SARS-CoV-2–Specific Vaccine Candidates; the Contribution of Structural Vaccinology
Abstract
:1. Introduction
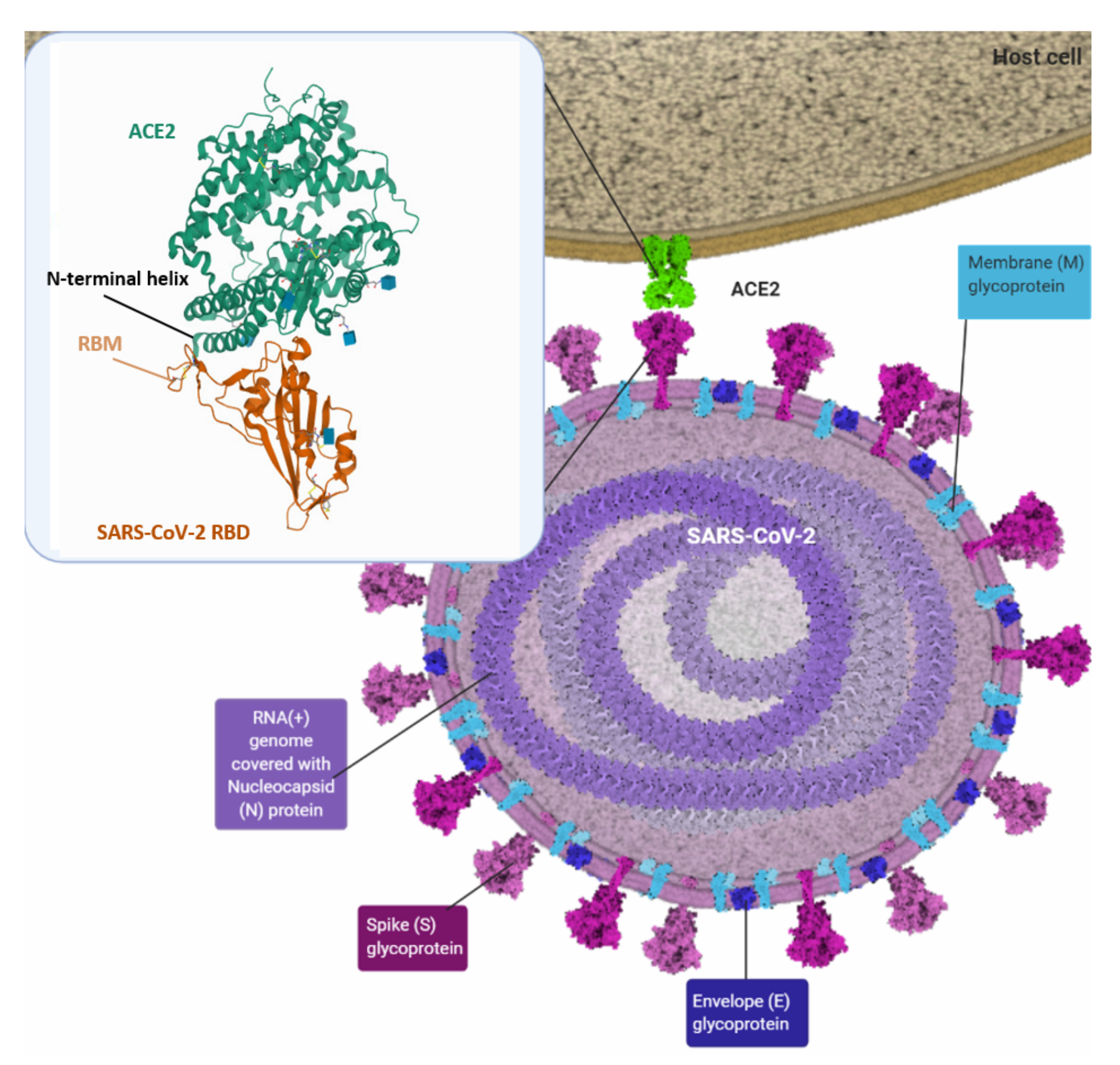
2. SARS-CoV-2 Infection
3. Structural Vaccinology
4. SARS-CoV-2 Spike Protein
5. Modern Vaccine Platforms
6. Alternative Candidates for SARS-CoV-2 Vaccines
7. Pan-Coronavirus Vaccine
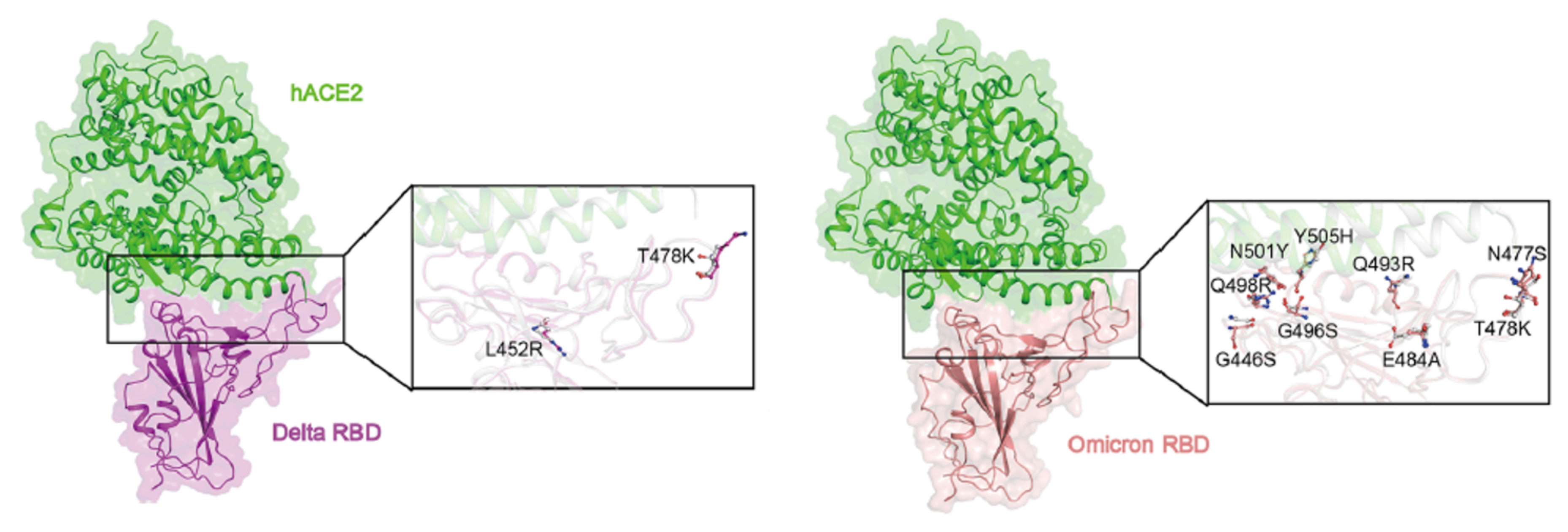
8. Emerging Variants and Vaccine Adaptation
9. Discussion
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drosten, C.; Günther, S.; Preiser, W.; Van Der Werf, S.; Brodt, H.-R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.M.; et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.D.; Tyrrell, D.A. The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J. Gen. Virol. 1967, 1, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.-j.; Ni, Z.-y.; Hu, Y.; Liang, W.-h.; Ou, C.-q.; He, J.-x.; Liu, L.; Shan, H.; Lei, C.-l.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; Krammer, F. SARS-CoV-2 Vaccines: Status Report. Immunity 2020, 52, 583–589. [Google Scholar] [CrossRef]
- Heymann, D.L.; Shindo, N. COVID-19: What is next for public health? Lancet 2020, 395, 542–545. [Google Scholar] [CrossRef] [Green Version]
- Rappuoli, R.; de Gregorio, E.; del Giudice, G.; Phogat, S.; Pecetta, S.; Pizza, M.; Hanon, E. Vaccinology in the post−COVID-19 era. Proc. Natl. Acad. Sci. USA 2021, 118, e2020368118. [Google Scholar] [CrossRef]
- Maxmen, A. One million coronavirus sequences: Popular genome site hits mega milestone. Nature 2021, 593, 21. [Google Scholar] [CrossRef]
- Pfizer and BioNTech Celebrate Historic First Authorization in the U.S. of Vaccine to Prevent COVID-19|Pfizer. Pfizer. 2020. Available online: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-celebrate-historic-first-authorization (accessed on 31 December 2021).
