Cell Lines for the Development of African Swine Fever Virus Vaccine Candidates: An Update
Abstract
:1. Introduction
2. Porcine Macrophages: The Main Target of African Swine Fever Virus
2.1. African Swine Fever Virus Entry and Receptor
2.2. Porcine Monocyte and Derived Macrophages
2.3. Bone-Marrow-Derived Macrophages
2.4. Porcine Alveolar Macrophages
2.5. Porcine Renal-Derived Macrophages
3. Other Porcine Primary Cell Lines
3.1. Dendritic Cells
3.2. Endothelial Cells
4. Monkey-Derived Continuous Cell Lines
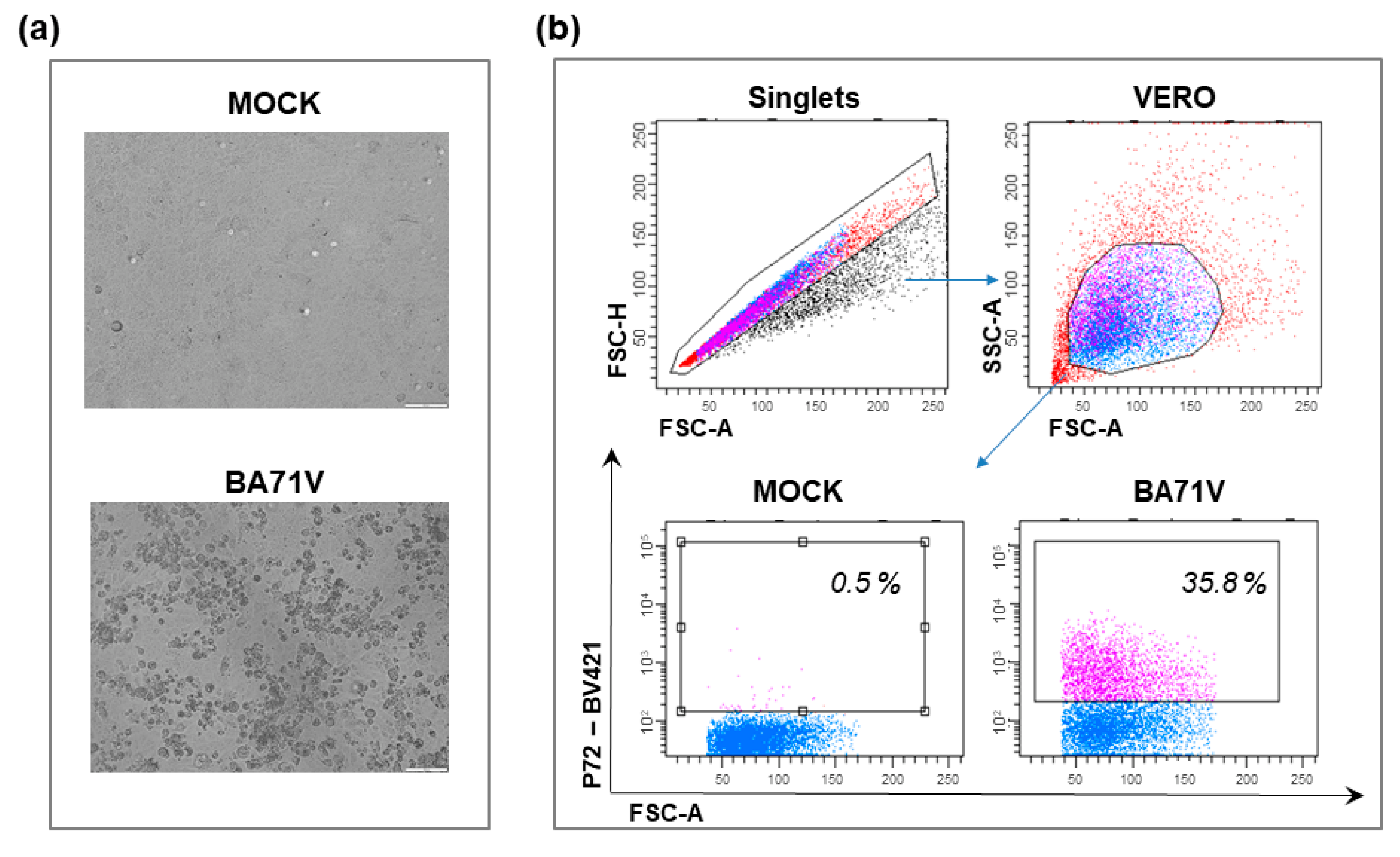
4.1. Vero Cells
4.2. COS Cells
4.3. MS Cells
4.4. CV1 Cells
4.5. Marc-145
4.6. MA-104 Cells
5. Human Continuous Cell Lines
HEK293T Cells
| Cell Line | Species of Cell Origin | Tissue of Cell Origin | Mechanism of Immortalization | Susceptibility to Field Isolates | Susceptibility to Adapted Isolates | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|
| VERO | African green monkey: Chlorocebus sabaeus [103] | Kidney epithelial cells [103] | Spontaneous, unknown process [104] | Low susceptibility to virulent isolates (BA71, Tangani, Hinde, Huganda, Lisbon60) [61,65,66,80] | - Adapted strain from Tengani [65] - BA71V [66,67] - ASFV-G/V [61] - Lisbon60V [80] | - Widely used to characterize function of several ASFV genes [68], proteomic analysis [68,69], mechanisms of viral entry [72,73,74], transcription and replication [69,70] - Titration by plaque formation [65,66] | - Genomic mutation during adaptation → reduction of virulence and immunogenicity in pigs [61,67,79,80,81] |
| COS | African green monkey: Chlorocebus sabaeus [105] | From CV1 (see below) [105] | From CV1 (transformation with a mutant strain of Simian Virus 40 (SV40), which codes for the wild-type T-antigen) [105] | - E70 [82] - Malawi 82 [82] - Uganda [82] - Lisbon57 [82] - Lisbon60 [20,82] - Mozam68 [82] - CC83 [82] | - BA71V [20,30,82] - BA71ΔCD2 [81] - NH/P68 [82] - ΔEP153R [20,30,82] | - Used for production of large amount of virus [20], and studies on virus entry mechanisms [87,88] - Plaque titration [66,82] - Construction of deleted ASFV mutants [81,89] - BA71ΔCD2 –> stability and integrity in its genome [81] - BA71 → no changes in virulence and immunogenity [81] | - NH/P68 derived mutants → genomic mutations during passages relevant to protection [89] |
| MS | African green monkey [62,106] | Kidney [62,106] | Unknown process | - Low susceptibility to virulent to field isolate E70 [62] | - E70MS14, E70MS44, E70MS81 [62,63,90] | - Not plaque for titration [62] - Genomic mutation during adaptation → reduction of virulence and immunogenicity in pigs [62,90] | |
| CV1 | African green monkey: Cercopithecus Aethiops [107] | Fibroblast-like cells derived from kidney tissue [107] | Transformation with a mutant strain of Simian Virus 40 (SV40), which codes for the wild-type T-antigen [107] | - E75 [91] - Stavropol 01/80 [93] | - E75CV1 [91] | - Plaque titration - E75CV1 → 100% protection against challenge with homologous E75 in pigs [91] but not against heterologous BA71 [92]. | - Stavropol 01/80 → Genomic mutation, during adaptation, with reduction of virulence and immunogenicity in pigs [93] |
| Marc-145 | African green monkey Chlorocebus aethiops [94,108] | Fetal kidney epithelial cells, subpopulation of MA-104 [94,108] | MA-104 derived [94] | -D/ASF/POT/Vietnam/2019, D/ASF/POB/Vietnam/2019, although only three passages were monitored [95]. | Unable to support the growth of ASFV Pol18/28298/Out111 [96] and ASFV-HLJ/18 [45]. | ||
| MA-104 | African green monkey: Cercopithecus aethiops [97,108] | Fetal kidney epithelial cells [97,108] | Spontaneously immortalized cell line [97] | - ASFV-G [97] - BA71 [97] - D/ASF/POT/Vietnam/2019, D/ASF/POB/Vietnam/2019 [95] - MW039157 [50] | - MW287337 [50] | - Suitable for ASFV isolation → able to detect ASFV with a TCID50 sensitivity comparable to that of primary swine macrophages [97] - Hemadsorption [97] - Genome stability during 15 passages of a genotype II ASFV isolates [98]. | More studies required its suitability to grow ASFV strains for large-scale vaccine production. |
| HEK293T | Human [45] | Kidney epithelial [45] | Transformation with sheared Adenovirus 5 DNA [109] | - Low susceptibility to OURT88/3 [102] and ASFV-HLJ/18 [45] | - Adapted strain from ASFV-HLJ/18 (ASFV-P121) [45] - OURT 88/3-ΔTK-GFP [102] | - Efficiently and high replication of the attenuated virus [45] - Hemadsorption [45] - Clear cytopathic effect [45] | ASFV-HLJ/18 → Genomic mutation during adaptation (mainly at the MGF genes) [45] → potential reduction of virulence and immunogenicity in pigs [45] |
