Neutralization of SARS-CoV-2 Variants by rVSV-ΔG-Spike-Elicited Human Sera
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Cells
2.3. Viruses
2.4. Vaccine Preparation
2.5. Whole-Genome Sequencing and Data Analysis
2.6. Plaque Reduction Neutralization Test (PRNT50)
2.7. Statistical Analysis
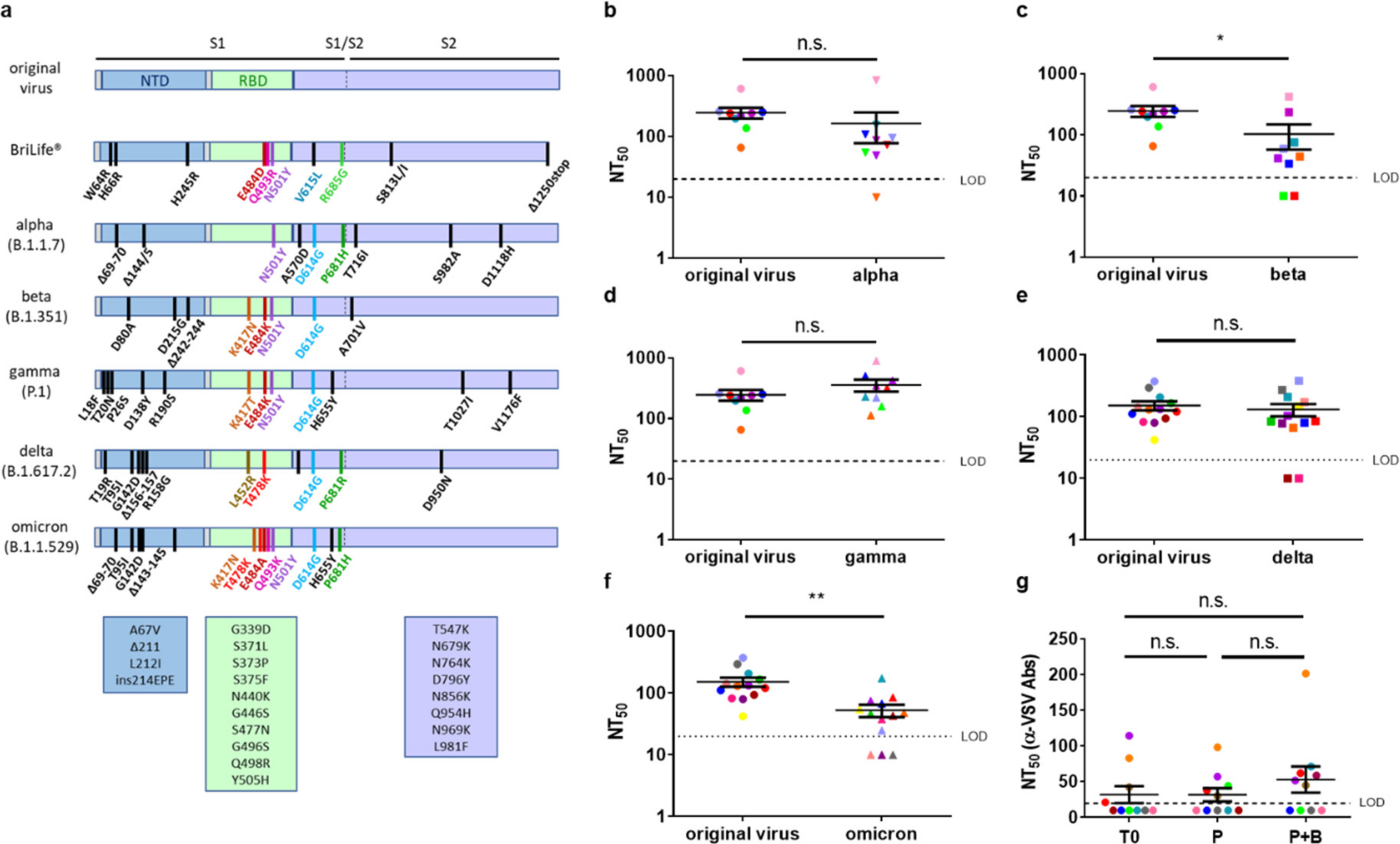
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whelan, S.P.J. Vesicular Stomatitis Virus. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2008; pp. 291–299. [Google Scholar]
- Yahalom-Ronen, Y.; Tamir, H.; Melamed, S.; Politi, B.; Shifman, O.; Achdout, H.; Vitner, E.B.; Israeli, O.; Milrot, E.; Stein, D.; et al. A single dose of recombinant VSV-G-spike vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020, 11, 6402. [Google Scholar] [CrossRef] [PubMed]
- Maor, Y.; Cohen, D.; Paran, N.; Israely, T.; Ezra, V.; Axelrod, O.; Shinar, E.; Izak, M.; Rahav, G.; Rahimi-Levene, N.; et al. Compassionate use of convalescent plasma for treatment of moderate and severe pneumonia in COVID-19 patients and association with IgG antibody levels in donated plasma. EClinicalMedicine 2020, 26, 100525. [Google Scholar] [CrossRef] [PubMed]
- Lustig, Y.; Zuckerman, N.; Nemet, I.; Atari, N.; Kliker, L.; Regev-Yochay, G.; Sapir, E.; Mor, O.; Alroy-Preis, S.; Mendelson, E.; et al. Neutralising capacity against Delta (B.1.617.2) and other variants of concern following Comirnaty (BNT162b2, BioNTech/Pfizer) vaccination in health care workers, Israel. Euro Surveill. 2021, 26, 2100557. [Google Scholar] [CrossRef] [PubMed]
- Lerer, E.; Oren, Z.; Kafri, Y.; Adar, Y.; Toister, E.; Cherry, L.; Lupu, E.; Monash, A.; Levy, R.; Dor, E.; et al. Highly Efficient Purification of Recombinant VSV-∆G-Spike Vaccine against SARS-CoV-2 by Flow-Through Chromatography. BioTech 2021, 10, 22. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Davis, C.; Logan, N.; Tyson, G.; Orton, R.; Harvey, W.T.; Perkins, J.S.; Mollett, G.; Blacow, R.M. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog. 2021, 17, e1010022. [Google Scholar] [CrossRef]
- Levine-Tiefenbrun, M.; Yelin, I.; Alapi, H.; Katz, R.; Herzel, E.; Kuint, J.; Chodick, G.; Gazit, S.; Patalon, T.; Kishony, R. Viral loads of Delta-variant SARS-CoV2 breakthrough infections following vaccination and booster with the BNT162b2 vaccine. Nat. Med. 2021, 27, 2108–2110. [Google Scholar] [CrossRef]
- Kuhlmann, C.; Mayer, C.K.; Claassen, M.; Maponga, T.; Burgers, W.A.; Keeton, R.; Riou, C.; Sutherland, A.D.; Suliman, T.; Shaw, M.L.; et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 2022. ahead of print. [Google Scholar] [CrossRef]