- COVID-19 Vaccine Tracker and Landscape. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 31 December 2021).
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Anasir, M.I.; Poh, C.L. Structural Vaccinology for Viral Vaccine Design. Front. Microbiol. 2019, 10, 738. [Google Scholar] [CrossRef]
- Tang, D.; Comish, P.; Kang, R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020, 16, e1008536. [Google Scholar] [CrossRef]
- Ong, E.; Wong, M.U.; Huffman, A.; He, Y. COVID-19 Coronavirus Vaccine Design Using Reverse Vaccinology and Machine Learning. Front. Immunol. 2020, 11, 1581. [Google Scholar] [CrossRef]
- Bruno, L.; Cortese, M.; Rappuoli, R.; Merola, M. Lessons from Reverse Vaccinology for viral vaccine design. Curr. Opin. Virol. 2015, 11, 89–97. [Google Scholar] [CrossRef]
- Enayatkhani, M.; Hasaniazad, M.; Faezi, S.; Gouklani, H.; Davoodian, P.; Ahmadi, N.; Einakian, M.A.; Karmostaji, A.; Ahmadi, K. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. J. Biomol. Struct. Dyn. 2020, 39, 2857–2872. [Google Scholar] [CrossRef] [Green Version]
- Pinto, D.; Park, Y.J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef]
- Juraszek, J.; Rutten, L.; Blokland, S.; Bouchier, P.; Voorzaat, R.; Ritschel, T.; Bakkers, M.J.; Renault, L.L.; Langedijk, J.P. Stabilizing the Closed SARS-CoV-2 Spike Trimer. Nat. Commun. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.; Walzer, K.; Hein, S.; Güler, A.; et al. A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. Science 2020, 370, 1022. [Google Scholar] [CrossRef]
- Parums, D.V. Editorial: First Full Regulatory Approval of a COVID-19 Vaccine, the BNT162b2 Pfizer-BioNTech Vaccine, and the Real-World Implications for Public Health Policy. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2021, 27, e934625-1–e934625-3. [Google Scholar] [CrossRef]
- Bangaru, S.; Ozorowski, G.; Turner, H.L.; Antanasijevic, A.; Huang, D.; Wang, X.; Torres, J.L.; Diedrich, J.K.; Tian, J.H.; Portnoff, A.D.; et al. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science 2020, 370, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Deng, Y.-Q.; Ye, Q.; Cao, L.; Sun, C.-Y.; Fan, C.; Huang, W.; Sun, S.; Sun, Y.; Zhu, L.; et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV 2 by a potent therapeutic antibody. Science 2020, 369, 1505–1509. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.-M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Shang, J.; Jiang, S.; Du, L. Subunit Vaccines Against Emerging Pathogenic Human Coronaviruses. Front. Microbiol. 2020, 11, 298. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- He, Y.; Zhou, Y.; Liu, S.; Kou, Z.; Li, W.; Farzan, M.; Jiang, S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: Implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004, 324, 773–781. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef]
