Leishmania infantum UBC1 in Metacyclic Promastigotes from Phlebotomus perniciosus, a Vaccine Candidate for Zoonotic Visceral Leishmaniasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Parasites and Whole Protein Extracts
2.2. Ethics
2.3. LiUBC1 Expression and Purification
2.4. Antibody Obtainment
2.5. Protein Expression Level Determination by Western Blot
2.6. Generation of L. infantum LiUBC1 Knock-In Promastigote Line
2.7. LiUBC1 Cellular Detection by Indirect Immunofluorescence (IIF)
2.8. Transmission Electron Microscopy (TEM) Detection of LiUBC1
2.9. Mass Spectrometry LC–MS/MS Analysis
2.10. L. infantum In Vitro Infection Experiments
2.11. Animal Challenge Experiments
2.12. Parasite Burden Evaluation and Immune IgG Response Determination in M. auratus Model
2.13. Relative IFN-γ Expression Levels in Splenocytes by qRT-PCR
2.14. Sequence Alignment and Molecular Modeling
3. Results
3.1. Primary Structure of LiUBC1
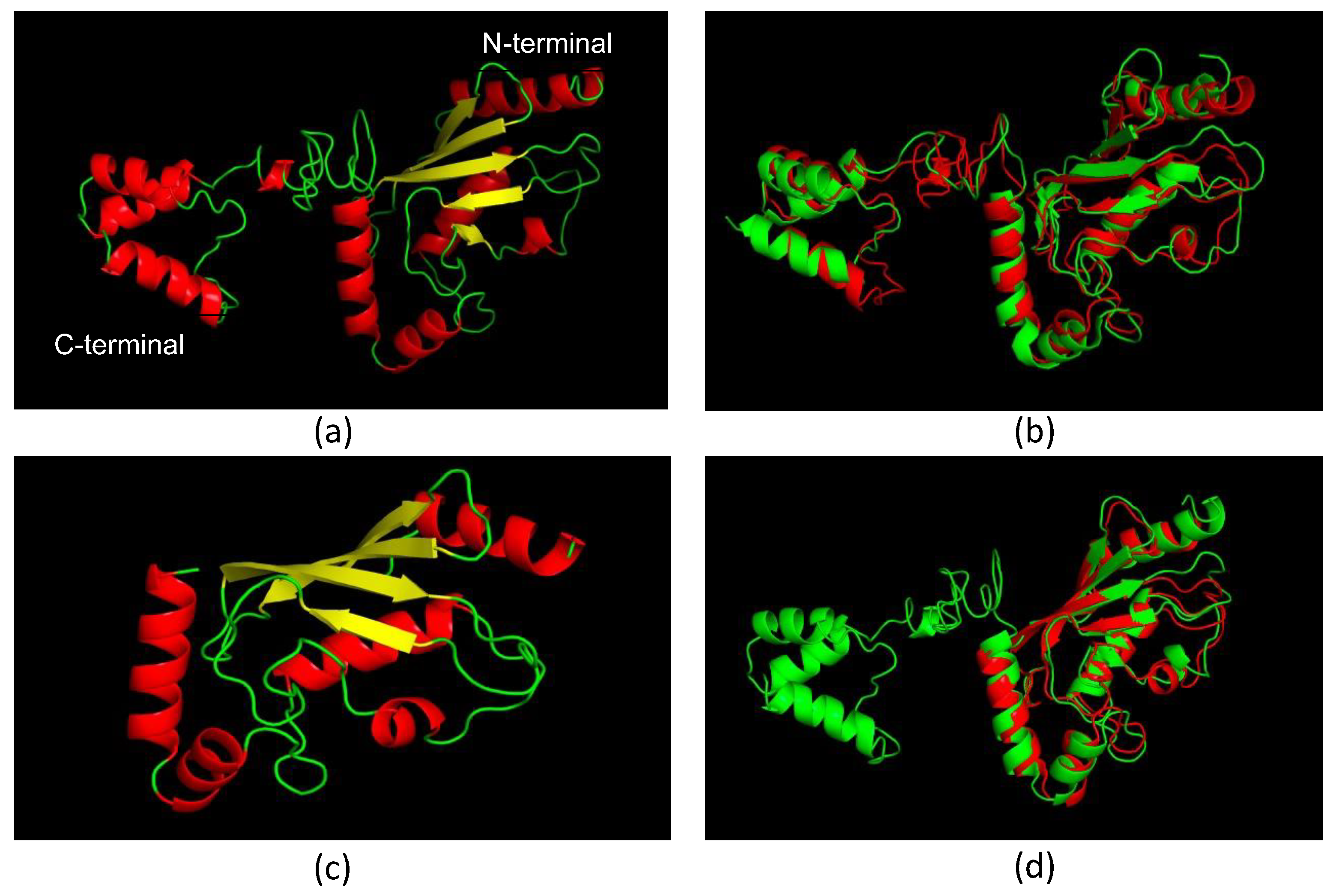
3.2. Molecular Models of LiUBC1
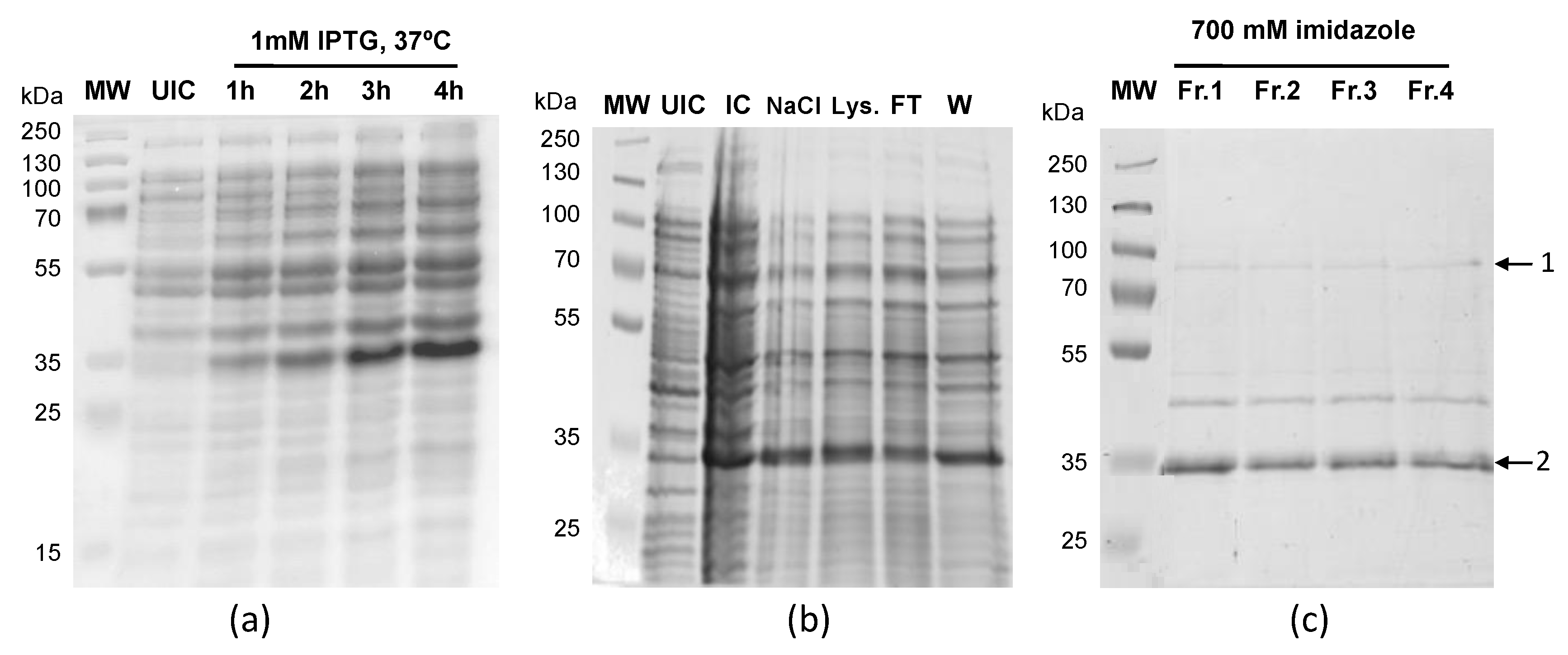
3.3. LiUBC1 Purification
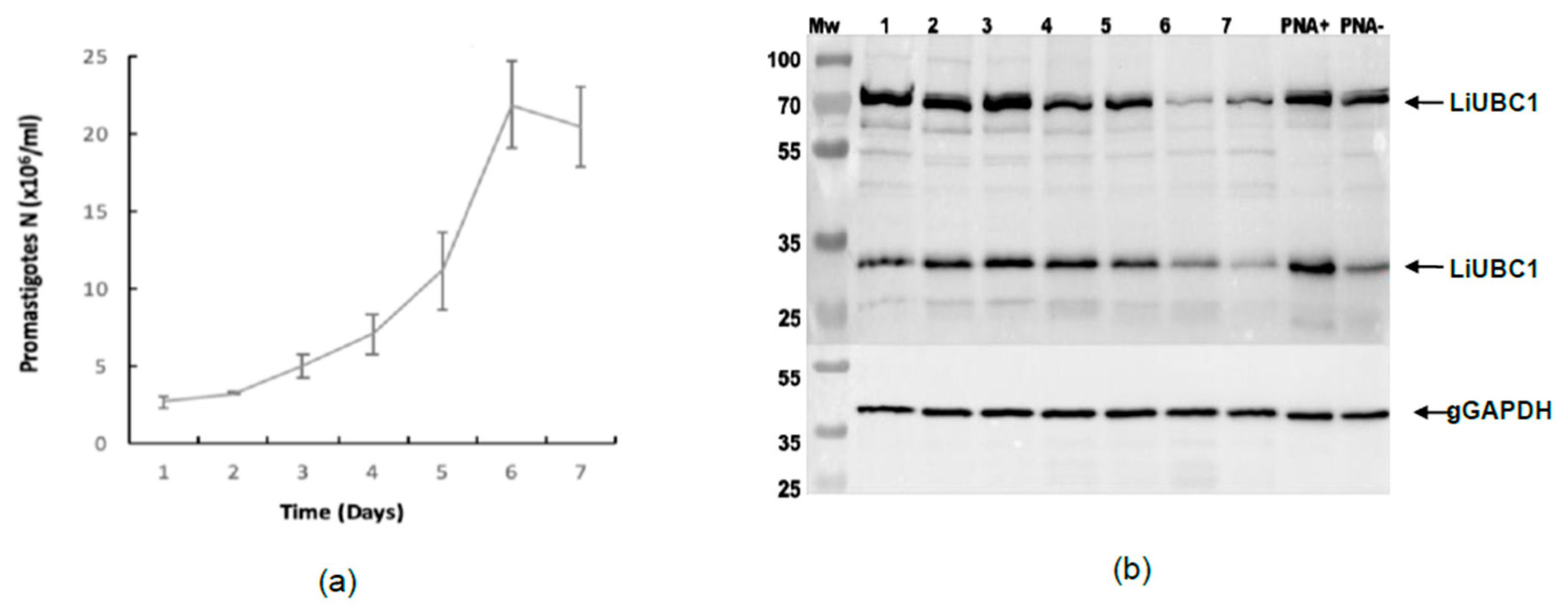
