Design of a Multi-Epitope Vaccine against Tropheryma whipplei Using Immunoinformatics and Molecular Dynamics Simulation Techniques
Abstract
1. Introduction
2. Material and Methods
2.1. Pan-Genome Analysis and Retrieval of Targeted Proteins
2.2. Choosing the Potential Vaccine Candidates
2.3. Epitope Mapping
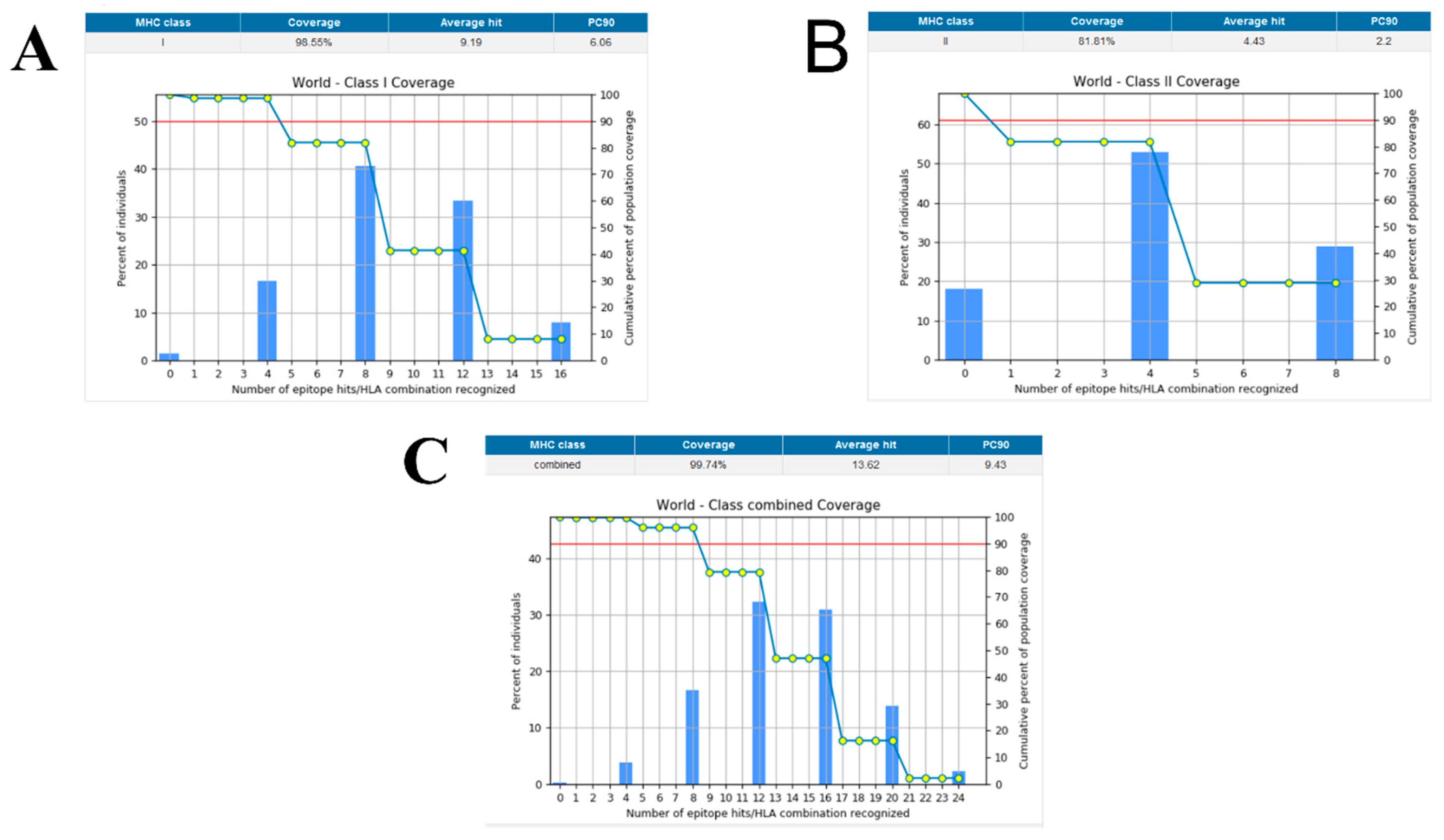
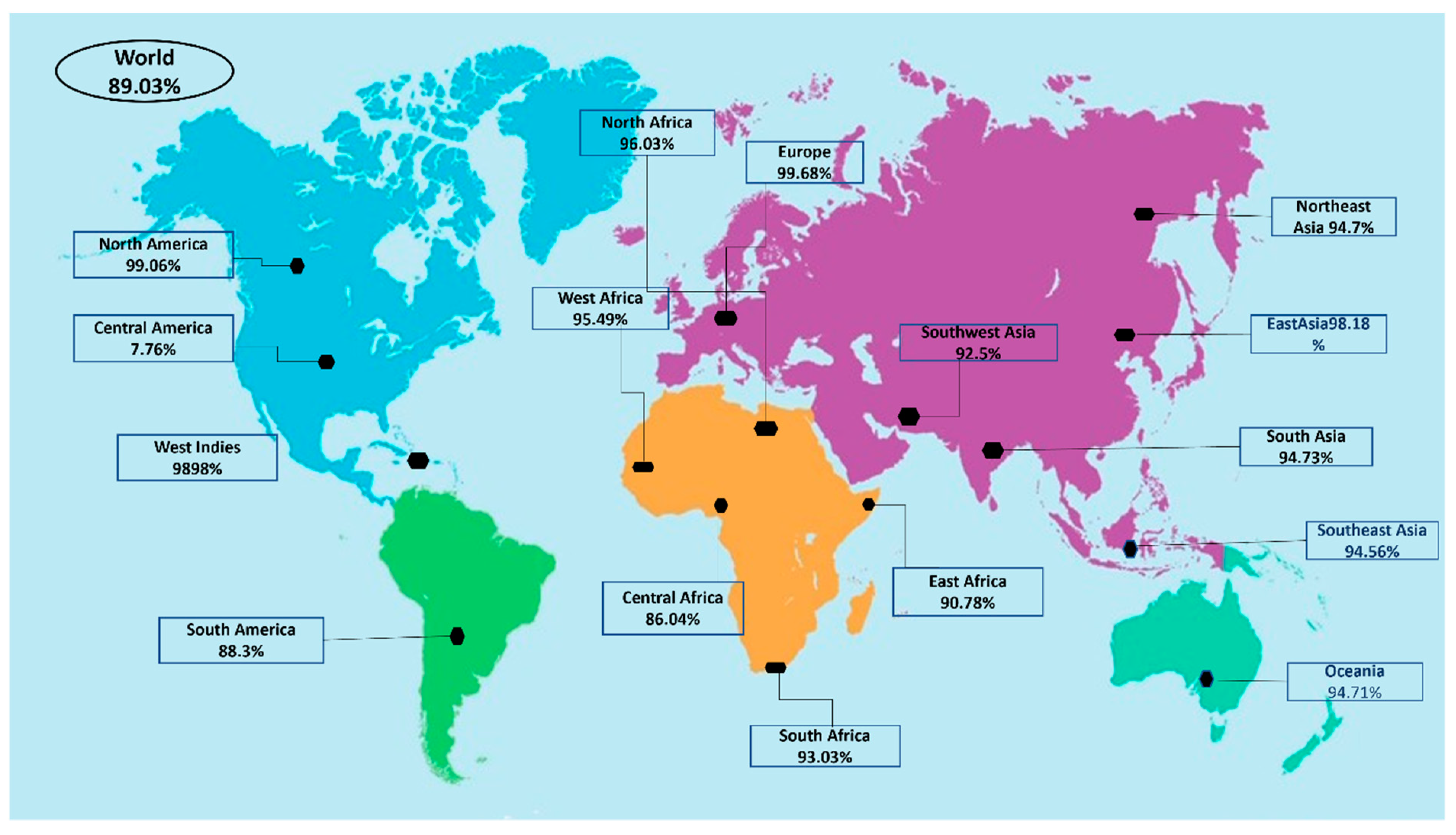
2.4. Population Coverage
2.5. Multi-Epitope Vaccine Design
2.6. Disulfide Engineering and In Silico Cloning
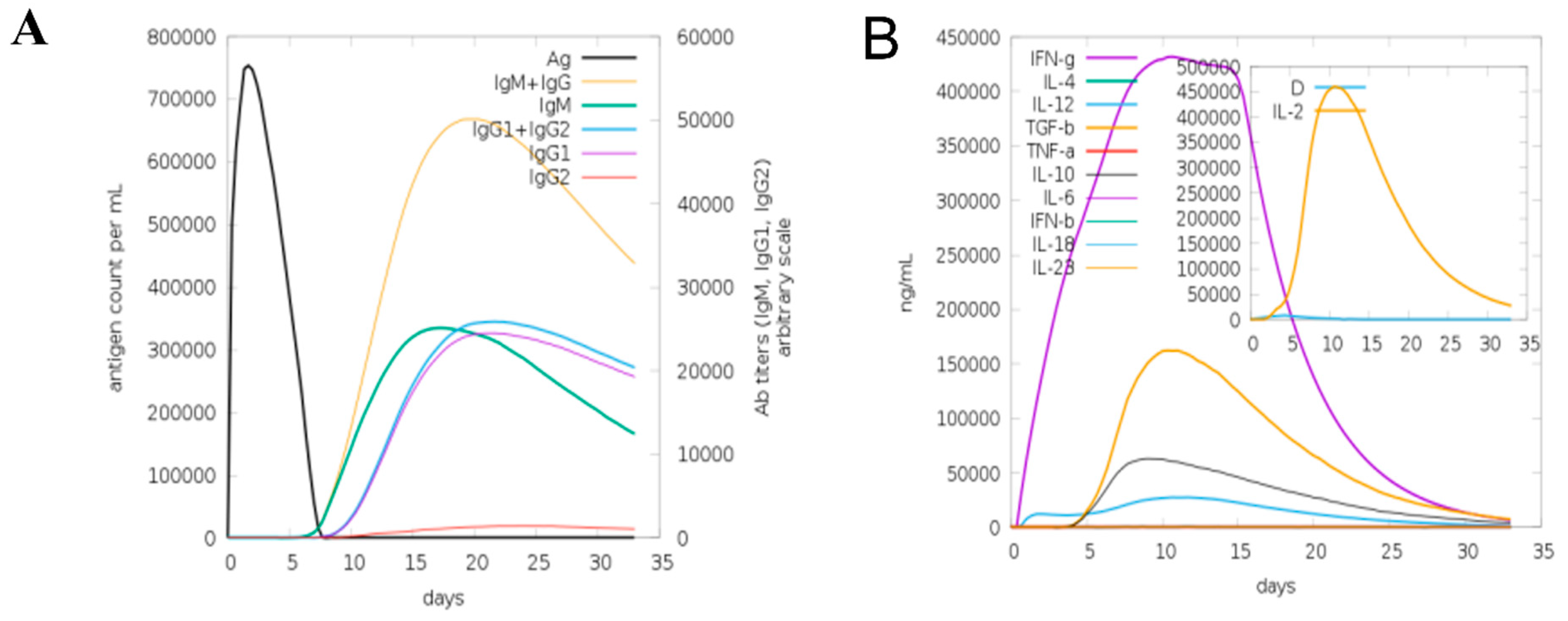
2.7. Computational Immune Simulation
2.8. Molecular Docking
2.9. Molecular Dynamics Simulation
2.10. MM-PB/GBSA Studies
3. Results
3.1. Core Proteome Retrieval
3.2. Determination of Potential Vaccine Candidates
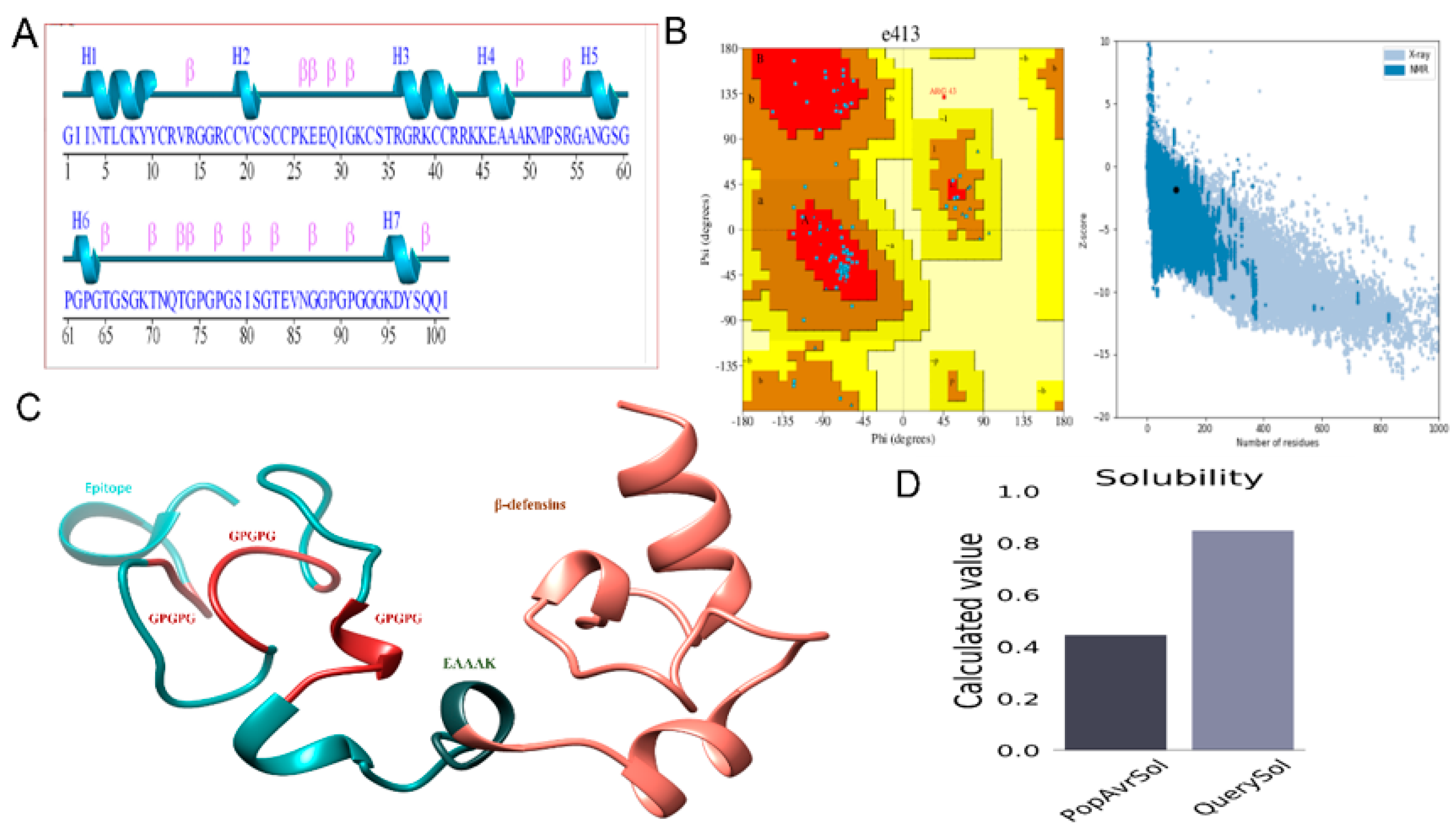
3.3. Epitope Prediction and Prioritization Leading to Vaccine Construct Formation
3.4. Structure Modeling
3.5. Population Coverage
3.6. Disulfide Engineering and In Silico Cloning
3.7. Computational Immune Simulation
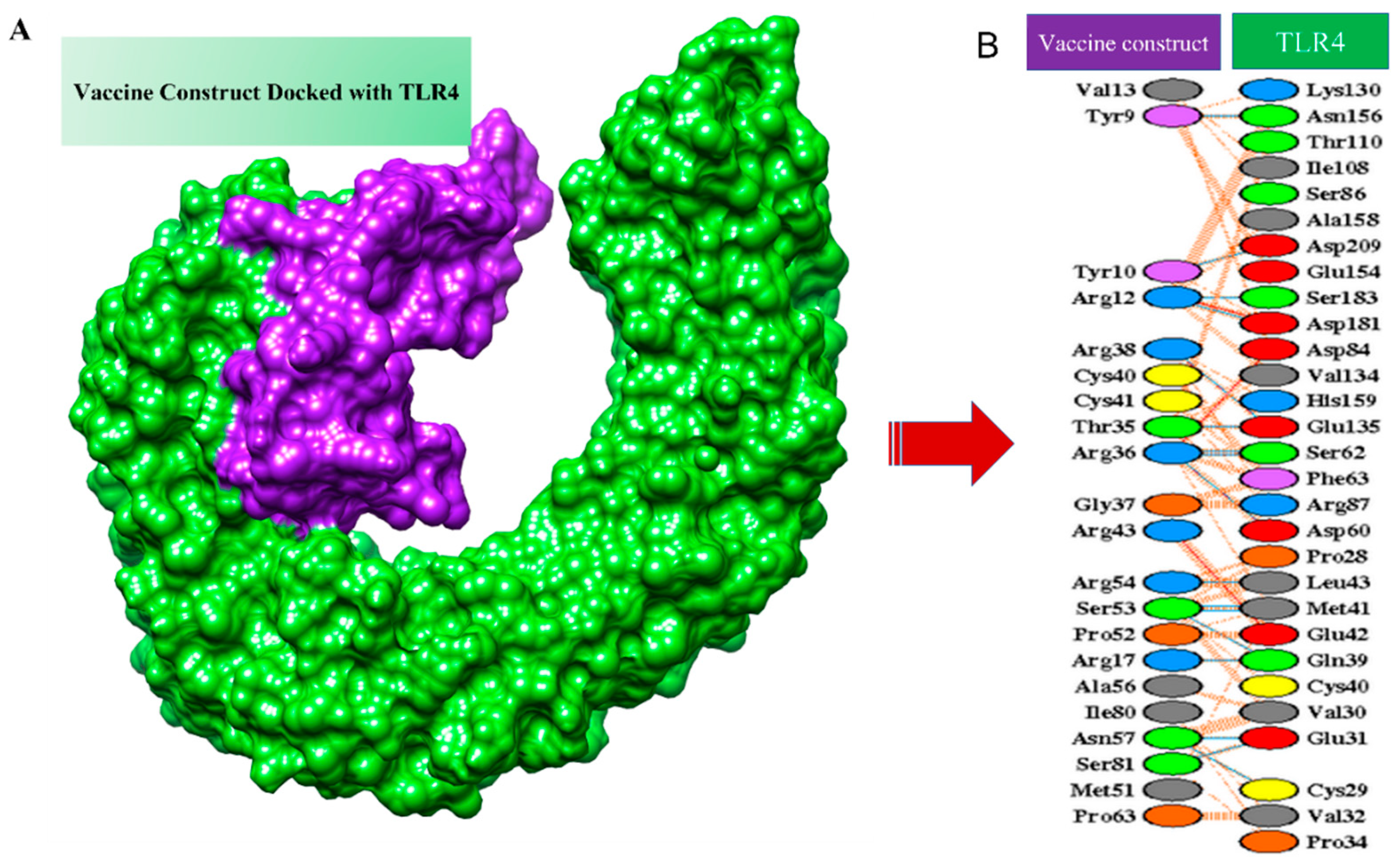
3.8. Molecular Docking of MEPV
3.9. Molecular Dynamics Simulation (MD Simulation)
3.10. Binding Free Energies
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMBER | Assisted Model Building with Energy Refinement tool |
| BPGA | Bacterial pan-genome analysis tool |
| CAI | Codon adaptation index |
| CD-HIT | Cluster database at high identity with tolerance |
| C-Immsimm | Computational immune simulation |
| CTL | Cytotoxic T lymphocytes |
| GRAVY | Grand average of hydropathicity |
| H.B | Hydrogen bonding |
| HTL | Helper T lymphocytes |
| IEDB | Immune epitope database resource |
| IFNg | Interferon gamma |
| IRMSD | Interface root-mean-square deviation |
| JCAT | Java codon adaptation tool |
| MD | Molecular dynamics |
| MEPTWV | Multi-epitope peptide T. whipplei vaccine |
| MMGBSA | Molecular mechanics generalized born surface area |
| MMPBSA | Molecular mechanics Poisson–Boltzmann surface area |
| MW | Molecular weight |
| NCBI | National center of biotechnology information |
| PCR | Polymerase chain reaction |