6. Porcine Continuous Cell Lines
6.1. PK Cell Lines
6.2. WSL Cells
6.3. Immortalized Porcine Alveolar Macrophages
6.4. Immortalized Porcine Kidney Macrophages
6.5. Plum Island Porcine Epithelial Cells
6.6. Zuckerman Macrophage-4
6.7. A4C2 and A4C2/9k Cells
6.8. PSGK-60 and PPK-66b
| Cell Line | Species of Cell Origin | Tissue of Cell Origin | Mechanism of Immortalization | Susceptibility to Field Isolates | Susceptibility to Adapted Isolates | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|
| PK | Pig [110,112,129] | Kidney [110,112,129] | Spontaneously immortalized cell line [129] | - PK2a: extremely low susceptibility to several ASFV strains: Spencer, Portuguese, Gasson, Madrid n. 1, Madrid n. 2 [110] - PK/A/C/13: extremely low susceptibility to Uganda and Hinde: [112] - PK15: extremely low susceptibility to Tengani [111,114] | -PK2a: susceptibility to ASFV strains after several passages on PK2a (Spencer, Portuguese, Gasson, Madrid n. 1, Madrid n. 2) [110] - PK/A/C/13: susceptibility to Uganda and Hinde after several passages on PK cells [112] - PK15, PK9, and PK0809: Attenuated Uganda, BA71V [46]. - PK15: strain derived from Tengani through passages on PK15 [111,114] | - Plaque titration (with plaque size heterogeneity) [110,112]. - Clear cytopathic effects [110] - Hemadsorption [110] | - Plaque formation limited to culture-adapted ASFV strains [30,110]. - PK15 no able to support the growth of ASFV-HLJ/18 strain [45], Benin 97/1, Virulent Uganda [46]. - PK9, and PK0809 no susceptible to Benin 97/1, Virulent Uganda [46]. - PK15 no able to support the growth of D/ASF/POT/Vietnam/2019, D/ASF/POB/Vietnam/2019 [95] - Spencer, Portuguese, Gasson adapted to PK cells → genomic mutation during adaptation with reduction of virulence in pigs but only partial protection against challenge with homologous strain [110] - Hinde isolates passaged 75 times in PK2a → reduction of virulence in pigs but only partial protection to challenge to parental strain; survived pigs only partial protection to other ASFV isolates [65] |
| WSL | Wild boar (fetus) [20,30,116] | Lung [20,30,116] | Spontaneously immortalized cell line [130] | - CC83, Malawi 82, Uganda, Lisbon 57, E70 [30] -ASFV-Kenya1033-IXL [118] | - NH/P68 [18,115] - ΔEP153R, Hinde attenuated, Uganda attenuated NH/P68, BA71V [30] - NHV-dTK-EGFP [116] - ASFV-KeΔA238L strain (generated by deletion of the gene A238 by ASFV-Kenya1033- IXL) [119] - OUTR88/3 [102] | - ASFV-Kenya1033-IXL and ASFV-KeΔA238L → genomic stability during passages in WSL [118,119] - NH/P68 ASFV strain produced in WSL cells (10 passages) → in vivo in pigs induced a similar infection pattern to that of NH/P68 passaged in PAM [18,115] - ASFV-Kenya-1033-IX produced in WSL cells (more than 20 passages) → retained the virulence in vivo [118] | - Slight or unnoticeable cytopathic effects [20,30] - No ability to efficiently support the growth of E70 or Armenia/07 [18,115]. - Promising results, but in vivo studies are required to evaluate its suitability to grow ASFV strains for large-scale vaccine production (protection to challenge). |
| IPAM | Sus scrofa [30] | Porcine alveolar macrophages [30,122] | Porcine myeloid cell lines established by transfecting primary porcine alveolar macrophage cultures with plasmid pSV3neo, carrying genes for SV40 large T antigen [30,122] | - Low susceptibility to few virulent isolates, including E70, CC83 [30] - Low susceptibility to field isolateLillie SI/85 [122] | - Uganda attenuated, Hinde attenuated [30] - 3D4/2, 3D4/21, and 3D4/31: growth of the attenuated ASFV-Lisbon 61 supported in different degrees. [122] | - Unable to maintain replication of the ASFV-HLJ/18 strain [45] - Weak susceptibility to BA71V, NH/P68, Malawi82, Lisbon57 [30]. - No susceptible to infection with Armenia/07 or NH/P68 [18,115]. | |
| IPKM | Pig [123] | Primary porcine kidney-derived macrophages [123] | Immortalization by transfection with recombinant lentivirus vectors carrying the gene for SV40 large T antigen (SV40LT) in combination with the gene for porcine telomerase reverse transcriptase (pTERT) [123] | - Armenia07 [23] - Kenya05/Tk-1 [23] - E75 [23] | - Lisbon60V (VERO cell-adapted isolate) [23] | - Clear cytopathic effect [23] - Hemadsorption [23] - Plaques formation → rapid isolation and purification of different strains [23] -Armenia07 → good genomic stability with only one non-synonymous nucleotide replacement detected in the CP530R region at passages 10 and 15 [23] | Promising results, but in vivo studies are required to evaluate its suitability to grow ASFV strains for large-scale vaccine production. |