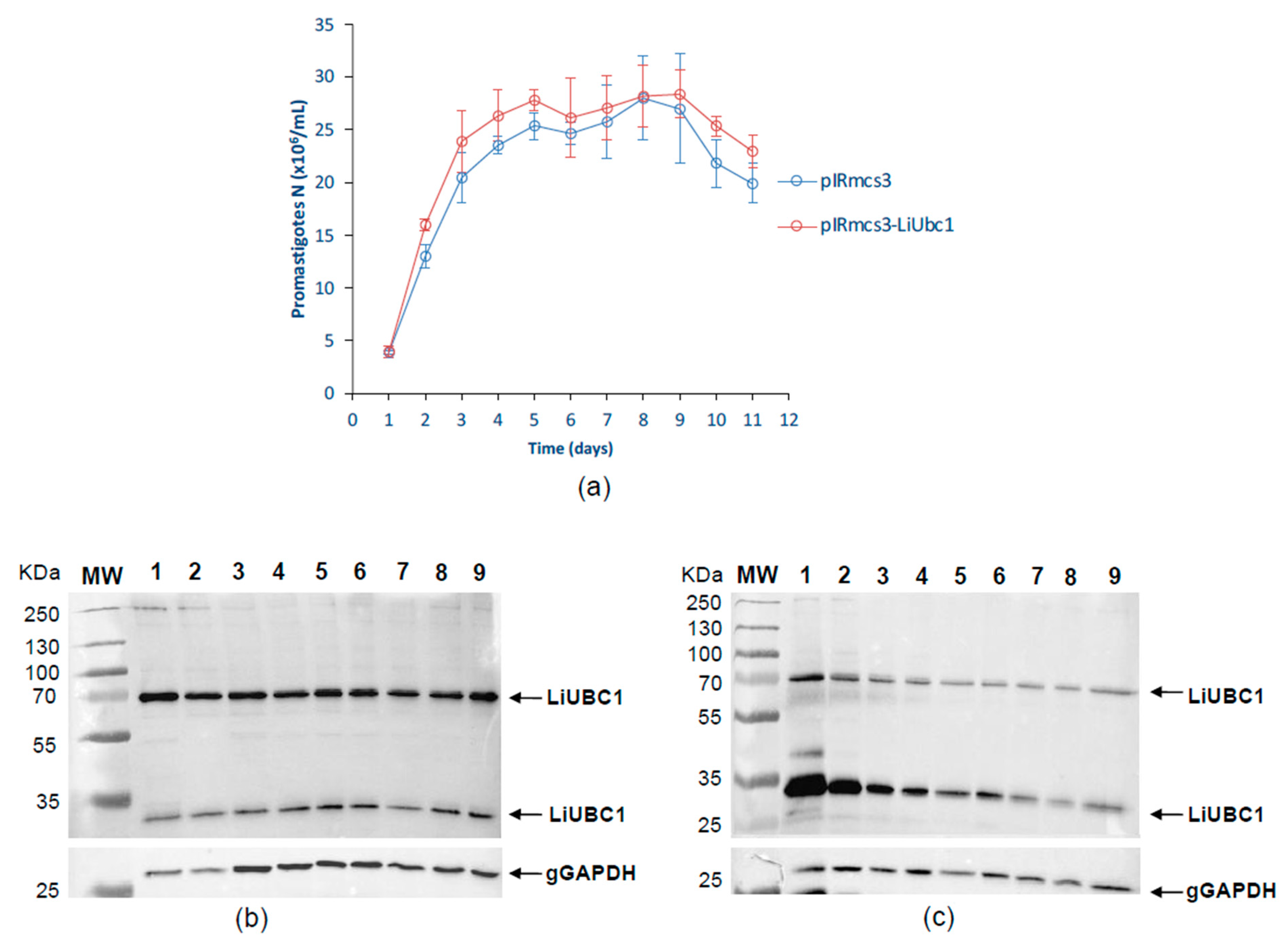
3.4. LiUBC1 Expression Levels throughout the Growth Curve of IPER/ES/2013/ATE1FL6 Strain of L. infantum Promastigotes in Axenic Culture
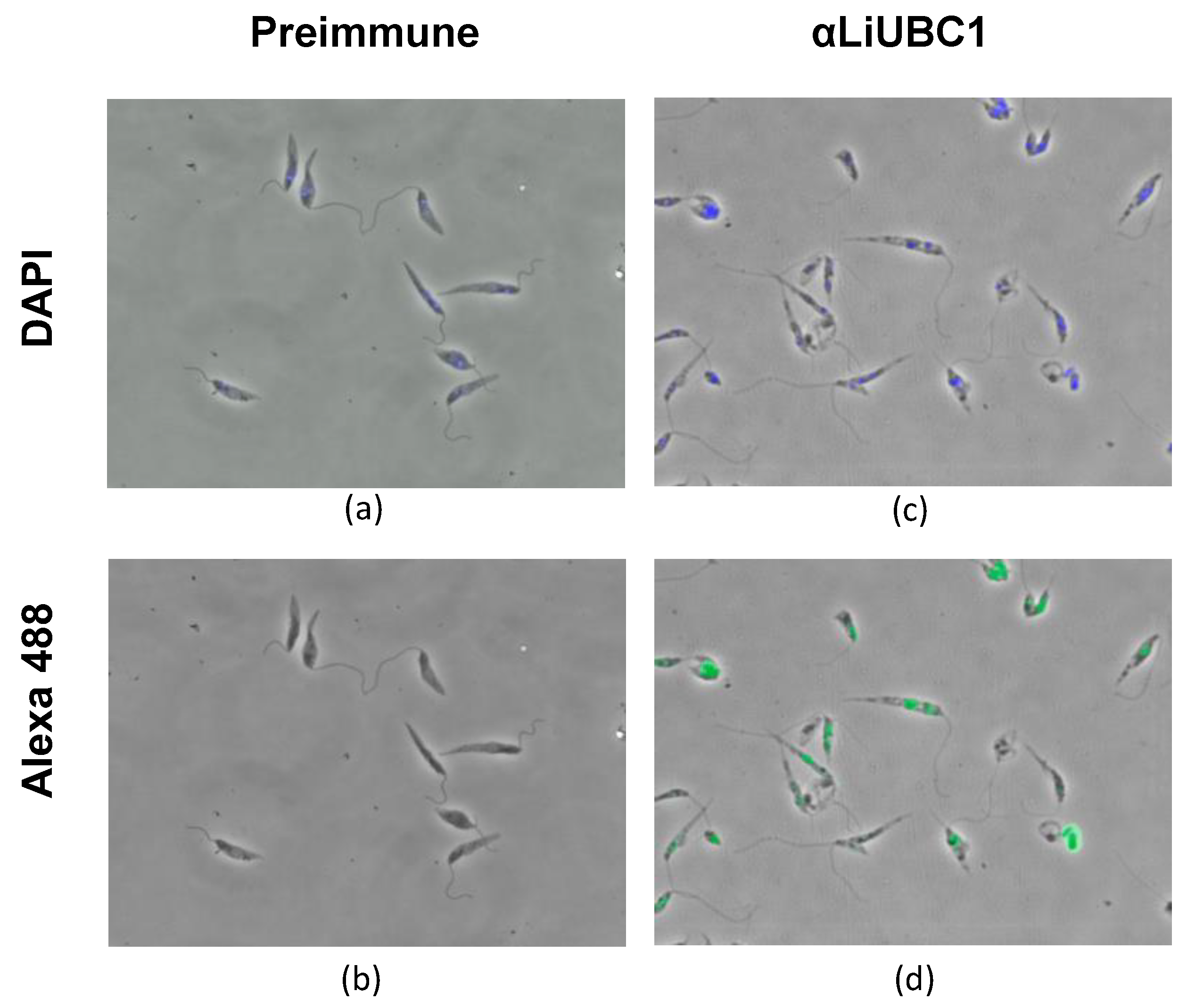
3.5. Subcellular Localization of LiUBC1 in the L. infantum Parasite
3.6. Characterization of an L. infantum Stable LiUBC1 Knock-In Promastigote Cell Line
| Gene | Description | Mean pIRmcs3-LiUBC1 vs. pIRmcs3 | ||
|---|---|---|---|---|
| Day 1 | Day 3 | Day 5 | ||
| LinJ.33.2910 | LiUBC1 Ubiquitin-conjugating enzyme E2, putative | 2.759 ± 0.30 | 0.031 ± 0.11 | 0.327 ± 0.51 |
| LinJ.23.0710 | Ubiquitin-conjugating enzyme E1, putative | 0.008 ± 0.01 | 0.062 ± 0.07 | 0.083 ± 0.06 |
| LinJ.35.1310 | Ubiquitin-conjugating enzyme E2, putative | 0.021 ± 0.01 | 0.095 ± 0.14 | 0.264 ± 0.25 |
| LinJ.13.1320 | Ubiquitin-conjugating enzyme, putative | 0.225 ± 0.29 | 0.317 ± 0,40 | 0.087 ± 0.59 |
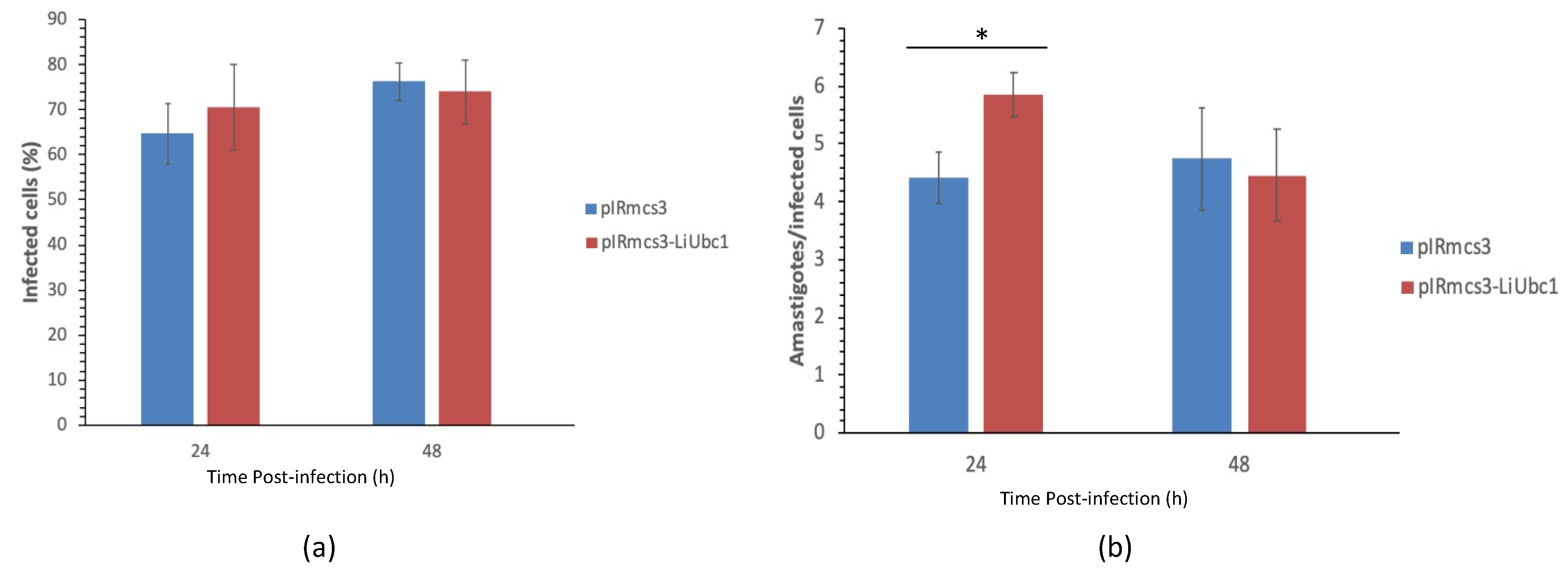
3.7. Infectivity of LiUBC1 Knock-In Promastigotes in U937 Cells In Vitro
3.8. Protective Effect of the LiUBC1 Recombinant Protein in M. auratus against L. infantum Challenge