| PDBsum | Protein databank structural summaries |
| PSORTB | Protein subcellular localization prediction tool |
| RMSD | Root-mean-square deviation |
| RMSF | Root-mean-square fluctuations |
| TLR4 | Toll-like receptor 4 |
| TMHMM | Transmembrane Helices; Hidden Markov Model |
References
- Marth, T.; Moos, V.; Müller, C.; Biagi, F.; Schneider, T. Tropheryma whipplei Infection and Whipple’s Disease. Lancet Infect. Dis. 2016, 16, e13–e22. [Google Scholar] [CrossRef]
- McGee, M.; Brienesse, S.; Chong, B.; Levendel, A.; Lai, K. Tropheryma whipplei Endocarditis: Case Presentation and Review of the Literature. Open Forum Infect. Dis. 2019, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Fenollar, F.; Lagier, J.C.; Raoult, D. Tropheryma whipplei and Whipple’s Disease. J. Infect. 2014, 69, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Ruffer, N.; Holzer, M.-T.; Gkanatsas, Y.; Schinglerová, I.; Boro, D.; Krusche, M.; Kötter, I. Chronic Tropheryma whipplei Infection: An Important Differential Diagnosis of Refractory Polyarthritis. Z. Rheumatol. 2022. [Google Scholar] [CrossRef]
- Lagier, J.C.; Lepidi, H.; Raoult, D.; Fenollar, F. Systemic Tropheryma whipplei: Clinical Presentation of 142 Patients with Infections Diagnosed or Confirmed in a Reference Center. Medicine 2010, 89, 337–345. [Google Scholar] [CrossRef]
- Marth, T.; Raoult, D. Whipple’s Disease. Lancet 2003, 361, 239–246. [Google Scholar] [CrossRef]
- Keita, A.K.; Mediannikov, O.; Ratmanov, P.; Diatta, G.; Bassene, H.; Roucher, C.; Tall, A.; Sokhna, C.; Trape, J.F.; Raoult, D.; et al. Looking for Tropheryma whipplei Source and Reservoir in Rural Senegal. Am. J. Trop. Med. Hyg. 2013, 88, 339. [Google Scholar] [CrossRef]
- Schöniger-Hekele, M.; Petermann, D.; Weber, B.; Müller, C. Tropheryma whipplei in the Environment: Survey of Sewage Plant Influxes and Sewage Plant Workers. Appl. Environ. Microbiol. 2007, 73, 2033–2035. [Google Scholar] [CrossRef]
- Fenollar, F.; Trani, M.; Davoust, B.; Salle, B.; Birg, M.L.; Rolain, J.M.; Raoult, D. Prevalence of Asymptomatic Tropheryma whipplei Carriage among Humans and Nonhuman Primates. J. Infect. Dis. 2008, 197, 880–887. [Google Scholar] [CrossRef]
