Elucidating the 3D Structure of a Surface Membrane Antigen from Trypanosoma cruzi as a Serodiagnostic Biomarker of Chagas Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
2.2. Crystallization
2.3. X-ray Data Collection and Structure Determination
2.4. Human Sera Sample Collection
2.5. Polyclonal Antibody Production
2.6. Dissociation-Enhanced Lanthanide Fluoroscence ImmunoAssays (DELFIA)
2.7. Protein Microarrays
2.8. Peptide Microarrays
2.9. Molecular Dynamics Simulations
2.10. Prediction of Epitopes: MLCE
3. Results and Discussion
3.1. Expression and Purification of the Extracellular Domain of TcSMP
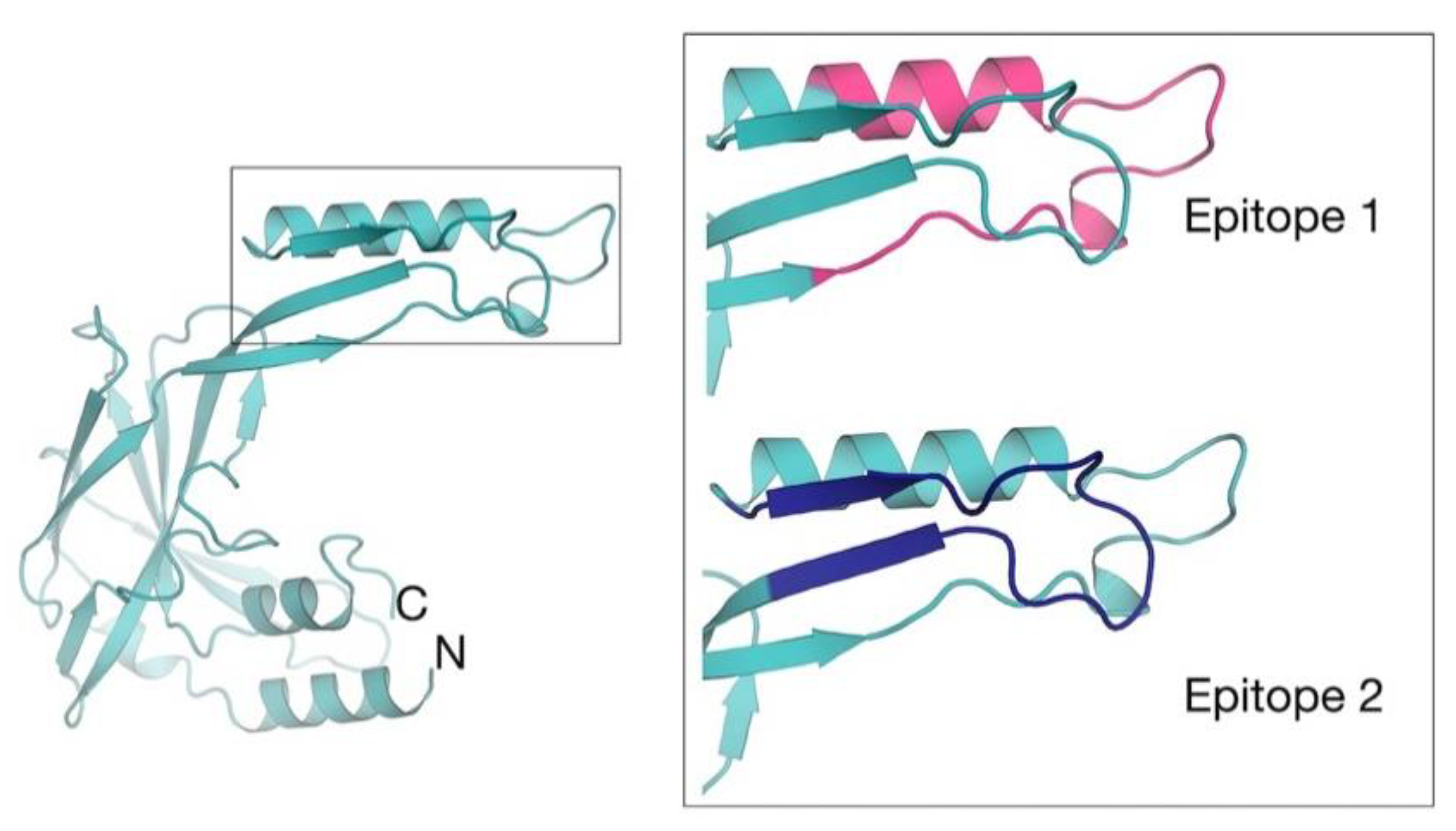
3.2. 3D Structure of TcSMP
3.3. In Silico Epitope Predictions
3.4. Epitope Peptide Synthesis