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- OMS. Report of a Meeting of the WHO Expert Committee on the Control of Leishmaniases; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Alvar, J.; Canavate, C.; Gutierrez-Solar, B.; Jimenez, M.; Laguna, F.; Lopez-Velez, R.; Molina, R.; Moreno, J. Leishmania and human immunodeficiency virus coinfection: The first 10 years. Clin. Microbiol. Rev. 1997, 10, 298–319. [Google Scholar] [CrossRef] [PubMed]
- Desjeux, P. Leishmania/HIV co-infections. Afr. Health 1995, 18, 20–22. [Google Scholar] [PubMed]
- Desjeux, P.; Piot, B.; O’Neill, K.; Meert, J.P. Co-infections of leishmania/HIV in south Europe. Med. Trop. Rev. Corps Sante Colon. 2001, 61, 187–193. [Google Scholar]
- Jiménez, M.; Alvar, J.; Tibayrenc, M. Leishmania infantum is clonal in AIDS patients too: Epidemiological implications. AIDS 1997, 11, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Wolday, D.; Berhe, N.; Akuffo, H.; Desjeux, P.; Britton, S. Emerging Leishmania/HIV co-infection in Africa. Med. Microbiol. Immunol. 2001, 190, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Desjeux, P. Leishmaniasis: Public health aspects and control. Clin. Dermatol. 1996, 14, 417–423. [Google Scholar] [CrossRef]