- Freeman, H.J. Tropheryma whipplei Infection. World J. Gastroenterol. 2009, 15, 2078. [Google Scholar] [CrossRef]
- Fenollar, F.; Laouira, S.; Lepidi, H.; Rolain, J.M.; Raoult, D. Value of Tropheryma whipplei Quantitative Polymerase Chain Reaction Assay for the Diagnosis of Whipple Disease: Usefulness of Saliva and Stool Specimens for First-Line Screening. Clin. Infect. Dis. 2008, 47, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Fenollar, F.; Puéchal, X.; Raoult, D. Whipple’s Disease. N. Engl. J. Med. 2009, 356, 55–66. [Google Scholar] [CrossRef]
- Bentley, S.D.; Maiwald, M.; Murphy, L.D.; Pallen, M.J.; Yeats, C.A.; Dover, L.G.; Norbertczak, H.T.; Besra, G.S.; Quail, M.A.; Harris, D.E.; et al. Sequencing and Analysis of the Genome of the Whipple’s Disease Bacterium Tropheryma whipplei. Lancet 2003, 361, 637–644. [Google Scholar] [CrossRef]
- Boumaza, A.; Azzouz, E.B.; Arrindell, J.; Lepidi, H.; Mezouar, S.; Desnues, B. Whipple’s Disease and Tropheryma whipplei Infections: From Bench to Bedside. Lancet Infect. Dis. 2022. [Google Scholar] [CrossRef]
- al Moussawi, K.; Ghigo, E.; Kalinke, U.; Alexopoulou, L.; Mege, J.L.; Desnues, B. Type I Interferon Induction Is Detrimental during Infection with the Whipple’s Disease Bacterium, Tropheryma whipplei. PLOS Pathog. 2010, 6, e1000722. [Google Scholar] [CrossRef]
- Bulut, K.; Markova, A.; Canbay, A.E.; Schmidt, W.E.; Kahraman, A. Whipple’s Disease—A Rare and Challenging Complication in a Patient with Crohn’s Disease. Z. Gastroenterol. 2022. [Google Scholar] [CrossRef]
- Santos, J.G.; Costa, P.; Galzerano, A.; Matos, C.; Lourenço, J. Challenging Case of Whipple’s Disease: The Contribution of Radiology. Radiol. Case Rep. 2022, 17, 1008–1012. [Google Scholar] [CrossRef]
- Raoult, D.; la Scola, B.; Lecocq, P.; Lepidi, H.; Fournier, P.E. Culture and Immunological Detection of Tropheryma whipplei from the Duodenum of a Patient with Whipple Disease. JAMA 2001, 285, 1039–1043. [Google Scholar] [CrossRef][Green Version]