| PIPEC | Pig (fetus) [124] | Kidney [124] | 60 passages from the LFPKaVb6 (a porcine fetal kidney cell line [124]). | - ASFV-G-ΔI177L → generation of ASFV-G- ΔI177LΔLVR after several passages [125] | - ASFV-G- ΔI177LΔLVR efficiently replicated in PIPEC and after 30 passages no additional genomic mutation [125]. - ASFV-G-ΔI177LΔLVR → safe and protective (100% protection against challenge with parental virulent ASFV-G) [125]. | Promising results, but more studies are required to evaluate its suitability to grow ASFV strains for large-scale vaccine production. | |
| ZMAC-4 | Pig (fetus) [126] | Lung macrophages [126]. | Spontaneously immortalized cell line [126]. | - Benin 1997/1, - Georgia 2007/1, - Malawi LIL20/1, - Tengani, - MOZ 94/1, - ZOM 2/84, - Dominican Republic [126] | - OUTR88/3 - NH/P68 [126] | - Efficiently supported the growth of Georgia 2007/1 to levels similar to bone-marrow-derived macrophages [126]. - 12 passages of the attenuated OURT88/3 ASFV in ZMAC-4 cells → safe and protective (able to induce a 100% protective response in pigs against challenge with virulent ASFV) [126] | Promising results, but more studies are required to evaluate its suitability to grow ASFV strains for large-scale vaccine production. |
| A4C2 and A4C2/9k | Pig [93] | Hybrid cell lines of SPEV TK with swine lymphocytes [93] | Not described [93] | -Stavropol 01/08 [93] - Sveromorsk 2010, Volgograd/Kalach 2012, Tver/Zavidovo [93] | - Hemadsorption was visible after adding 0.5% suspension of pig erythrocytes [93]. | - Stavropol at passage 14 in the A4C2/9k maintained the proper virulent titers strain [93] - Stavropol at passages 24 and 33 → loss of pathogenicity in pigs but no protection to challenge with parental virulent strain [93]. | |
| PSGK-60/PPK-66b | Pig [93] | Kidney [93] | Not described [93] | - Stavropol 01/08 in PSGK-60 and PPK-66b [93] - TSP-080 and TS-7 (two ASFV strains derived from ASFV Kiravira-67) in PPK-66b [128] - Uganda in PPK-66b [128] | - TSP-080/300 (derived from TSP-080) in PPK-66b [128] - TS-7/230 (derived from TS-7) in PPK-66b [128] - Uk-50 (derived from Uganda): PPK-66b [128] | - Stavropol 01/08: maintained virulent properties → infected pigs died [93] - TSP-080/300 and TS-7/230 →Low-reactogenic and able to protect pigs against challenge with the corresponding parental strain [128]. | - No hemadsorption [93] -Promising results, but more studies are required to evaluate its suitability to grow ASFV strains for large-scale vaccine production. |
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Martins, C.; Boinas, F.; Iacolina, L.; Ruiz-Fons, F.; Gavier-Widén, D. African Swine Fever (ASF), the pig health challenge of the century. In Understanding and Combatting African Swine Fever; Wageningen Iacolina, L., Penrith, M.-L., Bellini, S., Chenais, E., Jori, F., Montoya, M., Ståhl, K., Gavier-Widén, D., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2021; pp. 11–24. [Google Scholar]
- Montgomery, R.E. On A Form of swine fever occurring in British East Africa (Kenya Colony). J. Comp. Pathol. Ther. 1921, 34, 159–191. [Google Scholar] [CrossRef] [Green Version]
- Turlewicz-Podbielska, H.; Kuriga, A.; Niemyjski, R.; Tarasiuk, G.; Pomorska-Mól, M. African swine fever virus as a difficult opponent in the fight for a vaccine—Current data. Viruses 2021, 13, 1212. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Sun, H.; Roberts, H. African swine fever. Antivir. Res. 2019, 165, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Netherton, C.L.; Goatley, L.C.; Reis, A.L.; Portugal, R.; Nash, R.H.; Morgan, S.B.; Gault, L.; Nieto, R.; Norlin, V.; Gallardo, C.; et al. Identification and immunogenicity of African swine fever virus antigens. Front. Immunol. 2019, 10, 1318. [Google Scholar] [CrossRef] [Green Version]
- Quembo, C.J.; Jori, F.; Vosloo, W.; Heath, L. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transbound. Emerg. Dis. 2018, 65, 420–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achenbach, J.E.; Gallardo, C.; Pelegrin, E.N.; Rivera-Arroyo, B.; Degefa-Negi, T.; Arias, M.; Jenberie, S.; Mulisa, D.D.; Gizaw, D.; Gelaye, E.; et al. Identification of a new genotype of African swine fever virus in domestic pigs from Ethiopia. Transbound. Emerg. Dis. 2017, 64, 1393–1404. [Google Scholar] [CrossRef]
- OIE. WAHIS Interface. Available online: https://wahis.oie.int/#//dashboards/country-or-disease-dashboard (accessed on 4 January 2022).