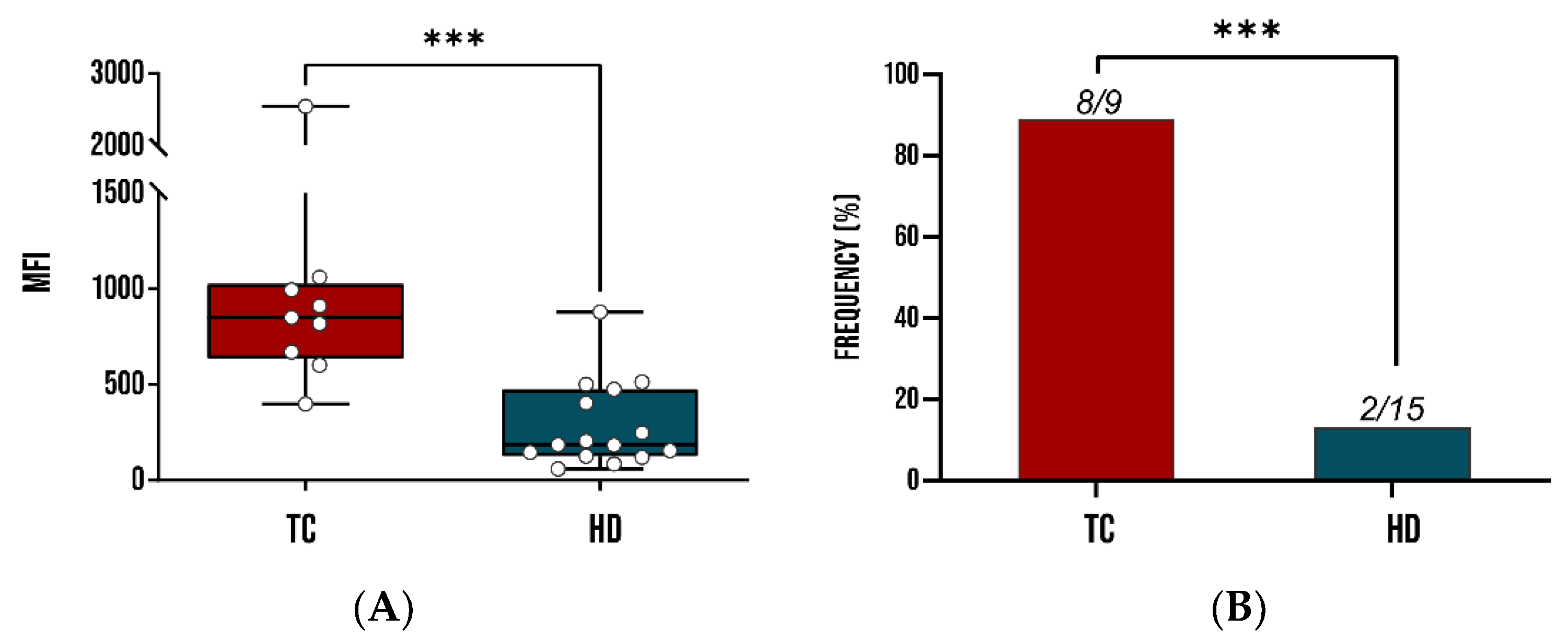
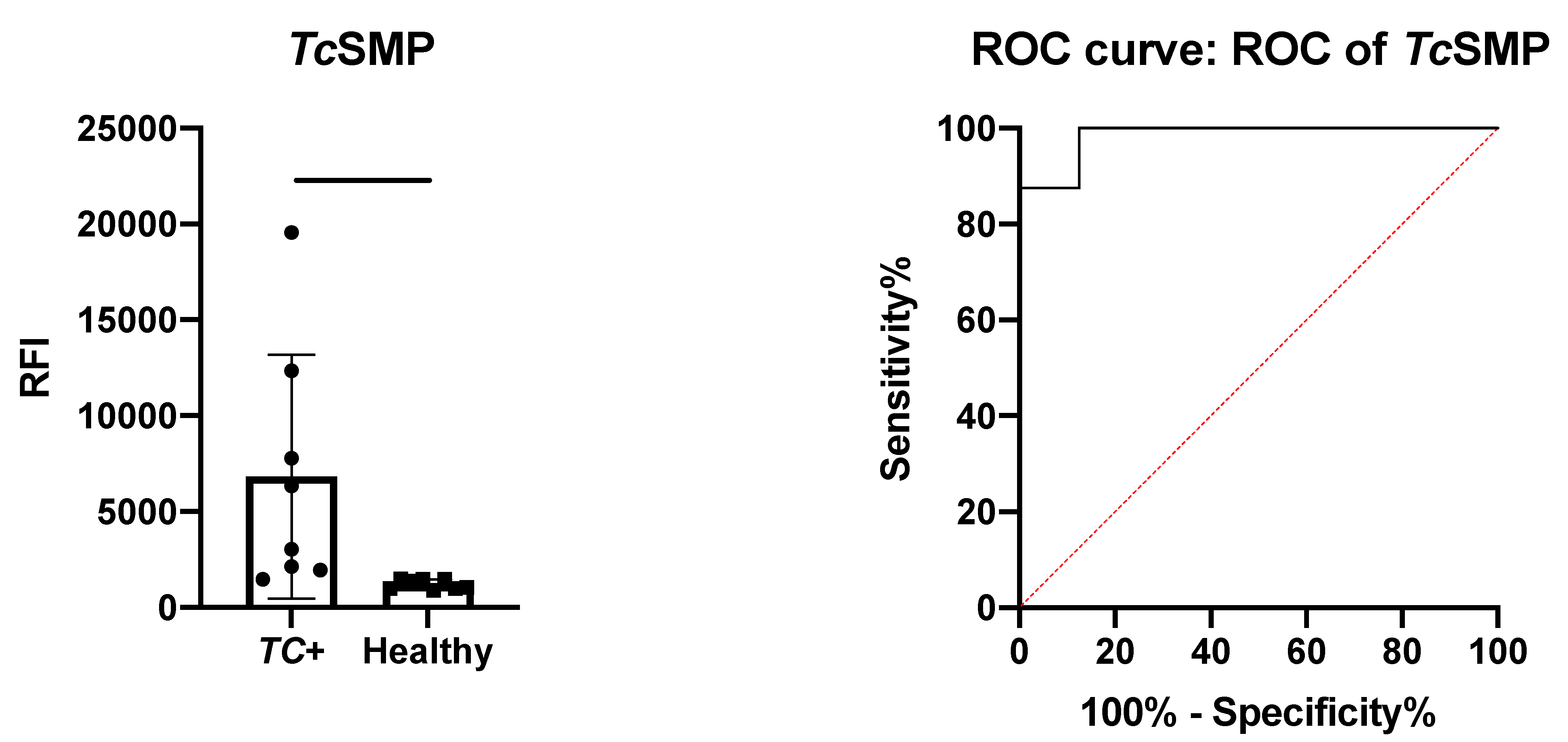
3.4.1. Immune Sera Reactivity Tests Via DELFIA Assay
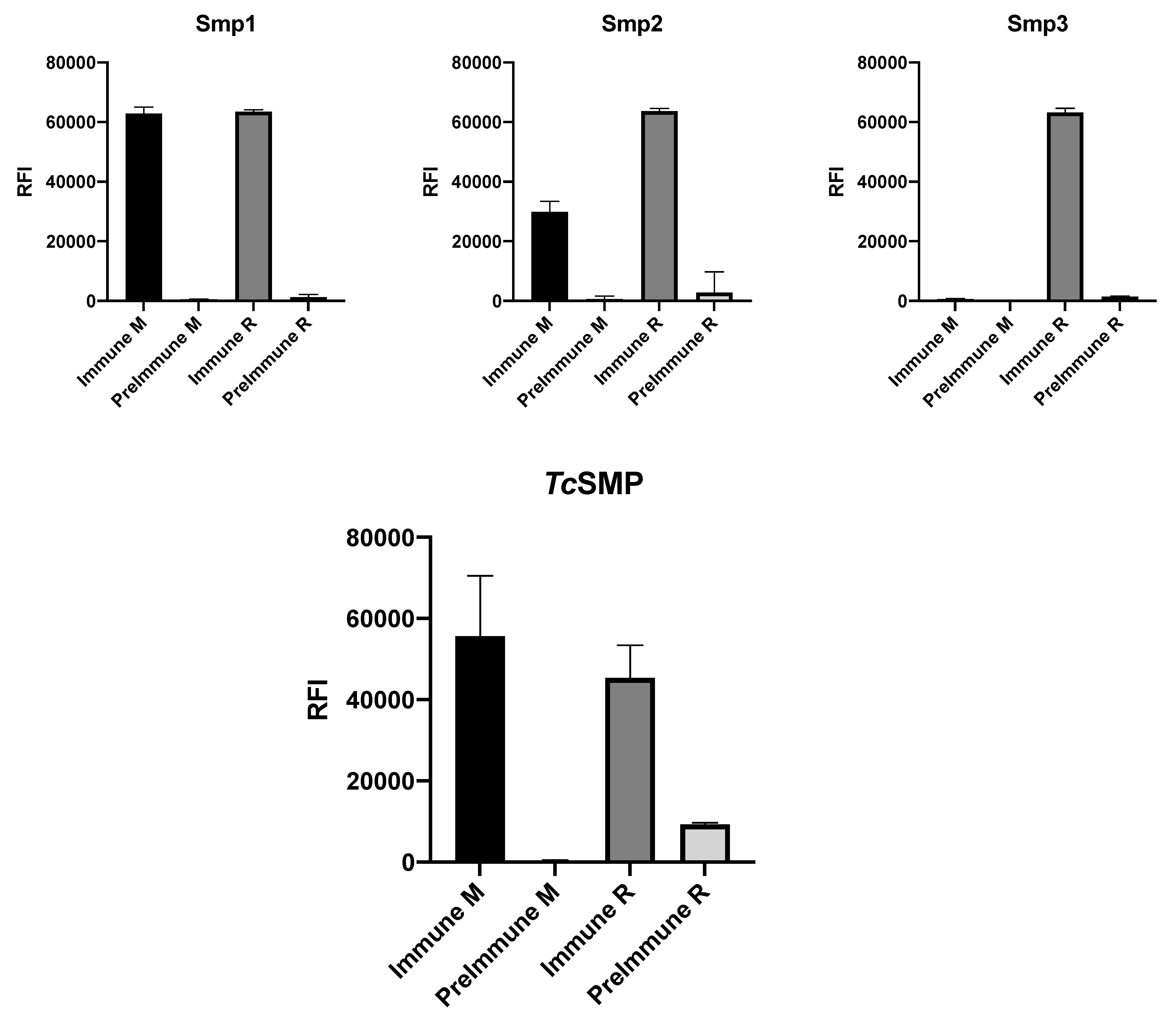
3.4.2. Polyclonal Antibody Recognition and Immune Sera Reactivity Tests Via Microarray
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schofield, C.J.; Jannin, J.; Salvatella, R. The future of Chagas disease control. Trends Parasitol. 2006, 22, 583–588. [Google Scholar] [CrossRef]
- WHO. Chagas Disease (American Trypanosomiasis). Available online: https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis) (accessed on 11 May 2021).
- CDC. American Trypanosomiasis. Available online: https://www.cdc.gov/parasites/chagas/ (accessed on 16 February 2021).