- Dolmans, R.A.V.; Edwin Boel, C.H.; Lacle, M.M.; Kusters, J.G. Clinical Manifestations, Treatment, and Diagnosis of Tropheryma whipplei Infections. Clin. Microbiol. Rev. 2017, 30, 529–555. [Google Scholar] [CrossRef]
- Dutly, F.; Altwegg, M. Whipple’s Disease and “Tropheryma whippelii”. Clin. Microbiol. Rev. 2001, 14, 561–583. [Google Scholar] [CrossRef]
- Marth, T. Systematic Review: Whipple’s Disease (Tropheryma whipplei Infection) and Its Unmasking by Tumour Necrosis Factor Inhibitors. Aliment. Pharmacol. Ther. 2015, 41, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Moro, L.; Pomari, E.; Leonardi, M.; La Marca, G.; Pajola, B.; Mazzi, C.; Piubelli, C.; Beltrame, A. Tropheryma whipplei, Helicobacter Pylori, and Intestinal Protozoal Co-Infections in Italian and Immigrant Populations: A Cross-Sectional Study. Microorganisms 2022, 10, 769. [Google Scholar] [CrossRef]
- Biagi, F.; Badulli, C.; Feurle, G.E.; Müller, C.; Moos, V.; Schneider, T.; Marth, T.; Mytilineos, J.; Garlaschelli, F.; Marchese, A.; et al. Cytokine Genetic Profile in Whipple’s Disease. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3145–3150. [Google Scholar] [CrossRef] [PubMed]
- Dobbins, W.O.I. The Diagnosis of Whipple’s Disease. N. Engl. J. Med. 2009, 332, 390–392. [Google Scholar] [CrossRef]
- Wang, S.; Xia, D.; Wu, J.; Jia, D.; Li, L.; Xu, S. Severe Pneumonia Caused by Infection with Tropheryma whipplei Complicated with Acinetobacter Baumannii Infection: A Case Report Involving a Young Woman. Front. Public Health 2021, 9, 1598. [Google Scholar] [CrossRef]
- Sayyahfar, S.; Latifian, M.; Esmaeili, P.; Baseri, N.; Bagheri Amiri, F.; Bakhshi, B.; Esteghamati, A.; Esmaeili, S. Tropheryma whipplei in the Stool Samples of Children with Acute Diarrhea: A Study from Tehran, Iran. BMC Infect. Dis. 2022, 22, 193. [Google Scholar] [CrossRef]
- Tison, A.; Preuss, P.; Leleu, C.; Robin, F.; le Pluart, A.; Vix, J.; le Mélédo, G.; Goupille, P.; Gervais, E.; Cormier, G.; et al. Rheumatological Features of Whipple Disease. Sci. Rep. 2021, 11, 12278. [Google Scholar] [CrossRef]