- Bosch-Camós, L.; López, E.; Rodriguez, F. African swine fever vaccines: A promising work still in progress. Porc. Health Manag. 2020, 6, 17. [Google Scholar] [CrossRef]
- Nielsen, S.S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Depner, K.; Drewe, J.A.; Garin-Bastuji, B.; Rojas, J.L.G.; Schmidt, C.G.; European Food Safety Authority (EFSA); et al. Research priorities to fill knowledge gaps on ASF seasonality that could improve the control of ASF. EFSA J. 2021, 19, e06550. [Google Scholar] [CrossRef]
- Mauroy, A.; Depoorter, P.; Saegerman, C.; Cay, B.; De Regge, N.; Filippitzi, M.; Fischer, C.; Laitat, M.; Maes, D.; Morelle, K.; et al. Semi-quantitative risk assessment by expert elicitation of potential introduction routes of African swine fever from wild reservoir to domestic pig industry and subsequent spread during the Belgian outbreak (2018–2019). Transbound. Emerg. Dis. 2021, 68, 2761–2773. [Google Scholar] [CrossRef]
- Loi, F.; Cappai, S.; Coccollone, A.; Rolesu, S. Standardized risk analysis approach aimed to evaluate the last African swine fever eradication program performance, in Sardinia. Front. Vet. Sci. 2019, 6, 299. [Google Scholar] [CrossRef]
- Qiu, Z.; Li, Z.; Yan, Q.; Li, Y.; Xiong, W.; Wu, K.; Li, X.; Fan, S.; Zhao, M.; Ding, H.; et al. Development of diagnostic tests provides technical support for the control of African swine fever. Vaccines 2021, 9, 343. [Google Scholar] [CrossRef]
- Busch, F.; Haumont, C.; Penrith, M.-L.; Laddomada, A.; Dietze, K.; Globig, A.; Guberti, V.; Zani, L.; Depner, K. Evidence-based African swine fever policies: Do we address virus and host adequately? Front. Vet. Sci. 2021, 8, 224. [Google Scholar] [CrossRef]
- Arias, M.; De La Torre, A.; Dixon, L.; Gallardo, C.; Jori, F.; Laddomada, A.; Martins, C.; Parkhouse, R.M.; Revilla, Y.; Rodriguez, F.a.J.-M.; et al. Approaches and perspectives for development of African swine fever virus vaccines. Vaccines 2017, 5, 35. [Google Scholar] [CrossRef]
- Blome, S.; Franzke, K.; Beer, M. African swine fever—A review of current knowledge. Virus Res. 2020, 287, 198099. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Madden, D.W.; Wilson, W.C.; Trujillo, J.D.; Richt, J.A. African swine fever virus: An emerging DNA arbovirus. Front. Vet. Sci. 2020, 7, 215. [Google Scholar] [CrossRef]
- Sánchez, E.G.; Pérez-Núñez, D.; Revilla, Y. Mechanisms of entry and endosomal pathway of African swine fever virus. Vaccines 2017, 5, 42. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Villamandos, J.; Bautista, M.; Cordón, P.S.; Carrasco, L. Pathology of African swine fever: The role of monocyte-macrophage. Virus Res. 2013, 173, 140–149. [Google Scholar] [CrossRef]
- Carrascosa, A.L.; Bustos, M.J.; de Leon, P. Methods for growing and titrating African swine fever virus: Field and laboratory samples. Curr. Protoc. Cell Biol. 2011, 53, 26.14.1–26.14.25. [Google Scholar] [CrossRef]
- Franzoni, G.; Giudici, S.D.; Oggiano, A. Infection, modulation and responses of antigen-presenting cells to African swine fever viruses. Virus Res. 2018, 258, 73–80. [Google Scholar] [CrossRef]
- Montoya, M.; Franzoni, G.; Pérez-Nuñez, D.; Revilla, Y.; Galindo, I.; Alonso, C.; Netherton, C.; Blohm, U. 3. Immune responses against African swine fever virus infection. In Understanding and Combatting African Swine Fever; Wageningen Academic Publishers: Wageningen, The Netherlands, 2021; pp. 11–24. [Google Scholar]
- Masujin, K.; Kitamura, T.; Kameyama, K.-I.; Okadera, K.; Nishi, T.; Takenouchi, T.; Kitani, H.; Kokuho, T. An immortalized porcine macrophage cell line competent for the isolation of African swine fever virus. Sci. Rep. 2021, 11, 4759. [Google Scholar] [CrossRef]
- Gómez-Villamandos, J.; Hervás, J.; Mendéz, A.; Carrasco, L.; Villeda, C.; Wilkinson, P.; Sierra, M. A pathological study of the perisinusoidal unit of the liver in acute African swine fever. Res. Vet. Sci. 1995, 59, 146–151. [Google Scholar] [CrossRef]
- Carrasco, L.; Sierra, M.A.; Hervas, J.; Gomez-Villamandos, J.C.; Mendez, A. The lesional changes and pathogenesis in the kidney in African swine fever. Vet. Res. Commun. 1996, 20, 285–299. [Google Scholar] [CrossRef]
- Mínguez, I.; Rueda, A.; Domínguez, J.; Sánchez-Vizcaíno, J.M. Double labeling immunohistological study of African swine fever virus-infected spleen and lymph nodes. Vet. Pathol. 1988, 25, 193–198. [Google Scholar] [CrossRef]
- Carrasco, L.; de Lara, F.C.-M.; Gómez-Villamandos, J.; Bautista, M.; Villeda, C.; Wilkinson, P.; Sierra, M. The pathogenic role of pulmonary intravascular macrophages in acute African swine fever. Res. Vet. Sci. 1996, 61, 193–198. [Google Scholar] [CrossRef]
- Gómez-Villamandos, J.C.; Hervás, J.; Mendez, A.; Carrasco, L.; Villeda, C.J.; Wilkinson, P.J.; Sierra, M.A. Ultrastructural study of the renal tubular system in acute experimental African swine fever: Virus replication in glomerular mesangial cells and in the collecting ducts. Arch. Virol. 1995, 140, 581–589. [Google Scholar] [CrossRef]
- Salguero, F.J.; Ruiz-Villamor, E.; Bautista, M.; Sánchez-Cordón, P.; Carrasco, L.; Gómez-Villamandos, J. Changes in macrophages in spleen and lymph nodes during acute African swine fever: Expression of cytokines. Vet. Immunol. Immunopathol. 2002, 90, 11–22. [Google Scholar] [CrossRef]
- de Leon, P.; Bustos, M.J.; Carrascosa, A.L. Laboratory methods to study African swine fever virus. Virus Res. 2013, 173, 168–179. [Google Scholar] [CrossRef]
- Galindo, I.; Alonso, C. African swine fever virus: A review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [Green Version]
- Galindo, I.; Cuesta-Geijo, M.A.; Hlavova, K.; Muñoz-Moreno, R.; Barrado-Gil, L.; Dominguez, J.; Alonso, C. African swine fever virus infects macrophages, the natural host cells, via clathrin- and cholesterol-dependent endocytosis. Virus Res. 2015, 200, 45–55. [Google Scholar] [CrossRef]
- Ballester, M.; Rodríguez-Cariño, C.; Pérez, M.; Gallardo, C.; Rodríguez, J.M.; Salas, M.L.; Rodriguez, F. Disruption of nuclear organization during the initial phase of african swine fever virus infection. J. Virol. 2011, 85, 8263–8269. [Google Scholar] [CrossRef] [Green Version]
