7 April 2021).
- Chung, M.K.; Karnik, S.; Saef, J.; Bergmann, C.; Barnard, J.; Lederman, M.M.; Tilton, J.; Cheng, F.; Harding, C.V.; Young, J.B.; et al. SARS-CoV-2 and ACE2: The biology and clinical data settling the ARB and ACEI controversy. EBioMedicine 2020, 58, 102907. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, G.R. SARS-CoV-2 spike and its adaptable furin cleavage site. Lancet Microbe 2021, 2, e488–e489. [Google Scholar] [CrossRef]
- Ge, X.Y.; Li, J.L.; Yang, X.L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Zajac, V.; Matelova, L.; Liskova, A.; Mego, M.; Holec, V.; Adamcikova, Z.; Stevurkova, V.; Shahum, A.; Krcmery, V. Confirmation of HIV-like sequences in respiratory tract bacteria of Cambodian and Kenyan HIV-positive pediatric patients. Med. Sci. Monit. 2011, 17, CR154–CR158. [Google Scholar] [CrossRef] [Green Version]
- Brogna, C.; Cristoni, S.; Petrillo, M.; Querci, M.; Piazza, O.; Van den Eede, G. Toxin-like peptides in plasma, urine and faecal samples from COVID-19 patients. F1000Research 2021, 10, 550. [Google Scholar] [CrossRef]
- Petrillo, M.; Brogna, C.; Cristoni, S.; Querci, M.; Piazza, O.; Van den Eede, G. Increase of SARS-CoV-2 RNA load in faecal samples prompts for rethinking of SARS-CoV-2 biology and COVID-19 epidemiology. F1000Research 2021, 10, 370. [Google Scholar] [CrossRef]
- Relationship between the Gut Microbiome and Diseases, Including COVID-19. Available online: https://ec.europa.eu/jrc/en/news/covid-19-and-our-gut-microbiome-evidence-close-relationship (accessed on 20 October 2021).
- Wang, H.; Wang, H.; Sun, Y.; Ren, Z.; Zhu, W.; Li, A.; Cui, G. Potential Associations between Microbiome and COVID-19. Front. Med. 2021, 8, 785496. [Google Scholar] [CrossRef]
- Haiminen, N.; Utro, F.; Seabolt, E.; Parida, L. Functional profiling of COVID-19 respiratory tract microbiomes. Sci. Rep. 2021, 11, 6433. [Google Scholar] [CrossRef]
- Honarmand Ebrahimi, K. SARS-CoV-2 spike glycoprotein-binding proteins expressed by upper respiratory tract bacteria may prevent severe viral infection. FEBS Lett. 2020, 594, 1651–1660. [Google Scholar] [CrossRef]
- Dragelj, J.; Mroginski, M.A.; Ebrahimi, K.H. Hidden in Plain Sight: Natural Products of Commensal Microbiota as an Environmental Selection Pressure for the Rise of New Variants of SARS-CoV-2. Chembiochem 2021, 22, 2946–2950. [Google Scholar] [CrossRef]
- Podlacha, M.; Grabowski, Ł.; Kosznik-Kawśnicka, K.; Zdrojewska, K.; Stasiłojć, M.; Węgrzyn, G.; Węgrzyn, A. Interactions of Bacteriophages with Animal and Human Organisms-Safety Issues in the Light of Phage Therapy. Int. J. Mol. Sci. 2021, 22, 8937. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Liu, Z.; Zhu, Y.; Lu, J.; Ge, C.; Zhang, C.; Li, N.; Jin, N.; Li, Y.; Tian, M.; et al. SARS-CoV-2 Causes Mitochondrial Dysfunction and Mitophagy Impairment. Front. Microbiol. 2022, 12, 780768. [Google Scholar] [CrossRef] [PubMed]
- Kohda, K.; Li, X.; Soga, N.; Nagura, R.; Duerna, T.; Nakajima, S.; Nakagawa, I.; Ito, M.; Ikeuchi, A. An In Vitro Mixed Infection Model With Commensal and Pathogenic Staphylococci for the Exploration of Interspecific Interactions and Their Impacts on Skin Physiology. Front. Cell Infect. Microbiol. 2021, 11, 712360. [Google Scholar] [CrossRef] [PubMed]
- Kameli, N.; Borman, R.; Lpez-Iglesias, C.; Savelkoul, P.; Stassen, F.R.M. Characterization of Feces-Derived Bacterial Membrane Vesicles and the Impact of Their Origin on the Inflammatory Response. Front. Cell Infect. Microbiol. 2021, 11, 667987. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ni, C.; Gao, R.; Wang, Y.; Yang, L.; Wei, J.; Lv, T.; Liang, J.; Zhang, Q.; Xu, W. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell 2020, 11, 771–775. [Google Scholar] [CrossRef] [Green Version]
- Cristoni, S.; Rubini, S.; Bernardi, L.R. Development and applications of surface-activated chemical ionization. Mass Spectrom. Rev. 2007, 26, 645–656. [Google Scholar] [CrossRef]
- Chen, X.; Yu, L.; Steill, J.D.; Oomens, J.; Polfer, N.C. Effect of peptide fragment size on the propensity of cyclization in collision-induced dissociation: Oligoglycine b(2)-b(8). J. Am. Chem. Soc. 2009, 131, 18272–18282. [Google Scholar] [CrossRef]
- Bertrand, K. Survival of exfoliated epithelial cells: A delicate balance between anoikis and apoptosis. J. Biomed. Biotechnol. 2011, 2011, 534139. [Google Scholar] [CrossRef] [Green Version]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [Green Version]