- Edwards, M.; Hamilton, R.; Oliver, N.; Fitzgibbon, S.; Samarasekera, R. Antibiotic Resistance: Modelling the Impact on Mortality and Morbidity A Report by the Antibiotic Resistance Working Party; Institute and Faculty of Actuaries: London, UK, 2019. [Google Scholar]
- Delany, I.; Rappuoli, R.; de Gregorio, E. Vaccines for the 21st Century. EMBO Mol. Med. 2014, 6, 708–720. [Google Scholar] [CrossRef]
- Donati, C.; Rappuoli, R. Reverse Vaccinology in the 21st Century: Improvements over the Original Design. Ann. N. Y. Acad. Sci. 2013, 1285, 115–132. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 24 February 2022).
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA—An Ultra-Fast Pan-Genome Analysis Pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Gardy, J.L.; Spencer, C.; Wang, K.; Ester, M.; Tusnády, G.E.; Simon, I.; Hua, S.; deFays, K.; Lambert, C.; Nakai, K.; et al. PSORT-B: Improving Protein Subcellular Localization Prediction for Gram-Negative Bacteria. Nucleic Acids Res. 2003, 31, 3613. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, I.; Flower, D.R.; Doytchinova, I. AllerTOP—A Server for in Silico Prediction of Allergens. BMC Bioinform. 2013, 14, S4. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xiang, Z.; Mobley, H.L.T. Vaxign: The First Web-Based Vaccine Design Program for Reverse Vaccinology and Applications for Vaccine Development. J. Biomed. Biotechnol. 2010, 2010, 297505. [Google Scholar] [CrossRef] [PubMed]
- Rappuoli, R.; Pizza, M.; Masignani, V.; Vadivelu, K. Meningococcal B Vaccine (4CMenB): The Journey from Research to Real World Experience. Expert Rev. Vaccines 2018, 17, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- BLAST—Ian Korf, Mark Yandell, Joseph Bedell—Google Books. Available online: https://books.google.com.pk/books?hl=en&lr=&id=zDqksXUaoBAC&oi=fnd&pg=PT7&dq=Korf,+I.,+Yandell,+M.+and+Bedell,+J.,+2003.%C2%A0Blast.+%22+O%27Reilly+Media,+Inc.%22.&ots=L2AHqdChVP&sig=jxbMGgnUb_frAqggRXJJQaQGRPI&redir_esc=y#v=onepage&q=Korf%2C%20I.%2C%20Yandell%2C%20M.%20and%20Bedell%2C%20J.%2C%202003.%C2%A0Blast.%20%22%20O’Reilly%20Media%2C%20Inc.%22.&f=false (accessed on 7 April 2022).