- Fairbairn, L.; Kapetanovic, R.; Sester, D.P.; Hume, D.A. The mononuclear phagocyte system of the pig as a model for understanding human innate immunity and disease. J. Leukoc. Biol. 2011, 89, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Torres, C.; Gómez-Puertas, P.; Gómez-del-Moral, M.; Alonso, F.; Escribano, J.M.; Ezquerra, A.; Domínguez, J. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch. Virol. 2003, 148, 2307–2323. [Google Scholar] [CrossRef] [PubMed]
- Popescu, L.; Gaudreault, N.N.; Whitworth, K.M.; Murgia, M.V.; Nietfeld, J.C.; Mileham, A.; Samuel, M.; Wells, K.D.; Prather, R.S.; Rowland, R.R. Genetically edited pigs lacking CD163 show no resistance following infection with the African swine fever virus isolate, Georgia 2007/1. Virology 2017, 501, 102–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franzoni, G.; Graham, S.P.; Sanna, G.; Angioi, P.; Fiori, M.S.; Anfossi, A.; Amadori, M.; Giudici, S.D.; Oggiano, A. Interaction of porcine monocyte-derived dendritic cells with African swine fever viruses of diverse virulence. Vet. Microbiol. 2018, 216, 190–197. [Google Scholar] [CrossRef]
- Gao, Q.; Yang, Y.; Feng, Y.; Quan, W.; Luo, Y.; Wang, H.; Zheng, J.; Chen, X.; Huang, Z.; Chen, X.; et al. Effects of the NF-κB signaling pathway inhibitor BAY11-7082 in the Replication of ASFV. Viruses 2022, 14, 297. [Google Scholar] [CrossRef]
- McCullough, K.C.; Basta, S.; Knötig, S.; Gerber, H.; Schaffner, R.; Kim, Y.B.; Saalmüller, A.; Summerfield, A. Intermediate stages in monocyte-macrophage differentiation modulate phenotype and susceptibility to virus infection. Immunology 1999, 98, 203–212. [Google Scholar] [CrossRef]
- Basta, S.; Knoetig, S.M.; Spagnuolo-Weaver, M.; Allan, G.; McCullough, K.C. Modulation of monocytic cell activity and virus susceptibility during differentiation into macrophages . J. Immunol. 1999, 162, 3961–3969. [Google Scholar]
- Franzoni, G.; Bonelli, P.; Graham, S.; Anfossi, A.G.; Giudici, S.D.; Pilo, G.; Pittau, M.; Nicolussi, P.; Oggiano, A. Comparative phenotypic and functional analyses of the effects of autologous plasma and recombinant human macrophage-colony stimulating factor (M-CSF) on porcine monocyte to macrophage differentiation. Vet. Immunol. Immunopathol. 2017, 187, 80–88. [Google Scholar] [CrossRef]
- Sautter, C.A.; Auray, G.; Python, S.; Liniger, M.; Summerfield, A. Phenotypic and functional modulations of porcine macrophages by interferons and interleukin-4. Dev. Comp. Immunol. 2018, 84, 181–192. [Google Scholar] [CrossRef]
- Malmquist, W.A.; Hay, D. Hemadsorption and cytopathic effect produced by African Swine Fever virus in swine bone marrow and buffy coat cultures. Am. J. Vet. Res. 1960, 21, 104–108. [Google Scholar]
- World Organization for Animal Health (OIE). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2019; Chapter 3.8.1—African Swine Fever (Infection with African Swine Fever Virus); OIE: Paris, France, 2019. [Google Scholar]
- Wang, T.; Wang, L.; Han, Y.; Pan, L.; Yang, J.; Sun, M.; Zhou, P.; Sun, Y.; Bi, Y.; Qiu, H. Adaptation of African swine fever virus to HEK293T cells. Transbound. Emerg. Dis. 2021, 68, 2853–2866. [Google Scholar] [CrossRef]
- Lithgow, P.; Takamatsu, H.; Werling, D.; Dixon, L.; Chapman, D. Correlation of cell surface marker expression with African swine fever virus infection. Vet. Microbiol. 2014, 168, 413–419. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Zhao, D.; He, X.; Liu, R.; Wang, Z.; Zhang, X.; Li, F.; Shan, D.; Chen, H.; Zhang, J.; et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020, 63, 623–634. [Google Scholar] [CrossRef]
- Carrascosa, A.L.; Santarén, J.F.; Viñuela, E. Production and titration of African swine fever virus in porcine alveolar macrophages. J. Virol. Methods 1982, 3, 303–310. [Google Scholar] [CrossRef]
- Du Manoir, J.M.; Albright, B.N.; Stevenson, G.; Thompson, S.H.; Mitchell, G.B.; Clark, M.E.; Caswell, J.L. Variability of neutrophil and pulmonary alveolar macrophage function in swine. Vet. Immunol. Immunopathol. 2002, 89, 175–186. [Google Scholar] [CrossRef]
- Oh, T.; Do, D.T.; Van Vo, H.; Kwon, H.-I.; Lee, S.-C.; Kim, M.H.; Nguyen, D.T.T.; Le, Q.T.V.; Tran, T.M.; Nguyen, T.T.; et al. The isolation and replication of African swine fever virus in primary renal-derived swine macrophages. Front. Vet. Sci. 2021, 8, 251. [Google Scholar] [CrossRef]
- Takenouchi, T.; Suzuki, S.; Shinkai, H.; Tsukimoto, M.; Sato, M.; Uenishi, H.; Kitani, H. Extracellular ATP does not induce P2X7 receptor-dependent responses in cultured renal- and liver-derived swine macrophages. Results Immunol. 2014, 4, 62–67. [Google Scholar] [CrossRef] [Green Version]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Franzoni, G.; Graham, S.P.; Giudici, S.D.; Oggiano, A. Porcine Dendritic Cells and Viruses: An Update. Viruses 2019, 11, 445. [Google Scholar] [CrossRef] [Green Version]
- Gregg, D.A.; Mebus, C.A.; Schlafer, D.H. Early infection of interdigitating dendritic cells in the pig lymph node with African swine fever viruses of high and low virulence: Immunohistochemical and ultrastructural studies. J. Vet. Diagn. Investig. 1995, 7, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Sehl, J.; Pikalo, J.; Schäfer, A.; Franzke, K.; Pannhorst, K.; Elnagar, A.; Blohm, U.; Blome, S.; Breithaupt, A. Comparative pathology of domestic pigs and wild boar infected with the moderately virulent African swine fever virus strain “Estonia 2014”. Pathogens 2020, 9, 662. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, C.P.; Rigden, R.C.; Schaffner, R.; Gerber, H.; Neuhaus, V.; Inumaru, S.; Takamatsu, H.; Bertoni, G.; McCullough, K.C.; Summerfield, A. Porcine dendritic cells generated in vitro: Morphological, phenotypic and functional properties. Immunology 2001, 104, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Nash, R.H. Understanding the Interactions of African Swine Fever Virus with Dendritic Cells and the Adaptive Immune Response. Ph.D. Thesis, School of Veterinary Medicine, University of Surrey, Guilford, UK, 31 March 2021. [Google Scholar]