- Marzi, A.; Feldmann, F.; Geisbert, T.W.; Feldmann, H.; Safronetz, D. Vesicular stomatitis virus-based vaccines against Lassa and Ebola viruses. Emerg. Infect. Dis. 2015, 21, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ginn, H.M.; Dejnirattisai, W.; Supasa, P.; Wang, B.; Tuekprakhon, A.; Nutalai, R.; Zhou, D.; Mentzer, A.J.; Zhao, Y.; et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021, 184, 4220–4236. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2021. unedited. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef]
- Klimstra, W.B.; Tilston-Lunel, N.L.; Nambulli, S.; Boslett, J.; McMillen, C.M.; Gilliland, T.; Dunn, M.D.; Sun, C.; Wheeler, S.E.; Wells, A.; et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 2020, 101, 1156–1169. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Pegu, A.; O’Connell, S.E.; Schmidt, S.D.; O’Dell, S.; Talana, C.A.; Lai, L.; Albert, J.; Anderson, E.; Bennett, H.; Corbett, K.S.; et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science 2021, 373, 1372–1377. [Google Scholar] [CrossRef]
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393. [Google Scholar] [CrossRef]
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; da Silva Filipe, A.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021, 184, 1171–1187. [Google Scholar] [CrossRef]
- Puranik, A.; Lenehan, P.J.; Silvert, E.; Niesen, M.J.M.; Corchado-Garcia, J.; O’Horo, J.C.; Virk, A.; Swift, M.D.; Halamka, J.; Badley, A.D.; et al. Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of Alpha and Delta variant prevalence. medRxiv 2021. preprint. [Google Scholar] [CrossRef]
- Noori, M.; Nejadghaderi, S.A.; Arshi, S.; Carson-Chahhoud, K.; Ansarin, K.; Kolahi, A.A.; Safiri, S. Potency of BNT162b2 and mRNA-1273 vaccine-induced neutralizing antibodies against severe acute respiratory syndrome-CoV-2 variants of concern: A systematic review of in vitro studies. Rev. Med. Virol. 2021. preprint. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ginn, H.M.; Dejnirattisai, W.; Supasa, P.; Wang, B.; Tuekprakhon, A.; Nutalai, R.; Zhou, D.; Mentzer, A.J.; Zhao, Y.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484. [Google Scholar]
- Nemet, I.; Kliker, L.; Lustig, Y.; Zuckerman, N.; Erster, O.; Cohen, C.; Kreiss, Y.; Alroy-Preis, S.; Regev-Yochay, G.; Mendelson, E.; et al. Third BNT162b2 Vaccination Neutralization of SARS-CoV-2 Omicron Infection. N. Engl. J. Med. 2021, 386, 492–494. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, N.; Hoshino, A.; Higuchi, Y.; Taminishi, S.; Inaba, T.; Matoba, S. SARS-CoV-2 Omicron variant escapes neutralization by vaccinated and convalescent sera and therapeutic monoclonal antibodies. medRxiv 2021. preprint. [Google Scholar]