- Fleri, W.; Paul, S.; Dhanda, S.K.; Mahajan, S.; Xu, X.; Peters, B.; Sette, A. The Immune Epitope Database and Analysis Resource in Epitope Discovery and Synthetic Vaccine Design. Front. Immunol. 2017, 8, 278. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Doytchinova, I.A.; Zygouri, C.; Flower, D.R. MHCPred: A Server for Quantitative Prediction of Peptide–MHC Binding. Nucleic Acids Res. 2003, 31, 3621–3624. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Hebditch, M.; Carballo-Amador, M.A.; Charonis, S.; Curtis, R.; Warwicker, J. Protein–Sol: A Web Tool for Predicting Protein Solubility from Sequence. Bioinformatics 2017, 33, 3098–3100. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, P.; Kim, Y.; Haste-Andersen, P.; Beaver, J.; Bourne, P.E.; Bui, H.-H.; Buus, S.; Frankild, S.; Greenbaum, J.; et al. Immune Epitope Database Analysis Resource (IEDB-AR). Nucleic Acids Res. 2008, 36, W513–W518. [Google Scholar] [CrossRef]
- Saadi, M.; Karkhah, A.; Nouri, H.R. Development of a Multi-Epitope Peptide Vaccine Inducing Robust T Cell Responses against Brucellosis Using Immunoinformatics Based Approaches. Infect. Genet. Evol. 2017, 51, 227–234. [Google Scholar] [CrossRef]
- Arai, R.; Ueda, H.; Kitayama, A.; Kamiya, N.; Nagamune, T. Design of the Linkers Which Effectively Separate Domains of a Bifunctional Fusion Protein. Protein Eng. Des. Sel. 2001, 14, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xing, Y.; Guo, L.; Lv, X.; Song, H.; Xi, T. Oral Immunization with Recombinant Lactococcus Lactis Delivering a Multi-Epitope Antigen CTB-UE Attenuates Helicobacter Pylori Infection in Mice. Pathog. Dis. 2014, 72, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Umar, A.; Haque, A.; Alghamdi, Y.S.; Mashraqi, M.M.; Rehman, A.; Shahid, F.; Khurshid, M.; Ashfaq, U.A. Development of a Candidate Multi-Epitope Subunit Vaccine against Klebsiella Aerogenes: Subtractive Proteomics and Immuno-Informatics Approach. Vaccines 2021, 9, 1373. [Google Scholar] [CrossRef] [PubMed]
- Tani, K.; Murphy, W.J.; Chertov, O.; Salcedo, R.; Koh, C.Y.; Utsunomiya, I.; Funakoshi, S.; Asai, O.; Herrmann, S.H.; Wang, J.M.; et al. Defensins Act as Potent Adjuvants That Promote Cellular and Humoral Immune Responses in Mice to a Lymphoma Idiotype and Carrier Antigens. Int. Immunol. 2000, 12, 691–700. [Google Scholar] [CrossRef] [PubMed]
- ProtParam—SIB Swiss Institute of Bioinformatics|Expasy. Available online: https://www.expasy.org/resources/protparam (accessed on 24 February 2022).
- Cheng, J.; Randall, A.Z.; Sweredoski, M.J.; Baldi, P. SCRATCH: A Protein Structure and Structural Feature Prediction Server. Nucleic Acids Res. 2005, 33, W72–W76. [Google Scholar] [CrossRef]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB Server for Protein Structure Prediction and Refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef]
- Craig, D.B.; Dombkowski, A.A. Disulfide by Design 2.0: A Web-Based Tool for Disulfide Engineering in Proteins. BMC Bioinform. 2013, 14, 346. [Google Scholar] [CrossRef]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A Novel Tool to Adapt Codon Usage of a Target Gene to Its Potential Expression Host. Nucleic Acids Res. 2005, 33, W526. [Google Scholar] [CrossRef]
- SnapGene|Software for Everyday Molecular Biology. Available online: https://www.snapgene.com/ (accessed on 24 February 2022).
- Rapin, N.; Lund, O.; Bernaschi, M.; Castiglione, F. Computational Immunology Meets Bioinformatics: The Use of Prediction Tools for Molecular Binding in the Simulation of the Immune System. PLoS ONE 2010, 5, e9862. [Google Scholar] [CrossRef]
- ClusPro 2.0: Protein-Protein Docking. Available online: https://cluspro.bu.edu/login.php?redir=/queue.php (accessed on 24 February 2022).
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Naz, K.; Ullah, N.; Naz, A.; Irum, S.; Dar, H.A.; Zaheer, T.; Shahid, F.; Ali, A. The Epidemiological and Pangenome Landscape of Staphylococcus Aureus and Identification of Conserved Novel Candidate Vaccine Antigens. Curr. Proteom. 2022, 19, 114–126. [Google Scholar] [CrossRef]
- Moxon, R.; Reche, P.A.; Rappuoli, R. Editorial: Reverse Vaccinology. Front. Immunol. 2019, 10, 2776. [Google Scholar] [CrossRef] [PubMed]
- Naz, A.; Awan, F.M.; Obaid, A.; Muhammad, S.A.; Paracha, R.Z.; Ahmad, J.; Ali, A. Identification of Putative Vaccine Candidates against Helicobacter Pylori Exploiting Exoproteome and Secretome: A Reverse Vaccinology Based Approach. Infect. Genet. Evol. 2015, 32, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Awate, S.; Babiuk, L.A.; Mutwiri, G. Mechanisms of Action of Adjuvants. Front. Immunol. 2013, 4, 114. [Google Scholar] [CrossRef]
- Jamroz, M.; Kolinski, A. Modeling of Loops in Proteins: A Multi-Method Approach. BMC Struct. Biol. 2010, 10, 5. [Google Scholar] [CrossRef]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein Structure Refinement Driven by Side-Chain Repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural Summaries of PDB Entries. Protein Sci. A Publ. Protein Soc. 2018, 27, 129. [Google Scholar] [CrossRef]