- Zajac, V.; Mego, M.; Martinický, D.; Stevurková, V.; Cierniková, S.; Ujházy, E.; Gajdosík, A.; Gajdosíková, A. Testing of bacteria isolated from HIV/AIDS patients in experimental models. Neuro Endocrinol. Lett. 2006, 27 (Suppl. S2), 61–64. [Google Scholar]
- Xu, H.; Wang, X.; Veazey, R.S. Mucosal immunology of HIV infection. Immunol. Rev. 2013, 254, 10–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diallo, B.; Sissoko, D.; Loman, N.J.; Bah, H.A.; Bah, H.; Worrell, M.C.; Conde, L.S.; Sacko, R.; Mesfin, S.; Loua, A.; et al. Resurgence of Ebola Virus Disease in Guinea Linked to a Survivor With Virus Persistence in Seminal Fluid for More Than 500 Days. Clin. Infect. Dis. 2016, 63, 1353–1356. [Google Scholar] [CrossRef]
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Brogna, B.; Brogna, C.; Petrillo, M.; Conte, A.M.; Benincasa, G.; Montano, L.; Piscopo, M. SARS-CoV-2 Detection in Fecal Sample from a Patient with Typical Findings of COVID-19 Pneumonia on CT but Negative to Multiple SARS-CoV-2 RT-PCR Tests on Oropharyngeal and Nasopharyngeal Swab Samples. Medicina 2021, 57, 290. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, K.; Kapikian, A.Z.; Turner, H.C.; Hartley, J.W.; Parrott, R.H.; Chanock, R.M. Seroepidemiologic studies of coronavirus infection in adults and children. Am. J. Epidemiol. 1970, 91, 585–592. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Liang, X.; Lou, F.; Oglesbee, M.; Krakowka, S.; Li, J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio 2015, 6, e00064. [Google Scholar] [CrossRef] [Green Version]
- Sabin, A.B.; Ward, R. The natural history of human poliomyelitis: II. Elimination of the virus. J. Exp. Med. 1941, 74, 519–529. [Google Scholar] [CrossRef]
- Drinker, P.; Shaw, L.A. An apparatus for the prolonged administration of artificial respiration: I. A design for adults and children. J. Clin. Investig. 1929, 7, 229–247. [Google Scholar] [CrossRef] [Green Version]
- Sabin, A.B. Pathogenesis of poliomyelitis; reappraisal in the light of new data. Science 1956, 123, 1151–1157. [Google Scholar] [CrossRef]
- Sabin, A.B. Behaviour of Chimpanzee-avirulent Poliomyelitis Viruses in Experimentally Infected Human Volunteers. Br. Med. J. 1955, 2, 160–162. [Google Scholar] [CrossRef] [Green Version]
- Sabin, A.B. Present status of attenuated live virus poliomyelitis vaccine. Bull. N. Y. Acad. Med. 1957, 33, 17–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabin, A.B. Oral Poliovirus Vaccine: Recent results and recommendations for optimum use. R. Soc. Health J. 1962, 82, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Likar, M.; Bartley, E.O.; Wilson, D.C. Observations on the interaction of poliovirus and host cells in vitro. III. The effect of some bacterial metabolites and endotoxins. Br. J. Exp. Pathol. 1959, 40, 391–397. [Google Scholar] [PubMed]
- Petrillo, M.; Querci, M.; Brogna, C.; Ponti, J.; Cristoni, S.; Markov, P.V.; Valsesia, A.; Leoni, G.; Benedetti, A.; Wiss, T.; et al. Evidence of SARS-CoV-2 bacteriophage potential in human gut microbiota. F1000Research 2022, 11, 292. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.; Zhang, Z.; Lee, X.; Wu, W.; Huang, Z.; Lei, Z.; Xu, W.; Chen, D.; Wu, X. Associationbetween the nasopharyngealmicrobiome and metabolome in patients with COVID-19. Synth. Syst. Biotechnol. 2021, 6, 135–143. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brogna, C.; Brogna, B.; Bisaccia, D.R.; Lauritano, F.; Marino, G.; Montano, L.; Cristoni, S.; Prisco, M.; Piscopo, M. Could SARS-CoV-2 Have Bacteriophage Behavior or Induce the Activity of Other Bacteriophages? Vaccines 2022, 10, 708. https://doi.org/10.3390/vaccines10050708
Brogna C, Brogna B, Bisaccia DR, Lauritano F, Marino G, Montano L, Cristoni S, Prisco M, Piscopo M. Could SARS-CoV-2 Have Bacteriophage Behavior or Induce the Activity of Other Bacteriophages? Vaccines. 2022; 10(5):708. https://doi.org/10.3390/vaccines10050708
Chicago/Turabian StyleBrogna, Carlo, Barbara Brogna, Domenico Rocco Bisaccia, Francesco Lauritano, Giuliano Marino, Luigi Montano, Simone Cristoni, Marina Prisco, and Marina Piscopo. 2022. "Could SARS-CoV-2 Have Bacteriophage Behavior or Induce the Activity of Other Bacteriophages?" Vaccines 10, no. 5: 708. https://doi.org/10.3390/vaccines10050708
APA StyleBrogna, C., Brogna, B., Bisaccia, D. R., Lauritano, F., Marino, G., Montano, L., Cristoni, S., Prisco, M., & Piscopo, M. (2022). Could SARS-CoV-2 Have Bacteriophage Behavior or Induce the Activity of Other Bacteriophages? Vaccines, 10(5), 708. https://doi.org/10.3390/vaccines10050708