- Salguero, F.J. Comparative pathology and pathogenesis of African swine fever infection in swine. Front. Vet. Sci. 2020, 7, 282. [Google Scholar] [CrossRef] [PubMed]
- Vallée, I.; Tait, S.W.G.; Powell, P.P. African swine fever virus infection of porcine aortic endothelial cells leads to inhibition of inflammatory responses, activation of the thrombotic state, and apoptosis. J. Virol. 2001, 75, 10372–10382. [Google Scholar] [CrossRef] [Green Version]
- Carrillo, A.; Chamorro, S.; Rodrı́guez-Gago, M.; Álvarez, B.; Molina, M.J.; Rodrı́guez-Barbosa, J.I.; Sánchez, A.; Ramı́rez, P.; Muñoz, A.; Domı́nguez, J.; et al. Isolation and characterization of immortalized porcine aortic endothelial cell lines. Vet. Immunol. Immunopathol. 2002, 89, 91–98. [Google Scholar] [CrossRef]
- Krug, P.W.; Holinka, L.G.; O’Donnell, V.; Reese, B.; Sanford, B.; Fernandez-Sainz, I.; Gladue, D.P.; Arzt, J.; Rodriguez, L.; Risatti, G.R.; et al. The Progressive adaptation of a Georgian isolate of African swine fever virus to Vero cells leads to a gradual attenuation of virulence in swine corresponding to major modifications of the viral genome. J. Virol. 2015, 89, 2324–2332. [Google Scholar] [CrossRef] [Green Version]
- Santurde, G.; Gonzalvo, F.R.; Carnero, M.E. Genetic stability of African swine fever virus grown in monkey kidney cells. Arch. Virol. 1988, 98, 117–122. [Google Scholar] [CrossRef]
- Tabarés, E.; Olivares, I.; Santurde, G.; Garcia, M.J.; Martin, E.; Carnero, M.E. African swine fever virus DNA: Deletions and additions during adaptation to growth in monkey kidney cells. Arch. Virol. 1987, 97, 333–346. [Google Scholar] [CrossRef]
- Sánchez, E.G.; Pérez-Núñez, D.; Revilla, Y. Development of vaccines against African swine fever virus. Virus Res. 2019, 265, 150–155. [Google Scholar] [CrossRef]
- Hess, W.R. African swine fever virus. Virol. Monogr. 1971, 9, 1–33. [Google Scholar]
- Enjuanes, L.; Carrascosa, A.L.; Moreno, M.A.; Vinuela, E. Titration of African Swine Fever (ASF) virus. J. Gen. Virol. 1976, 32, 471–477. [Google Scholar] [CrossRef]
- Rodriguez, J.M.; Moreno, L.T.; Alejo, A.; Lacasta, A.; Rodríguez, F.; Salas, M.L. Genome sequence of African Swine Fever Virus BA71, the virulent parental strain of the nonpathogenic and tissue-culture adapted BA71V. PLoS ONE 2015, 10, e0142889. [Google Scholar] [CrossRef] [Green Version]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic atlas of the African swine fever virus particle. J. Virol. 2018, 92, e01293–e01318. [Google Scholar] [CrossRef] [Green Version]
- Cackett, G.; Matelska, D.; Sýkora, M.; Portugal, R.; Malecki, M.; Bähler, J.; Dixon, L.; Werner, F. The African swine fever virus transcriptome. J. Virol. 2020, 94, e00119–e00220. [Google Scholar] [CrossRef] [Green Version]
- Matamoros, T.; Alejo, A.; Rodríguez, J.M.; Hernáez, B.; Guerra, M.; Fraile-Ramos, A.; Andrés, G. African swine fever virus protein pE199L mediates virus entry by enabling membrane fusion and core penetration. mBio 2020, 11, e00789–e00820. [Google Scholar] [CrossRef]
- Nogal, M.L.; de Buitrago, G.G.; Rodrı́guez, C.; Cubelos, B.; Carrascosa, A.L.; Salas, M.L.; Revilla, Y. african swine fever virus IAP homologue inhibits caspase activation and promotes cell survival in mammalian cells. J. Virol. 2001, 75, 2535–2543. [Google Scholar] [CrossRef] [Green Version]
- Alonso, C.I.; Cuesta-Geijo, M.A.; Cabezas, M.; Hernaez, B.; Munoz-Moreno, R. African swine fever virus-cell interactions: From virus entry to cell survival. Virus Res. 2013, 173, 42–57. [Google Scholar] [CrossRef]
- Alcamí, A.; Carrascosa, A.L.; Viñuela, E. The entry of African swine fever virus into Vero cells. Virology 1989, 171, 68–75. [Google Scholar] [CrossRef]
- Sánchez, E.G.; Quintas, A.; Pérez-Núñez, D.; Nogal, M.; Barroso, S.; Carrascosa, Á.L.; Revilla, Y. African swine fever virus uses macropinocytosis to enter host cells. PLoS Pathog. 2012, 8, e1002754. [Google Scholar] [CrossRef] [Green Version]
- Hakobyan, A.; Galindo, I.; Nañez, A.; Arabyan, E.; Karalyan, Z.; Chistov, A.; Streshnev, P.P.; Korshun, V.A.; Alonso, C.; Zakaryan, H. Rigid amphipathic fusion inhibitors demonstrate antiviral activity against African swine fever virus. J. Gen. Virol. 2018, 99, 148–156. [Google Scholar] [CrossRef]
- Arabyan, E.; Hakobyan, A.; Kotsinyan, A.; Karalyan, Z.; Arakelov, V.; Arakelov, G.; Nazaryan, K.; Simonyan, A.; Aroutiounian, R.; Ferreira, F.; et al. Genistein inhibits African swine fever virus replication in vitro by disrupting viral DNA synthesis. Antivir. Res. 2018, 156, 128–137. [Google Scholar] [CrossRef]
- Hakobyan, A.; Arabyan, E.; Kotsinyan, A.; Karalyan, Z.; Sahakyan, H.; Arakelov, V.; Nazaryan, K.; Ferreira, F.; Zakaryan, H. Inhibition of African swine fever virus infection by genkwanin. Antivir. Res. 2019, 167, 78–82. [Google Scholar] [CrossRef]
- Mottola, C.; de Freitas, F.B.; Simões, M.P.; Martins, C.; Leitao, A.; Ferreira, F. In vitro antiviral activity of fluoroquinolones against African swine fever virus. Vet. Microbiol. 2013, 165, 86–94. [Google Scholar] [CrossRef]
- Sanford, B.; Holinka, L.; O’Donnell, V.; Krug, P.; Carlson, J.; Alfano, M.; Carrillo, C.; Wu, P.; Lowe, A.; Risatti, G.; et al. Deletion of the thymidine kinase gene induces complete attenuation of the Georgia isolate of African swine fever virus. Virus Res. 2016, 213, 165–171. [Google Scholar] [CrossRef]
- Pires, S.; Ribeiro, G.; Costa, J.V. Sequence and organization of the left multigene family 110 region of the Vero-adapted L60V strain of African swine fever virus. Virus Genes 1997, 15, 271–274. [Google Scholar] [CrossRef]
- Monteagudo, P.L.; Lacasta, A.; Lopez, L.; Bosch, J.; Collado, J.; Pina-Pedrero, S.; Correa-Fiz, F.; Accensi, F.; Navas, M.J.; Vidal, E.; et al. BA71deltaCD2: A new recombinant live attenuated African swine fever virus with cross-protective capabilities. J. Virol. 2017, 91, e1058–e1117. [Google Scholar] [CrossRef] [Green Version]