- Dar, H.A.; Ismail, S.; Waheed, Y.; Ahmad, S.; Jamil, Z.; Aziz, H.; Hetta, H.F.; Muhammad, K. Designing a Multi-Epitope Vaccine against Mycobacteroides abscessus by Pangenome-Reverse Vaccinology. Sci. Rep. 2021, 11, 11197. [Google Scholar] [CrossRef]
- Gao, X.; Dong, X.; Li, X.; Liu, Z.; Liu, H. Prediction of Disulfide Bond Engineering Sites Using a Machine Learning Method. Sci. Rep. 2020, 10, 10330. [Google Scholar] [CrossRef]
- Castro, F.; Cardoso, A.P.; Gonçalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef] [PubMed]
- Molteni, M.; Gemma, S.; Rossetti, C. The Role of Toll-Like Receptor 4 in Infectious and Noninfectious Inflammation. Mediat. Inflamm. 2016, 2016, 6978936. [Google Scholar] [CrossRef] [PubMed]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro Web Server for Protein-Protein Docking. Nat. Protoc. 2017, 12, 255. [Google Scholar] [CrossRef]
- Keita, A.K.; Raoult, D.; Fenollar, F. Tropheryma whipplei as a Commensal Bacterium. Future Microbiol. 2013, 8, 57–71. [Google Scholar] [CrossRef]
- Marth, T. Tropheryma whipplei, Immunosuppression and Whipple’s Disease: From a Low-Pathogenic, Environmental Infectious Organism to a Rare, Multifaceted Inflammatory Complex. Dig. Dis. 2015, 33, 190–199. [Google Scholar] [CrossRef]
- Stroher, C.K.; Santos, A.L.M.; Ceron, L.; Fabrin, L.L. Whipple’s Disease: A Case Report in Santa Catarina, Brazil. Le Infez. Med. 2022, 30, 139. [Google Scholar]
- Ding, W.; Baumdicker, F.; Neher, R.A. PanX: Pan-Genome Analysis and Exploration. Nucleic Acids Res. 2018, 46, e5. [Google Scholar] [CrossRef]
- Ma, J.; Qiu, J.; Wang, S.; Ji, Q.; Xu, D.; Wang, H.; Wu, Z.; Liu, Q. A Novel Design of Multi-Epitope Vaccine Against Helicobacter Pylori by Immunoinformatics Approach. Int. J. Pept. Res. Ther. 2021, 27, 1027–1042. [Google Scholar] [CrossRef]
- Alharbi, M.; Alshammari, A.; Alasmari, A.F.; Alharbi, S.M.; ul Qamar, M.; Ullah, A.; Ahmad, S.; Irfan, M.; Khalil, A.A.K. Designing of a Recombinant Multi-Epitopes Based Vaccine against Enterococcus Mundtii Using Bioinformatics and Immunoinformatics Approaches. Int. J. Environ. Res. Public Health 2022, 19, 3729. [Google Scholar] [CrossRef]
- Ojha, R.; Gupta, N.; Naik, B.; Singh, S.; Verma, V.K.; Prusty, D.; Prajapati, V.K. High Throughput and Comprehensive Approach to Develop Multiepitope Vaccine against Minacious COVID-19. Eur. J. Pharm. Sci. 2020, 151, 105375. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Bui, H.H.; Sidney, J.; Li, W.; Fusseder, N.; Sette, A. Development of an Epitope Conservancy Analysis Tool to Facilitate the Design of Epitope-Based Diagnostics and Vaccines. BMC Bioinform. 2007, 8, 361. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.S.; Prabhakaran, V.; Verma, R.J. Design of Non-Viral Vector with Improved Regulatory Features towards Therapeutic Application. Bioinformation 2020, 16, 307. [Google Scholar] [CrossRef]
- Parvizpour, S.; Pourseif, M.M.; Razmara, J.; Rafi, M.A.; Omidi, Y. Epitope-Based Vaccine Design: A Comprehensive Overview of Bioinformatics Approaches. Drug Discov. Today 2020, 25, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Rappuoli, R. Reverse Vaccinology: Developing Vaccines in the Era of Genomics. Immunity 2010, 33, 530–541. [Google Scholar] [CrossRef] [PubMed]



| Core Protein Gene Ids | TMHMM | Molecular Weight | T. PI | Instability Index | Gravy | Allergenicity | Antigenicity | ||
|---|---|---|---|---|---|---|---|---|---|
| core/655/1/Org1_Gene691 Length: 140 | 0 | 15.07 | 9.27 | 31.58 | Stable | −0.539 | Non-Allergen | 0.6879 | Selected |
| core/655/1/Org1_Gene691 Length: 140 | 0 | 15.07 | 9.27 | 31.58 | Stable | −0.539 | Non-Allergen | 0.6879 | Selected |
| core/796/1/Org1_Gene758 Length: 49 | 0 | 5.257 | 8.1 | 27.51 | Stable | −0.678 | Non-Allergen | 0.5907 | selected |
| Proteins | B-Cell Epitopes | MHC-II | MHC I | MHC Pred | IC50 Value | Allergenicity | Antigenicity | Toxicity | Solubility | |
|---|---|---|---|---|---|---|---|---|---|---|
| >core/711/1/Org1_Gene771 | MPSRGANGSDTFLY | MPSRGANGSDT | MPSRGANGS | 2.9 | MPSRGANGS | 22.18 | Nonallergen | 2.1437 | NON-TOXIN | Soluble |
| SNTWTYTGSGKTNQTQG | TGSGKTNQTQG | SGKTNQTQG | 13 | SGKTNQTQG | 76.21 | Nonallergen | 2.7169 | NON-TOXIN | Soluble | |
| TGSGKTNQTQ | 12 | TGSGKTNQT | 58.88 | Nonallergen | 2.6625 | NON-TOXIN | Soluble | |||
| >core/655/1/Org1_Gene691 | DVLTKGGKDYSQQITT | KGGKDYSQQIT | GGKDYSQQI | 5.1 | GGKDYSQQI | 53.09 | Nonallergen | 0.9704 | NON-TOXIN | Soluble |
| Model | Global Distance Test—High Accuracy (GDT-HA) | Root Mean Square Deviation (RMSD) | MolProbity | Clash Score | Poor Rotamers | Rama Favored |
|---|---|---|---|---|---|---|
| Initial | 1 | 0 | 3.594 | 167.3 | 1.3 | 72.7 |
| MODEL 1 | 0.9653 | 0.36 | 2.605 | 30 | 0 | 85.9 |
| MODEL 2 | 0.9629 | 0.383 | 2.53 | 29.3 | 0 | 88.9 |
| MODEL 3 | 0.9653 | 0.359 | 2.604 | 31.4 | 0 | 86.9 |
| MODEL 4 | 0.9629 | 0.386 | 2.585 | 30 | 0 | 86.9 |
| MODEL 5 | 0.953 | 0.409 | 2.582 | 31.4 | 0 | 87.9 |