- Mauricio, I.L.; Stothard, J.R.; Miles, M.A. The Strange Case of Leishmania chagasi. Parasitol. Today 2000, 16, 188–189. [Google Scholar] [CrossRef]
- Arce, A.; Estirado, A.; Ordobas, M.; Sevilla, S.; García, N.; Moratilla, L.; De La Fuente, S.; Martínez, A.M.; Pérez, A.M.; Aránguez, E.; et al. Re-emergence of leishmaniasis in Spain: Community outbreak in Madrid, Spain, 2009 to 2012. Eurosurveillance 2013, 18, 20546. [Google Scholar] [CrossRef]
- Jiménez, M.; González, E.; Martin-Martin, I.; Hernández, S.; Molina, R. Could wild rabbits (Oryctolagus cuniculus) be reservoirs for Leishmania infantum in the focus of Madrid, Spain? Veter Parasitol. 2014, 202, 296–300. [Google Scholar] [CrossRef]
- Martin-Martin, I.; Molina, R.; Rohousova, I.; Drahota, J.; Volf, P.; Jiménez, M. High levels of anti-Phlebotomus perniciosus saliva antibodies in different vertebrate hosts from the re-emerging leishmaniosis focus in Madrid, Spain. Veter. Parasitol. 2014, 202, 207–216. [Google Scholar] [CrossRef]