- Hurtado, C.; Bustos, M.J.; Carrascosa, A.L. The use of COS-1 cells for studies of field and laboratory African swine fever virus samples. J. Virol. Methods 2010, 164, 131–134. [Google Scholar] [CrossRef] [Green Version]
- Gallardo, C.; Fernández-Pinero, J.; Arias, M. African swine fever (ASF) diagnosis, an essential tool in the epidemiological investigation. Virus Res. 2019, 271, 197676. [Google Scholar] [CrossRef]
- Carrascosa, A.L.; Bustos, M.J.; Galindo, I.; Viñuela, E. Virus-specific cell receptors are necessary, but not sufficient, to confer cell susceptibility to African swine fever virus. Arch. Virol. 1999, 144, 1309–1321. [Google Scholar] [CrossRef]
- Galindo, I.; Almazan, F.; Bustos, M.J.; Viñuela, E.; Carrascosa, A.L. African swine fever virus EP153R open reading frame encodes a glycoprotein involved in the hemadsorption of infected cells. Virology 2000, 266, 340–351. [Google Scholar] [CrossRef]
- Hurtado, C.; Granja, A.G.; Bustos, M.J.; Nogal, M.L.; de Buitrago, G.G.; de Yébenes, V.G.; Salas, M.L.; Revilla, Y.; Carrascosa, A.L. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology 2004, 326, 160–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernáez, B.; Alonso, C. Dynamin- C and clathrin-dependent endocytosis in African swine fever virus entry. J. Virol. 2010, 84, 2100–2109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernaez, B.; Guerra, M.; Salas, M.L.; Andrés, G. African swine fever virus undergoes outer envelope disruption, capsid disassembly and inner envelope fusion before core release from multivesicular endosomes. PLoS Pathog. 2016, 12, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Sánchez, E.G.; Pérez-Núñez, D.; Nogal, M.; de Leon, P.; Carrascosa, Á.L.; Nieto, R.; Soler, A.; Arias, M.L.; Revilla, Y. African swine fever virus (ASFV) protection mediated by NH/P68 and NH/P68 recombinant live-attenuated viruses. Vaccine 2018, 36, 2694–2704. [Google Scholar] [CrossRef]
- Alcaraz, C.; Brun, A.; Ruiz-Gonzalvo, F.; Escribano, J. Cell culture propagation modifies the African swine fever virus replication phenotype in macrophages and generates viral subpopulations differing in protein p54. Virus Res. 1992, 23, 173–182. [Google Scholar] [CrossRef]
- Ruiz Gonzalvo, F.; Carnero, E.; Bruvel, V. Immunological responses of pigs to partially attenuated African swine fever virus and their resistance to virulent homologous and heterologous viruses. In African Swine Fever; Proc EUR 8466, EN.; Wilkinson, P.J., Ed.; CEC/FAO Research Seminar: Sardinia, Italy, 1983; pp. 206–216. [Google Scholar]
- Lacasta, A.; Monteagudo, P.L.; Jiménez-Marín, Á.; Accensi, F.; Ballester, M.; Argilaguet, J.; Galindo-Cardiel, I.; Segalés, J.; Salas, M.L.; Domínguez, J.; et al. Live attenuated African swine fever viruses as ideal tools to dissect the mechanisms involved in viral pathogenesis and immune protection. Vet. Res. 2015, 46, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Balysheva, V.I.; Prudnikova, E.Y.; Galnbek, T.V.; Balyshev, V.M. Immunological properties of attenuated variants of African swine fever virus isolated in the Russian Federation. Russ. Agric. Sci. 2015, 41, 178–182. [Google Scholar] [CrossRef]
- Kim, H.S.; Kwang, J.; Yoon, I.J.; Joo, H.S.; Frey, M.L. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 1993, 133, 477–483. [Google Scholar] [CrossRef]
- Do, D.T.; Nguyen, T.T. Development of high-growth African Swine Fever Virus (ASFV) in MA-104 cells. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Woźniakowski, G.; Mazur-Panasiuk, N.; Walczak, M.; Juszkiewicz, M.; Frant, M.; Niemczuk, K. Attempts at the development of a recombinant African swine fever virus strain with abrogated EP402R, 9GL, and A238L gene structure using the CRISPR/Cas9 system. J. Vet. Res. 2020, 64, 197–205. [Google Scholar] [CrossRef]
- Rai, A.; Pruitt, S.; Ramirez-Medina, E.; Vuono, E.A.; Silva, E.; Velazquez-Salinas, L.; Carrillo, C.; Borca, M.V.; Gladue, D.P. Identification of a continuously stable and commercially available cell line for the identification of infectious African swine fever virus in clinical samples. Viruses 2020, 12, 820. [Google Scholar] [CrossRef]
- Kwon, H.; Do, D.T.; Vo, H.V.; Lee, S.; Kim, M.; Nguyen, D.T.T.; Tran, T.M.; Le, Q.T.V.; Ngo, T.T.; Nguyen, N.M.; et al. Development of optimized protocol for culturing African Swine Fever Virus (ASFV) field isolates in MA104 cells. Mol. Biol. Rep. (under review).
- Yang, B.; Zhang, D.; Shi, X.; Shen, C.; Hao, Y.; Zhang, T.; Yang, J.; Yuan, X.; Chen, X.; Zhao, D.; et al. Construction, identification and analysis of the interaction network of African swine fever virus MGF360-9L with host proteins. Viruses 2021, 13, 1804. [Google Scholar] [CrossRef]
- Zhao, G.; Li, T.; Liu, X.; Zhang, T.; Zhang, Z.; Kang, L.; Song, J.J.; Zhou, S.; Chen, X.; Wang, X.; et al. African swine fever virus cysteine protease pS273R inhibits pyroptosis by non-canonically cleaving gasdermin D. JBC 2022, 298, 101480. [Google Scholar] [CrossRef]
- Niederwerder, M.C.; Hefley, T.J. Diagnostic sensitivity of porcine biological samples for detecting African swine fever virus infection after natural consumption in feed and liquid. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Keßler, C.; Forth, J.H.; Keil, G.M.; Mettenleiter, T.C.; Blome, S.; Karger, A. The intracellular proteome of African swine fever virus. Sci. Rep. 2018, 8, 14714. [Google Scholar] [CrossRef] [Green Version]
- Ammerman, N.C.; Beier-Sexton, M.; Azad, A.F. Growth and maintenance of Vero cell lines. Curr. Protoc. Microbiol. 2008, 11, A.4E.1–A.4E.7. [Google Scholar] [CrossRef] [Green Version]
- Manohar, M.; Orrison, B.; Peden, K.; Lewis, A.M. Assessing the tumorigenic phenotype of Vero cells in adult and newborn nude mice. Biology 2008, 36, 65–72. [Google Scholar] [CrossRef]
- Hancock, J.F. COS cell expression. Methods Mol Biol. 1992, 8, 153–158. [Google Scholar]
- ECACC. General Cell Collection: 91070510 MS. Available online: https://culturecollections.org.uk (accessed on 7 March 2022).