| Cluster | Members | Representative | Weighted Score |
|---|---|---|---|
| 0 | 62 | Center | −819.7 |
| Lowest Energy | −916.3 | ||
| 1 | 62 | Center | −721.6 |
| Lowest Energy | −858.9 | ||
| 2 | 54 | Center | −720.2 |
| Lowest Energy | −821.7 | ||
| 3 | 50 | Center | −828.4 |
| Lowest Energy | −878 | ||
| 4 | 49 | Center | −738.2 |
| Lowest Energy | −871.3 | ||
| 5 | 49 | Center | −770.2 |
| Lowest Energy | −849.7 | ||
| 6 | 48 | Center | −722.7 |
| Lowest Energy | −778.8 | ||
| 7 | 46 | Center | −820.7 |
| Lowest Energy | −878.4 | ||
| 8 | 33 | Center | −725.7 |
| Lowest Energy | −805 | ||
| 9 | 29 | Center | −761.8 |
| Lowest Energy | −815.8 | ||
| 10 | 28 | Center | −742.8 |
| MM-GBSA | MM-PBSA | ||||||
|---|---|---|---|---|---|---|---|
| Complex | |||||||
| Energy Component | Average | Std. Dev. | Err. of Mean | Energy Component | Average | Std. Dev. | Err. of Mean |
| VDWAALS | −14,268.3 | 51.3606 | 5.1361 | VDWAALS | −14,268.3 | 51.3606 | 5.1361 |
| EEL | −125,777 | 160.9493 | 16.0949 | EEL | −125,777 | 160.9493 | 16.0949 |
| EGB | −23,027.7 | 128.2908 | 12.8291 | EPB | −22,314.4 | 132.7034 | 13.2703 |
| ESURF | 616.7839 | 3.4972 | 0.3497 | ENPOLAR | 410.6266 | 1.8671 | 0.1867 |
| G gas | −140,045 | 155.5427 | 15.5543 | G gas | −140,045 | 155.5427 | 15.5543 |
| G solv | −22,410.9 | 127.4705 | 12.7471 | G solv | −21,903.8 | 132.0576 | 13.2058 |
| TOTAL | −162,456 | 110.4932 | 11.0493 | TOTAL | −161,949 | 124.2808 | 12.4281 |
| Receptor: | |||||||
| Energy Component | Average | Std. Dev. | Err. of Mean | Energy Component | Average | Std. Dev. | Err. of Mean |
| VDWAALS | −12,136.3 | 50.2011 | 5.0201 | VDWAALS | −12,136.3 | 50.2011 | 5.0201 |
| EEL | −10,4629 | 162.6708 | 16.2671 | EEL | −104,629 | 162.6708 | 16.2671 |
| EGB | −18,516.2 | 136.2444 | 13.6244 | EPB | −17,941.6 | 136.4663 | 13.6466 |
| ESURF | 489.8555 | 3.2995 | 0.3299 | ENPOLAR | 330.3065 | 1.6312 | 0.1631 |
| G gas | −116,765 | 155.9755 | 15.5976 | G gas | −116,765 | 155.9755 | 15.5976 |
| G solv | −18,026.3 | 135.2806 | 13.5281 | G solv | −17,611.3 | 135.8915 | 13.5892 |
| TOTAL | −134,791 | 95.4058 | 9.5406 | TOTAL | −134,376 | 106.9958 | 10.6996 |
| Ligand: | |||||||
| Energy Component | Average | Std. Dev. | Err. of Mean | Energy Component | Average | Std. Dev. | Err. of Mean |
| VDWAALS | −1902.44 | 19.2818 | 1.9282 | VDWAALS | −1902.44 | 19.2818 | 1.9282 |
| EEL | −21,889.9 | 83.9904 | 8.399 | EEL | −21,889.9 | 83.9904 | 8.399 |
| EGB | −4010.19 | 70.0089 | 7.0009 | EPB | −3857.43 | 65.2142 | 6.5214 |
| ESURF | 156.9794 | 1.8144 | 0.1814 | ENPOLAR | 109.2052 | 1.3425 | 0.1342 |
| G gas | −23,792.4 | 88.4392 | 8.8439 | G gas | −23,792.4 | 88.4392 | 8.8439 |
| G solv | −3853.21 | 69.1563 | 6.9156 | G solv | −3748.22 | 64.88 | 6.488 |
| TOTAL | −27,645.6 | 41.7466 | 4.1747 | TOTAL | −27,540.6 | 46.1238 | 4.6124 |
| Differences (Complex) | |||||||
| Energy Component | Average | Std. Dev. | Err. of Mean | Energy Component | Average | Std. Dev. | Err. of Mean |
| VDWAALS | −239.588 | 8.5386 | 0.8539 | VDWAALS | −239.588 | 8.5386 | 0.8539 |
| EEL | 741.5124 | 66.0836 | 6.6084 | EEL | 741.5124 | 66.0836 | 6.6084 |
| EGB | −501.319 | 60.9302 | 6.093 | EPB | −515.362 | 57.6386 | 5.7639 |
| ESURF | −30.0511 | 1.1245 | 0.1125 | ENPOLAR | −28.8851 | 0.877 | 0.0877 |
| DELTA G gas | 521.9245 | 66.2058 | 6.6206 | DELTA G gas | 521.9245 | 66.2058 | 6.6206 |
| DELTA G solv | −531.37 | 60.2798 | 6.028 | DELTA G solv | −544.247 | 57.0585 | 5.7058 |
| DELTA TOTAL | −29.4452 | 9.8089 | 0.9809 | DELTA TOTAL | −42.3229 | 18.834 | 1.8834 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albekairi, T.H.; Alshammari, A.; Alharbi, M.; Alshammary, A.F.; Tahir ul Qamar, M.; Anwar, T.; Ismail, S.; Shaker, B.; Ahmad, S. Design of a Multi-Epitope Vaccine against Tropheryma whipplei Using Immunoinformatics and Molecular Dynamics Simulation Techniques. Vaccines 2022, 10, 691. https://doi.org/10.3390/vaccines10050691
Albekairi TH, Alshammari A, Alharbi M, Alshammary AF, Tahir ul Qamar M, Anwar T, Ismail S, Shaker B, Ahmad S. Design of a Multi-Epitope Vaccine against Tropheryma whipplei Using Immunoinformatics and Molecular Dynamics Simulation Techniques. Vaccines. 2022; 10(5):691. https://doi.org/10.3390/vaccines10050691
Chicago/Turabian StyleAlbekairi, Thamer H., Abdulrahman Alshammari, Metab Alharbi, Amal F. Alshammary, Muhammad Tahir ul Qamar, Tasneem Anwar, Saba Ismail, Bilal Shaker, and Sajjad Ahmad. 2022. "Design of a Multi-Epitope Vaccine against Tropheryma whipplei Using Immunoinformatics and Molecular Dynamics Simulation Techniques" Vaccines 10, no. 5: 691. https://doi.org/10.3390/vaccines10050691
APA StyleAlbekairi, T. H., Alshammari, A., Alharbi, M., Alshammary, A. F., Tahir ul Qamar, M., Anwar, T., Ismail, S., Shaker, B., & Ahmad, S. (2022). Design of a Multi-Epitope Vaccine against Tropheryma whipplei Using Immunoinformatics and Molecular Dynamics Simulation Techniques. Vaccines, 10(5), 691. https://doi.org/10.3390/vaccines10050691