- Carreno, J.M.; Alshammary, H.; Tcheou, J.; Singh, G.; Raskin, A.; Kawabata, H.; Sominsky, L.; Clark, J.; Adelsberg, D.C.; Bielak, D.; et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 2021. unedited. [Google Scholar] [CrossRef]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.G.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2021. unedited. [Google Scholar] [CrossRef]
- Wang, B.; Goh, Y.S.; Prince, T.; Ngoh, E.Z.X.; Salleh, S.N.M.; Hor, P.X.; Loh, C.Y.; Fong, S.W.; Hartley, C.; Tan, S.Y.; et al. Resistance of SARS-CoV-2 variants to neutralization by convalescent plasma from early COVID-19 outbreak in Singapore. NPJ Vaccines 2021, 6, 125. [Google Scholar] [CrossRef]
- Geisbert, T.W.; Feldmann, H. Recombinant vesicular stomatitis virus-based vaccines against Ebola and Marburg virus infections. J. Infect. Dis. 2011, 204 (Suppl. S3), S1075–S1081. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Choi, A.; Koch, M.; Wu, K.; Chu, L.; Ma, L.; Hill, A.; Nunna, N.; Huang, W.; Oestreicher, J.; Colpitts, T.; et al. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: An interim analysis. Nat. Med. 2021, 27, 2025–2031. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yahalom-Ronen, Y.; Erez, N.; Fisher, M.; Tamir, H.; Politi, B.; Achdout, H.; Melamed, S.; Glinert, I.; Weiss, S.; Cohen-Gihon, I.; et al. Neutralization of SARS-CoV-2 Variants by rVSV-ΔG-Spike-Elicited Human Sera. Vaccines 2022, 10, 291. https://doi.org/10.3390/vaccines10020291
Yahalom-Ronen Y, Erez N, Fisher M, Tamir H, Politi B, Achdout H, Melamed S, Glinert I, Weiss S, Cohen-Gihon I, et al. Neutralization of SARS-CoV-2 Variants by rVSV-ΔG-Spike-Elicited Human Sera. Vaccines. 2022; 10(2):291. https://doi.org/10.3390/vaccines10020291
Chicago/Turabian StyleYahalom-Ronen, Yfat, Noam Erez, Morly Fisher, Hadas Tamir, Boaz Politi, Hagit Achdout, Sharon Melamed, Itai Glinert, Shay Weiss, Inbar Cohen-Gihon, and et al. 2022. "Neutralization of SARS-CoV-2 Variants by rVSV-ΔG-Spike-Elicited Human Sera" Vaccines 10, no. 2: 291. https://doi.org/10.3390/vaccines10020291
APA StyleYahalom-Ronen, Y., Erez, N., Fisher, M., Tamir, H., Politi, B., Achdout, H., Melamed, S., Glinert, I., Weiss, S., Cohen-Gihon, I., Israeli, O., Izak, M., Mandelboim, M., Caraco, Y., Madar-Balakirski, N., Mechaly, A., Shinar, E., Zichel, R., Cohen, D., ... Paran, N. (2022). Neutralization of SARS-CoV-2 Variants by rVSV-ΔG-Spike-Elicited Human Sera. Vaccines, 10(2), 291. https://doi.org/10.3390/vaccines10020291