- Lidani, K.C.F.; Andrade, F.A.; Bavia, L.; Damasceno, F.S.; Beltrame, M.H.; Messias-Reason, I.J.; Sandri, T.L. Chagas Disease: From Discovery to a Worldwide Health Problem. Front. Public Health 2019, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Stuart, K.; Brun, R.; Croft, S.; Fairlamb, A.; Gürtler, R.E.; McKerrow, J.; Reed, S.; Tarleton, R. Kinetoplastids: Related protozoan pathogens, different diseases. J. Clin. Investig. 2008, 118, 1301–1310. [Google Scholar] [CrossRef] [Green Version]
- Prata, A. Clinical and epidemiological aspects of Chagas disease. Lancet Infect. Dis. 2001, 1, 92–100. [Google Scholar] [CrossRef]
- Nunes, M.C.P.; Dones, W.; Morillo, C.A.; Encina, J.J.; Ribeiro, A.L. Chagas Disease: An Overview of Clinical and Epidemiological Aspects. J. Am. Coll. Cardiol. 2013, 62, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Rassi, A., Jr.; Rassi, A.; Marin-Neto, J.A. Chagas disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Vago, A.R.; Andrade, L.O.; Leite, A.A.; Reis, D.D.; Macedo, A.M.; Adad, S.J.; Tostes, S.; Moreira, M.D.C.V.; Filho, G.B.; Pena, S.D. Genetic Characterization of Trypanosoma cruzi Directly from Tissues of Patients with Chronic Chagas Disease. Am. J. Pathol. 2000, 156, 1805–1809. [Google Scholar] [CrossRef]
- Cançado, J.R. Long term evaluation of etiological treatment of chagas disease with benznidazole. Rev. Inst. Med. Trop. São Paulo 2002, 44, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Torrico, F.; Gascón, J.; Barreira, F.; Blum, B.; Almeida, I.C.; Alonso-Vega, C.; Barboza, T.; Bilbe, G.; Correia, E.; Garcia, W.; et al. New regimens of benznidazole monotherapy and in combination with fosravuconazole for treatment of Chagas disease (BENDITA): A phase 2, double-blind, randomised trial. Lancet Infect. Dis. 2021, 21, 1129–1140. [Google Scholar] [CrossRef]
- Moraes, C.B.; Witt, G.; Kuzikov, M.; Ellinger, B.; Calogeropoulou, T.; Prousis, K.C.; Mangani, S.; Di Pisa, F.; Landi, G.; Iacono, L.D.; et al. Accelerating Drug Discovery Efforts for Trypanosomatidic Infections Using an Integrated Transnational Academic Drug Discovery Platform. SLAS Discov. Adv. Life Sci. R&D 2019, 24, 346–361. [Google Scholar] [CrossRef] [Green Version]
- Di Pisa, F.; Landi, G.; Iacono, L.D.; Pozzi, C.; Borsari, C.; Ferrari, S.; Santucci, M.; Santarem, N.; Cordeiro-Da-Silva, A.; Moraes, C.B.; et al. Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity. Molecules 2017, 22, 426. [Google Scholar] [CrossRef] [Green Version]
- Panecka-Hofman, J.; Pöhner, I.; Spyrakis, F.; Zeppelin, T.; Di Pisa, F.; Iacono, L.D.; Bonucci, A.; Quotadamo, A.; Venturelli, A.; Mangani, S.; et al. Comparative mapping of on-targets and off-targets for the discovery of anti-trypanosomatid folate pathway inhibitors. Biochim. Biophys. Acta 2017, 1861, 3215–3230. [Google Scholar] [CrossRef]
- Linciano, P.; Pozzi, C.; Iacono, L.D.; di Pisa, F.; Landi, G.; Bonucci, A.; Gul, S.; Kuzikov, M.; Ellinger, B.; Witt, G.; et al. Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections. J. Med. Chem. 2019, 62, 3989–4012. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, E.S.; Shikanai-Yasuda, M.A.; Stolf, A.M. Changes in isotype composition and antigen recognition of anti-Trypanosoma cruzi antibodies from acute to chronic Chagas disease. J. Clin. Lab. Anal. 1996, 10, 407–413. [Google Scholar] [CrossRef]