- Polyiam, K.; Phoolcharoen, W.; Butkhot, N.; Srisaowakarn, C.; Thitithanyanont, A.; Auewarakul, P.; Hoonsuwan, T.; Ruengjitchatchawalya, M.; Mekvichitsaeng, P.; Roshorm, Y.M. Immunodominant linear B cell epitopes in the spike and membrane proteins of SARS-CoV-2 identified by immunoinformatics prediction and immunoassay. Sci. Rep. 2021, 11, 20383. [Google Scholar] [CrossRef]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, L.; Gao, G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2020, 21, 73–82. [Google Scholar] [CrossRef]
- Mendonça, S.A.; Lorincz, R.; Boucher, P.; Curiel, D.T. Adenoviral vector vaccine platforms in the SARS-CoV-2 pandemic. npj Vaccines 2021, 6, 97. [Google Scholar] [CrossRef]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Dicks, M.; Spencer, A.; Edwards, N.; Wadell, G.; Bojang, K.; Gilbert, S.; Hill, A.V.S.; Cottingham, M.G. A Novel Chimpanzee Adenovirus Vector with Low Human Seroprevalence: Improved Systems for Vector Derivation and Comparative Immunogenicity. PLoS ONE 2012, 7, e40385. [Google Scholar] [CrossRef] [Green Version]
- Vergauwen, L.; el Hajjami, N.; Brantner, M.; Mantri, S.; Cebi, B. Manufacturing Strategies for mRNA Vaccines and Therapeutics. Merck. Available online: https://www.sigmaaldrich.com/NL/en/technical-documents/technical-article/pharmaceutical-and-biopharmaceutical-manufacturing/vaccine-manufacturing/manufacturing-strategies-for-mrna-vaccines (accessed on 20 January 2022).
- Liu, M.A. A Comparison of Plasmid DNA and mRNA as Vaccine Technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Evans, C.F.; Dolter, K.E.; Song, J.; Luxembourg, A.; Bernard, R.; Drew, H. 240 Risk of Insertional Mutagenesis Following In Vivo Transfer of Plasmid DNA: Role of Host Immune Responses. Mol. Ther. 2008, 16, S91. [Google Scholar] [CrossRef]
- Uddin, M.N.; Roni, M.A. Challenges of Storage and Stability of mRNA-Based COVID-19 Vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef]
- Grudzien, E.; Kalek, M.; Jemielity, J.; Darzynkiewicz, E.; Rhoads, R.E. Differential Inhibition of mRNA Degradation Pathways by Novel Cap Analogs. J. Biol. Chem. 2006, 281, 1857–1867. [Google Scholar] [CrossRef] [Green Version]
- FDA. COVID-19 Vaccines|FDA. Available online: https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines (accessed on 31 December 2021).
- Ledford, H. Moderna COVID vaccine becomes second to get US authorization. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Pinkbook|Vaccine Storage and Handling|Epidemiology of VPDs|CDC. Available online: https://www.cdc.gov/vaccines/pubs/pinkbook/vac-storage.html (accessed on 24 January 2022).
- Pollet, J.; Chen, W.-H.; Strych, U. Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv. Drug Deliv. Rev. 2021, 170, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Dunkle, L.M.; Kotloff, K.L.; Gay, C.L.; Áñez, G.; Adelglass, J.M.; Hernández, A.Q.B.; Harper, W.L.; Duncanson, D.M.; McArthur, M.A.; Florescu, D.F.; et al. Efficacy and Safety of NVX-CoV2373 in Adults in the United States and Mexico. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef]
- European Medicines Agency. EMA Recommends Nuvaxovid for Authorisation in the EU|European Medicines Agency. 2021. Available online: https://www.ema.europa.eu/en/news/ema-recommends-nuvaxovid-authorisation-eu (accessed on 31 December 2021).