- Gluzman, Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell 1981, 23, 175–182. [Google Scholar] [CrossRef]
- Dotti, S.; Lombardo, T.; Villa, R.; Cacciamali, A.; Zanotti, C.; Andreani, N.A.; Cinotti, S.; Ferrari, M. Transformation and tumorigenicity testing of simian cell lines and evaluation of poliovirus replication. PLoS ONE 2017, 12, e0169391. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Boone, M.; Meuris, L.; Lemmens, I.; Van Roy, N.; Soete, A.; Reumers, J.; Moisse, M.; Plaisance, S.; Drmanac, R.T.; et al. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun. 2014, 5, 4767. [Google Scholar] [CrossRef] [Green Version]
- Greig, A.S.; Boulanger, P.; Bannister, G.L. African swine fever. V. Cultivation of the virus in primary pig kidney cells. Can. J. Comp. Med. Veter.-Sci. 1967, 31, 24–31. [Google Scholar]
- Dardiri, A.H.; Bachrach, H.L.; Heller, E. Inhibition by rifampin of African swine fever virus replication in tissue culture. Infect. Immun. 1971, 4, 34–36. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.; Plowright, W. Plaque formation by African swine fever virus. Nature 1968, 219, 524–525. [Google Scholar] [CrossRef]
- Calvert, J.G.; Slade, D.E.; Shields, S.L.; Jolie, R.; Mannan, R.M.; Ankenbauer, R.G.; Welch, S.-K.W. CD163 Expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 2007, 81, 7371–7379. [Google Scholar] [CrossRef] [Green Version]
- Arabyan, E.; Kotsynyan, A.; Hakobyan, A.; Zakaryan, H. Antiviral agents against African swine fever virus. Virus Res. 2019, 270, 197669. [Google Scholar] [CrossRef]
- Sánchez, E.G.; Riera, E.; Nogal, M.; Gallardo, C.; Fernández, P.; Bello-Morales, R.; López-Guerrero, J.A.; Chitko-McKown, C.G.; Richt, J.A.; Revilla, Y. Phenotyping and susceptibility of established porcine cells lines to African Swine Fever Virus infection and viral production. Sci. Rep. 2017, 7, 10369. [Google Scholar] [CrossRef] [PubMed]
- Portugal, R.; Martins, C.; Keil, G.M. Novel approach for the generation of recombinant African swine fever virus from a field isolate using GFP expression and 5-bromo-2′-deoxyuridine selection. J. Virol. Methods 2012, 183, 86–89. [Google Scholar] [CrossRef]
- Keil, G.M.; Giesow, K.; Portugal, R. A novel bromodeoxyuridine-resistant wild boar lung cell line facilitates generation of African swine fever virus recombinants. Arch. Virol. 2014, 159, 2421–2428. [Google Scholar] [CrossRef] [PubMed]
- Hemmink, J.D.; Abkallo, H.M.; Henson, S.P.; Khazalwa, E.M.; Oduor, B.; Lacasta, A.; Okoth, E.; Fuchs, W.; Bishop, R.P.; Steinaa, L. The African swine fever isolate ASFV-Kenya-1033-IX is highly virulent and stable after growth in the wild boar cell line WSL. bioRxiv 2021, 12, 472778. [Google Scholar]
- Abkallo, H.M.; Svitek, N.; Oduor, B.; Awino, E.; Henson, S.P.; Oyola, S.O.; Mwalimu, S.; Assad-Garcia, N.; Fuchs, W.; Vashee, S.; et al. Rapid CRISPR/Cas9 editing of genotype IX African swine fever virus circulating in eastern and central Africa. Front. Genet. 2021, 12, 1613. [Google Scholar] [CrossRef] [PubMed]
- Wöhnke, E.; Fuchs, W.; Hartmann, L.; Blohm, U.; Blome, S.; Mettenleiter, T.C.; Karger, A. Comparison of the proteomes of porcine macrophages and a stable porcine cell line after infection with African swine fever virus. Viruses 2021, 13, 2198. [Google Scholar] [CrossRef] [PubMed]
- Hübner, A.; Petersen, B.; Keil, G.M.; Niemann, H.; Mettenleiter, T.C.; Fuchs, W. Efficient inhibition of African swine fever virus replication by CRISPR/Cas9 targeting of the viral p30 gene (CP204L). Sci. Rep. 2018, 8, 1449. [Google Scholar] [CrossRef] [Green Version]
- Weingartl, H.; Sabara, M.; Pasick, J.; van Moorlehem, E.; Babiuk, L. Continuous porcine cell lines developed from alveolar macrophages: Partial characterization and virus susceptibility. J. Virol. Methods 2002, 104, 203–216. [Google Scholar] [CrossRef]
- Takenouchi, T.; Kitani, H.; Suzuki, S.; Nakai, M.; Fuchimoto, D.-I.; Tsukimoto, M.; Shinkai, H.; Sato, M.; Uenishi, H. Immortalization and characterization of porcine macrophages that had been transduced with lentiviral vectors encoding the SV40 large T antigen and porcine telomerase reverse transcriptase. Front. Vet. Sci. 2017, 4, 132. [Google Scholar] [CrossRef] [Green Version]
- LaRocco, M.; Krug, P.W.; Kramer, E.; Ahmed, Z.; Pacheco, J.M.; Duque, H.; Baxt, B.; Rodriguez, L.L. A continuous bovine kidney cell line constitutively expressing bovine avb6 integrin has increased susceptibility to foot-andmouth disease virus. J. Clin. Microbiol. 2013, 51, 1714–1720. [Google Scholar] [CrossRef] [Green Version]
- Borca, M.V.; Rai, A.; Ramirez-Medina, E.; Silva, E.; Velazquez-Salinas, L.; Vuono, E.; Pruitt, S.; Espinoza, N.; Gladue, D.P. A cell culture-adapted vaccine virus against the current African swine fever virus pandemic strain. J. Virol. 2021, 95, e00123–e00221. [Google Scholar] [CrossRef]
- Portugal, R.; Goatley, L.C.; Husmann, R.; Zuckermann, F.A.; Dixon, L.K. A porcine macrophage cell line that supports high levels of replication of OURT88/3, an attenuated strain of African swine fever virus. Emerg. Microbes Infect. 2020, 9, 1245–1253. [Google Scholar] [CrossRef]
- Calzada-Nova, G.; Husmann, R.J.; Schnitzlein, W.M.; Zuckermann, F.A. Effect of the host cell line on the vaccine efficacy of an attenuated porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2012, 148, 116–125. [Google Scholar] [CrossRef]
- Sereda, A.D.; Balyshev, V.M.; Kazakova, A.S.; Imatdinov, A.R.; Kolbasov, D.V. protective properties of attenuated strains of African swine fever virus belonging to seroimmunotypes I–VIII. Pathogen 2020, 9, 274. [Google Scholar] [CrossRef] [Green Version]
- Cellosaurus PK-2a. Available online: https://web.expasy.org/cellosaurus/CVCL_S287 (accessed on 7 March 2022).
- Cellosaurus WSL-R. Available online: https://web.expasy.org/cellosaurus/CVCL_0I66 (accessed on 7 March 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meloni, D.; Franzoni, G.; Oggiano, A. Cell Lines for the Development of African Swine Fever Virus Vaccine Candidates: An Update. Vaccines 2022, 10, 707. https://doi.org/10.3390/vaccines10050707
Meloni D, Franzoni G, Oggiano A. Cell Lines for the Development of African Swine Fever Virus Vaccine Candidates: An Update. Vaccines. 2022; 10(5):707. https://doi.org/10.3390/vaccines10050707
Chicago/Turabian StyleMeloni, Dionigia, Giulia Franzoni, and Annalisa Oggiano. 2022. "Cell Lines for the Development of African Swine Fever Virus Vaccine Candidates: An Update" Vaccines 10, no. 5: 707. https://doi.org/10.3390/vaccines10050707
APA StyleMeloni, D., Franzoni, G., & Oggiano, A. (2022). Cell Lines for the Development of African Swine Fever Virus Vaccine Candidates: An Update. Vaccines, 10(5), 707. https://doi.org/10.3390/vaccines10050707