- Carlier, Y.; Altcheh, J.; Angheben, A.; Freilij, H.; Luquetti, A.O.; Schijman, A.G.; Segovia, M.; Wagner, N.; Vinas, P.A. Congenital Chagas disease: Updated recommendations for prevention, diagnosis, treatment, and follow-up of newborns and siblings, girls, women of childbearing age, and pregnant women. PLoS Negl. Trop. Dis. 2019, 13, e0007694. [Google Scholar] [CrossRef] [PubMed]
- Bern, C.; Montgomery, S.P.; Herwaldt, B.L.; Rassi, A.; Marin-Neto, J.A.; Dantas, R.O.; Maguire, J.H.; Acquatella, H.; Morillo, C.; Kirchhoff, L.V.; et al. Evaluation and Treatment of Chagas Disease in the United States. JAMA J. Am. Med. Assoc. 2007, 298, 2171–2181. [Google Scholar] [CrossRef] [Green Version]
- Schijman, A.G.; Bisio, M.; Orellana, L.; Sued, M.; Duffy, T.; Mejía-Jaramillo, A.; Cura, C.; Auter, F.; Veron, V.; Qvarnstrom, Y.; et al. International Study to Evaluate PCR Methods for Detection of Trypanosoma cruzi DNA in Blood Samples from Chagas Disease Patients. PLoS Negl. Trop. Dis. 2011, 5, e931. [Google Scholar] [CrossRef]
- Gourlay, L.; Peri, C.; Bolognesi, M.; Colombo, G. Structure and Computation in Immunoreagent Design: From Diagnostics to Vaccines. Trends Biotechnol. 2017, 35, 1208–1220. [Google Scholar] [CrossRef] [Green Version]
- Dormitzer, P.R.; Grandi, G.; Rappuoli, R. Structural vaccinology starts to deliver. Nat. Rev. Genet. 2012, 10, 807–813. [Google Scholar] [CrossRef]
- Gourlay, L.J.; Peri, C.; Ferrer-Navarro, M.; Conchillo-Solé, O.; Gori, A.; Rinchai, D.; Thomas, R.J.; Champion, O.L.; Michell, S.L.; Kewcharoenwong, C.; et al. Exploiting the Burkholderia pseudomallei Acute Phase Antigen BPSL2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology. Chem. Biol. 2013, 20, 1147–1156. [Google Scholar] [CrossRef] [Green Version]
- De Benedetti, S.; Di Pisa, F.; Fassi, E.; Cretich, M.; Musicò, A.; Frigerio, R.; Mussida, A.; Bombaci, M.; Grifantini, R.; Colombo, G.; et al. Structure, Immunoreactivity, and In Silico Epitope Determination of SmSPI S. mansoni Serpin for Immunodiagnostic Application. Vaccines 2021, 9, 322. [Google Scholar] [CrossRef] [PubMed]
- Gori, A.; Sola, L.; Gagni, P.; Bruni, G.; Liprino, M.; Peri, C.; Colombo, G.; Cretich, M.; Chiari, M. Screening Complex Biological Samples with Peptide Microarrays: The Favorable Impact of Probe Orientation via Chemoselective Immobilization Strategies on Clickable Polymeric Coatings. Bioconjugate Chem. 2016, 27, 2669–2677. [Google Scholar] [CrossRef] [PubMed]
- Krishnarjuna, B.; Andrew, D.; MacRaild, C.; Morales, R.; Beeson, J.G.; Anders, R.F.; Richards, J.S.; Norton, R.S. Strain-transcending immune response generated by chimeras of the malaria vaccine candidate merozoite surface protein 2. Sci. Rep. 2016, 6, 20613. [Google Scholar] [CrossRef] [PubMed]
- Martins, N.O.; De Souza, R.T.; Cordero, E.M.; Maldonado, D.C.; Cortez, C.; Marini, M.M.; Ferreira, E.R.; Bayer-Santos, E.; De Almeida, I.C.; Yoshida, N.; et al. Molecular Characterization of a Novel Family of Trypanosoma cruzi Surface Membrane Proteins (TcSMP) Involved in Mammalian Host Cell Invasion. PLoS Negl. Trop. Dis. 2015, 9, e0004216. [Google Scholar] [CrossRef]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 760–763. [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular replacement withMOLREP. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 66, 22–25. [Google Scholar] [CrossRef]