- Molina, R.; Jiménez, M.I.; Cruz, I.; Iriso, A. The hare (Lepus granatensis) as potential sylvatic reservoir of Leishmania infantum in Spain. Vet. Parasitol. 2012, 190, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Abranches, P.; Silva-Pereira, M.C.D.; Conceicao-Silva, F.M.; Santos-Gomes, G.M.; Janz, J.G. Canine Leishmaniasis: Pathological and Ecological Factors Influencing Transmission of Infection. J. Parasitol. 1991, 77, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Semião-Santos, S.J.; El Harith, A.; Ferreira, E.; Pires, C.A.; Sousa, C.; Gusmão, R. Evora district as a new focus for canine leishmaniasis in Portugal. Parasitol. Res. 1995, 81, 235–239. [Google Scholar] [PubMed]
- Moody, S.F. Molecular variation in Leishmania. Acta Trop. 1993, 53, 185–204. [Google Scholar] [CrossRef]
- Sundar, S.; Rai, M. Advances in the treatment of leishmaniasis. Curr. Opin. Infect. Dis. 2002, 15, 593–598. [Google Scholar] [CrossRef]
- Singh, N.; Kumar, M.; Singh, R.K. Leishmaniasis: Current status of available drugs and new potential drug targets. Asian Pac. J. Trop. Med. 2012, 5, 485–497. [Google Scholar] [CrossRef]