- Nascimento, I.P.; Leite, L.C.C. Recombinant vaccines and the development of new vaccine strategies. Braz. J. Med. Biol. Res. 2012, 45, 1102–1111. [Google Scholar] [CrossRef] [Green Version]
- Hotez, P.J.; Bottazzi, M.E. Developing a low-cost and accessible COVID-19 vaccine for global health. PLoS Negl. Trop. Dis. 2020, 14, e0008548. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Noh, J.Y.; Jeong, H.W.; Kim, J.H.; Shin, E.-C. T cell-oriented strategies for controlling the COVID-19 pandemic. Nat. Rev. Immunol. 2021, 21, 687–688. [Google Scholar] [CrossRef]
- Tirado, S.M.C.; Yoon, K.-J. Antibody-Dependent Enhancement of Virus Infection and Disease. Viral Immunol. 2003, 16, 69–86. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Sun, S.; Tai, W.; Chen, J.; Geng, Q.; He, L.; Chen, Y.; Wu, J.; Shi, Z. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J. Virol. 2020, 94, e02015-19. [Google Scholar] [CrossRef] [Green Version]
- Yip, M.S.; Leung, N.H.L.; Cheung, C.Y.; Li, P.H.; Lee, H.H.Y.; Daëron, M.; Peiris, J.S.M.; Bruzzone, R.; Jaume, M. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 2014, 11, 82. [Google Scholar] [CrossRef] [Green Version]
- Maemura, T.; Kuroda, M.; Armbrust, T.; Yamayoshi, S.; Halfmann, P.J.; Kawaoka, Y. Antibody-Dependent Enhancement of SARS-CoV-2 Infection Is Mediated by the IgG Receptors FcγRIIA and FcγRIIIA but Does Not Contribute to Aberrant Cytokine Production by Macrophages. mBio 2021, 12, e01987-21. [Google Scholar] [CrossRef]
- CDC COVID-19 Response Team and Food and Drug Administration. Allergic Reactions Including Anaphylaxis After Receipt of the First Dose of Pfizer-BioNTech COVID-19 Vaccine—United States, December 14–23, 2020. MMWR Morb. Mortal Wkly. Rep. 2020, 70, 46–51. [Google Scholar]
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef]
- Dai, L.; Zheng, T.; Xu, K.; Han, Y.; Xu, L.; Huang, E.; An, Y.; Cheng, Y.; Li, S.; Liu, M.; et al. A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS. Cell 2020, 182, 722–733.e11. [Google Scholar] [CrossRef]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef]
- Grant, O.C.; Montgomery, D.; Ito, K.; Woods, R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020, 10, 14991. [Google Scholar] [CrossRef]
- Guo, Y.; Sun, S.; Wang, K.; Zhang, S.; Zhu, W.; Chen, Z. Elicitation of Immunity in Mice After Immunization with the S2 Subunit of the Severe Acute Respiratory Syndrome Coronavirus. DNA Cell Biol. 2005, 24, 510–515. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, G.; Liu, H.; Ding, P.; Liu, Y.; Tian, Y.; Wang, Y.; Wang, A. Epitope Profiling Reveals the Critical Antigenic Determinants in SARS-CoV-2 RBD-Based Antigen. Front. Immunol. 2021, 12, 3854. [Google Scholar] [CrossRef]
- Yang, S.; Li, Y.; Dai, L.; Wang, J.; He, P.; Li, C.; Fang, X.; Wang, C.; Zhao, X.; Huang, E.; et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD protein vaccine against COVID-19 in adults: Pooled analysis of two randomized, double-blind, placebo-controlled, phase 1 and 2 trials. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, L.; Schank, M.; Dang, X.; Lu, Z.; Cao, D.; Khanal, S.; Nguyen, L.N.; Nguyen, L.N.; Zhang, J.; et al. SARS-CoV-2 specific memory T cell epitopes identified in COVID-19-recovered subjects. Virus Res. 2021, 304, 198508. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, X.; Gai, W.; Wong, G.; Wang, H.; Jin, H.; Feng, N.; Zhao, Y.; Zhang, W.; Li, N.; et al. Novel chimeric virus-like particles vaccine displaying MERS-CoV receptor-binding domain induce specific humoral and cellular immune response in mice. Antivir. Res. 2017, 140, 55–61. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.; Chen, Z.; Lu, S.; Yang, F.; Bi, Z.; Bao, L.; Mo, F.; Li, X.; Huang, Y.; et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 2020, 586, 572–577. [Google Scholar] [CrossRef]