- Ramaswamy, R.; Nobary, S.G.; Eyford, B.A.; Pearson, T.W.; Boulanger, M.J. Structural characterization reveals a novel bilobed architecture for the ectodomains of insect stage expressed Trypanosoma brucei PSSA-2 and Trypanosoma congolense ISA. Protein Sci. A Publ. Protein Soc. 2016, 25, 2297–2302. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [Green Version]
- Langer, G.; Cohen, S.X.; Lamzin, V.; Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008, 3, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.I.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zingaretti, C.; Arigò, M.; Cardaci, A.; Moro, M.; Crosti, M.; Sinisi, A.; Sugliano, E.; Cheroni, C.; Marabita, F.; Nogarotto, R.; et al. Identification of New Autoantigens by Protein Array Indicates a Role for IL4 Neutralization in Autoimmune Hepatitis. Mol. Cell. Proteom. 2012, 11, 1885–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cretich, M.; DI Carlo, G.; Longhi, R.; Gotti, C.; Spinella, N.; Coffa, S.; Galati, C.; Renna, L.; Chiari, M. High Sensitivity Protein Assays on Microarray Silicon Slides. Anal. Chem. 2009, 81, 5197–5203. [Google Scholar] [CrossRef] [PubMed]
- Cretich, M.; Monroe, M.R.; Reddington, A.; Zhang, X.; Daaboul, G.G.; Damin, F.; Sola, L.; Ünlü, M.S.; Chiari, M. Interferometric silicon biochips for label and label-free DNA and protein microarrays. Proteomics 2012, 12, 2963–2977. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Loncharich, R.J.; Brooks, B.R.; Pastor, R.W. Langevin dynamics of peptides: The frictional dependence of isomerization rates ofN-acetylalanyl-N?-methylamide. Biopolymers 1992, 32, 523–535. [Google Scholar] [CrossRef]
- Åqvist, J.; Wennerström, P.; Nervall, M.; Bjelic, S.; Brandsdal, B.O. Molecular dynamics simulations of water and biomolecules with a Monte Carlo constant pressure algorithm. Chem. Phys. Lett. 2004, 384, 288–294. [Google Scholar] [CrossRef]
- Le Grand, S.; Götz, A.W.; Walker, R.C. SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013, 184, 374–380. [Google Scholar] [CrossRef]
- Darden, T.D.Y.; Pedersen, L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Scarabelli, G.; Morra, G.; Colombo, G. Predicting Interaction Sites from the Energetics of Isolated Proteins: A New Approach to Epitope Mapping. Biophys. J. 2010, 98, 1966–1975. [Google Scholar] [CrossRef] [Green Version]
- Peri, C.; Gagni, P.; Combi, F.; Gori, A.; Chiari, M.; Longhi, R.; Cretich, M.; Colombo, G. Rational Epitope Design for Protein Targeting. ACS Chem. Biol. 2012, 8, 397–404. [Google Scholar] [CrossRef]
- Montefiori, M.; Pilotto, S.; Marabelli, C.; Moroni, E.; Ferraro, M.; Serapian, S.A.; Mattevi, A.; Colombo, G. Impact of Mutations on NPAC Structural Dynamics: Mechanistic Insights from MD Simulations. J. Chem. Inf. Model. 2019, 59, 3927–3937. [Google Scholar] [CrossRef]
- Serapian, S.A.; Marchetti, F.; Triveri, A.; Morra, G.; Meli, M.; Moroni, E.; Sautto, G.A.; Rasola, A.; Colombo, G. The Answer Lies in the Energy: How Simple Atomistic Molecular Dynamics Simulations May Hold the Key to Epitope Prediction on the Fully Glycosylated SARS-CoV-2 Spike Protein. J. Phys. Chem. Lett. 2020, 11, 8084–8093. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Fragoso, C.M.; Burkard, G.S.; Oberle, M.; Renggli, C.K.; Hilzinger, K.; Roditi, I. PSSA-2, a Membrane-Spanning Phosphoprotein of Trypanosoma brucei, Is Required for Efficient Maturation of Infection. PLoS ONE 2009, 4, e7074. [Google Scholar] [CrossRef] [Green Version]