- Cotrina, J.F.; Iniesta, V.; Monroy, I.; Baz, V.; Hugnet, C.; Marañon, F.; Fabra, M.; Gómez-Nieto, L.C.; Alonso, C. A large-scale field randomized trial demonstrates safety and efficacy of the vaccine LetiFend(R) against canine leishmaniosis. Vaccine 2018, 36, 1972–1982. [Google Scholar] [CrossRef]
- Oliva, G.; Nieto, J.; Manzillo, V.F.; Cappiello, S.; Fiorentino, E.; Di Muccio, T.; Scalone, A.; Moreno, J.; Chicharro, C.; Carrillo, E.; et al. A Randomised, Double-Blind, Controlled Efficacy Trial of the LiESP/QA-21 Vaccine in Naïve Dogs Exposed to Two Leishmania infantum Transmission Seasons. PLOS Negl. Trop. Dis. 2014, 8, e3213. [Google Scholar] [CrossRef]
- Borja-Cabrera, G.P.; Mendes, A.C.; de Souza, E.P.; Okada, L.Y.H.; de A Trivellato, F.A.; Kawasaki, J.K.A.; Costa, A.C.; Reis, A.B.; Genaro, O.; Batista, L.M.M.; et al. Effective immunotherapy against canine visceral leishmaniasis with the FML-vaccine. Vaccine 2004, 22, 2234–2243. [Google Scholar] [CrossRef]
- Palatnik-De-Sousa, C.B. Vaccines for Canine Leishmaniasis. Front. Immunol. 2012, 3, 69. [Google Scholar] [CrossRef]