- Jiaming, L.; Yanfeng, Y.; Yao, D.; Yawei, H.; Linlin, B.; Baoying, H.; Jinghua, Y.; Gao, G.F.; Chuan, Q.; Wenjie, T. The recombinant N-terminal domain of spike proteins is a potential vaccine against Middle East respiratory syndrome coronavirus (MERS-CoV) infection. Vaccine 2017, 35, 10–18. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Cohen, K.W.; Linderman, S.L.; Moodie, Z.; Czartoski, J.; Lai, L.; Mantus, G.; Norwood, C.; Nyhoff, L.E.; Edara, V.V.; Floyd, K.; et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep. Med. 2021, 2, 100354. [Google Scholar] [CrossRef]
- Ravichandran, S.; Coyle, E.M.; Klenow, L.; Tang, J.; Grubbs, G.; Liu, S.; Wang, T.; Golding, H.; Khurana, S. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 2020, 12, eabc3539. [Google Scholar] [CrossRef] [PubMed]
- Wec, A.Z.; Wrapp, D.; Herbert, A.S.; Maurer, D.P.; Haslwanter, D.; Sakharkar, M.; Jangra, R.K.; Dieterle, M.E.; Lilov, A.; Huang, D.; et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020, 369, 731–736. [Google Scholar] [CrossRef]
- Poh, C.M.; Carissimo, G.; Wang, B.; Amrun, S.N.; Lee, C.Y.P.; Chee, R.S.L.; Fong, S.W.; Yeo, N.K.W.; Lee, W.H.; Torres-Ruesta, A.; et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020, 11, 2806. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G Variant Exhibits Enhanced Replication ex vivo and Earlier Transmission in vivo. bioRxiv 2020. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.; Russell, T.W.; Tully, D.C.; Washburne, A.D. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv 2021. [Google Scholar] [CrossRef]
- He, X.; Hong, W.; Pan, X.; Lu, G.; Wei, X. SARS-CoV-2 Omicron variant: Characteristics and prevention. MedComm 2021, 2, 838–845. [Google Scholar] [CrossRef]
- Corum, J.; Zimmer, C. Tracking Omicron and Other Coronavirus Variants. The New York Times. 2022. Available online: https://www.nytimes.com/interactive/2021/health/coronavirus-variant-tracker.html (accessed on 25 January 2022).
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 Covid-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef]
- Shinde, V.; Bhikha, S.; Hoosain, Z.; Archary, M.; Bhorat, Q.; Fairlie, L.; Lalloo, U.; Masilela, M.S.; Moodley, D.; Hanley, S.; et al. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1899–1909. [Google Scholar] [CrossRef]
- Han, P.; Li, L.; Liu, S.; Wang, Q.; Zhang, D.; Xu, Z.; Han, P.; Li, X.; Peng, Q.; Su, C.; et al. Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and Delta SARS-CoV-2. Cell, 2022; in press. [Google Scholar] [CrossRef]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef]
- Collie, S.; Champion, J.; Moultrie, H.; Bekker, L.-G.; Gray, G. Effectiveness of BNT162b2 Vaccine against Omicron Variant in South Africa. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef]
- Callaway, E. The coronavirus is mutating—Does it matter? Nature 2020, 585, 174–177. [Google Scholar] [CrossRef]
- Walensky, R. CDC Expands COVID-19 Booster Recommendations|CDC Online Newsroom|CDC. Centers for Disease Control and Prevention; 2021. Available online: https://www.cdc.gov/media/releases/2021/s1129-booster-recommendations.html (accessed on 25 January 2022).