- McNicholas, S.; Potterton, E.; Wilson, K.; Noble, M. Presenting your structures: The CCP4mgmolecular-graphics software. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 386–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holm, L. Benchmarking fold detection by DaliLite v.5. Bioinformation 2019, 35, 5326–5327. [Google Scholar] [CrossRef]
- Lassaux, P.; Peri, C.; Ferrer-Navarro, M.; Gourlay, L.J.; Gori, A.; Conchillo-Solé, O.; Rinchai, D.; Lertmemongkolchai, G.; Longhi, R.; Daura, X.; et al. A Structure-Based Strategy for Epitope Discovery in Burkholderia pseudomallei OppA Antigen. Structure 2013, 21, 167–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, S.; Cretich, M.; Gagni, P.; Ahrens, B.; Grishina, G.; Sampson, H.A.; Niggemann, B.; Chiari, M.; Beyer, K. Performance of a polymer coated silicon microarray for simultaneous detection of food allergen-specific IgE and IgG4. Clin. Exp. Allergy 2017, 47, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Sola, L.; Gagni, P.; D’Annessa, I.; Capelli, R.; Bertino, C.; Romanato, A.; Damin, F.; Bergamaschi, G.; Marchisio, E.; Cuzzocrea, A.; et al. Enhancing Antibody Serodiagnosis Using a Controlled Peptide Coimmobilization Strategy. ACS Infect. Dis. 2018, 4, 998–1006. [Google Scholar] [CrossRef]
- Gori, A.; Peri, C.; Quilici, G.; Nithichanon, A.; Gaudesi, D.; Longhi, R.; Gourlay, L.; Bolognesi, M.; Lertmemongkolchai, G.; Musco, G.; et al. Flexible vs Rigid Epitope Conformations for Diagnostic- and Vaccine-Oriented Applications: Novel Insights from the Burkholderia pseudomallei BPSL2765 Pal3 Epitope. ACS Infect. Dis. 2016, 2, 221–230. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pisa, F.; De Benedetti, S.; Fassi, E.M.A.; Bombaci, M.; Grifantini, R.; Musicò, A.; Frigerio, R.; Pontillo, A.; Rigo, C.; Abelli, S.; et al. Elucidating the 3D Structure of a Surface Membrane Antigen from Trypanosoma cruzi as a Serodiagnostic Biomarker of Chagas Disease. Vaccines 2022, 10, 71. https://doi.org/10.3390/vaccines10010071
Di Pisa F, De Benedetti S, Fassi EMA, Bombaci M, Grifantini R, Musicò A, Frigerio R, Pontillo A, Rigo C, Abelli S, et al. Elucidating the 3D Structure of a Surface Membrane Antigen from Trypanosoma cruzi as a Serodiagnostic Biomarker of Chagas Disease. Vaccines. 2022; 10(1):71. https://doi.org/10.3390/vaccines10010071
Chicago/Turabian StyleDi Pisa, Flavio, Stefano De Benedetti, Enrico Mario Alessandro Fassi, Mauro Bombaci, Renata Grifantini, Angelo Musicò, Roberto Frigerio, Angela Pontillo, Cinzia Rigo, Sandra Abelli, and et al. 2022. "Elucidating the 3D Structure of a Surface Membrane Antigen from Trypanosoma cruzi as a Serodiagnostic Biomarker of Chagas Disease" Vaccines 10, no. 1: 71. https://doi.org/10.3390/vaccines10010071
APA StyleDi Pisa, F., De Benedetti, S., Fassi, E. M. A., Bombaci, M., Grifantini, R., Musicò, A., Frigerio, R., Pontillo, A., Rigo, C., Abelli, S., Grande, R., Zanchetta, N., Mileto, D., Mancon, A., Rizzo, A., Gori, A., Cretich, M., Colombo, G., Bolognesi, M., & Gourlay, L. J. (2022). Elucidating the 3D Structure of a Surface Membrane Antigen from Trypanosoma cruzi as a Serodiagnostic Biomarker of Chagas Disease. Vaccines, 10(1), 71. https://doi.org/10.3390/vaccines10010071