- Barda, N.; Dagan, N.; Cohen, C.; Hernán, M.A.; Lipsitch, M.; Kohane, I.S.; Reis, B.Y.; Balicer, R.D. Effectiveness of a third dose of the BNT162b2 mRNA COVID-19 vaccine for preventing severe outcomes in Israel: An observational study. Lancet 2021, 398, 2093–2100. [Google Scholar] [CrossRef]
- UK Health Security Agency. SARS-CoV-2 Variants of Concern and Variants under Investigation in England; UK Health Security Agency: London, UK, 2021. [Google Scholar]
- Ford, C.T.; Machado, D.J.; Janies, D.A. Predictions of the SARS-CoV-2 Omicron Variant (B.1.1.529) Spike Protein Receptor-Binding Domain Structure and Neutralizing Antibody Interactions. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ni, D.; Lau, K.; Turelli, P.; Raclot, C.; Beckert, B.; Nazarov, S.; Pojer, F.; Myasnikov, A.; Stahlberg, H.; Trono, D. Structural analysis of the Spike of the Omicron SARS-COV-2 variant by Cryo-EM and implications for immune evasion. bioRxiv 2021. [Google Scholar] [CrossRef]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science 2022, eabn7760. [Google Scholar] [CrossRef]
- Rutten, L.; Lai, Y.-T.; Blokland, S.; Truan, D.; Bisschop, I.J.; Strokappe, N.M.; Koornneef, A.; van Manen, D.; Chuang, G.-Y.; Farney, S.K.; et al. A Universal Approach to Optimize the Folding and Stability of Prefusion-Closed HIV-1 Envelope Trimers. Cell Rep. 2018, 23, 584–595. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Cao, R.-Y.; Chen, R.; Mo, L.; Han, J.-F.; Wang, X.; Xu, X.; Jiang, T.; Deng, Y.-Q.; Lyu, K.; et al. Rational design of thermostable vaccines by engineered peptide-induced virus self-biomineralization under physiological conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 7619–7624. [Google Scholar] [CrossRef] [Green Version]
- Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R.H.; Peters, B.; Sette, A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe 2020, 27, 671–680.e2. [Google Scholar] [CrossRef]

| WHO-Approved Vaccines (All against the Spike Protein) | Modern Vaccine Platform | Vaccine Design | |
|---|---|---|---|
| Pharmaceutical Company | Research Name | ||
| Pfizer-BioNTech | BNT162b2 | mRNA | Full-length S protein stabilised with two proline substitution (K986P and V987P) encapsulated in lipid nanoparticle |
| Moderna | mRNA-1273 | mRNA | Full-length S protein stabilised with two proline substitution (K986P and V987P) encapsulated in lipid nanoparticle |
| Novavax | NVX-CoV2373 | Protein subunit | Full-length S protein stabilised with two proline substitution (K986P and V987P) and 682-QQAQ-685 mutation at S1/S2 cleavage site formulated with Matrix-M adjuvant |
| Janssen | JNJ-78436735 (Ad26.COV2.S) | Non-replicating viral vector (Adenovirus) | Full-length S protein stabilised with two proline substitution (K986P and V987P) contained in replication-deficient adenovirus vector Ad26 |
| Oxford-AstraZeneca | AZD1222 (ChAdOx1) | Non-replicating viral vector (Adenovirus) | Full-length, wild-type S protein with the human tissue plasminogen activator gene leader added at the N-terminus contained in replication-deficient chimpanzee adenovirus vector ChAdOx1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pack, S.M.; Peters, P.J. SARS-CoV-2–Specific Vaccine Candidates; the Contribution of Structural Vaccinology. Vaccines 2022, 10, 236. https://doi.org/10.3390/vaccines10020236
Pack SM, Peters PJ. SARS-CoV-2–Specific Vaccine Candidates; the Contribution of Structural Vaccinology. Vaccines. 2022; 10(2):236. https://doi.org/10.3390/vaccines10020236
Chicago/Turabian StylePack, Su Min, and Peter J. Peters. 2022. "SARS-CoV-2–Specific Vaccine Candidates; the Contribution of Structural Vaccinology" Vaccines 10, no. 2: 236. https://doi.org/10.3390/vaccines10020236
APA StylePack, S. M., & Peters, P. J. (2022). SARS-CoV-2–Specific Vaccine Candidates; the Contribution of Structural Vaccinology. Vaccines, 10(2), 236. https://doi.org/10.3390/vaccines10020236