- Alcolea, P.J.; Alonso, A.; Larraga, V. The antibiotic resistance-free mammalian expression plasmid vector pPAL for development of third generation vaccines. Plasmid 2018, 101, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Gifawesen, C.; Farrell, J.P. Comparison of T-cell responses in self-limiting versus progressive visceral Leishmania donovani infections in golden hamsters. Infect. Immun. 1989, 57, 3091–3096. [Google Scholar] [CrossRef] [PubMed]
- Vasconcellos, R.C.; Urago, K.P.; Bunn-Moreno, M.M.; Madeira, E.D. Suppressor activity in Leishmania donovani-infected hamster serum: Reversion by delipidated bovine serum albumin and role in cell cycle events. Braz. J. Med. Biol. Res. 1996, 29, 615–622. [Google Scholar] [PubMed]
- Requena, J.M.; Soto, M.; Doria, M.D.; Alonso, C. Immune and clinical parameters associated with Leishmania infantum infection in the golden hamster model. Veter. Immunol. Immunopathol. 2000, 76, 269–281. [Google Scholar] [CrossRef]
- Sacks, D.B.; Noben-Trauth, N. The immunology of susceptibility and resistance to Leishmania major in mice. Nat. Rev. Immunol. 2002, 2, 845–858. [Google Scholar] [CrossRef]
- Alcolea, P.J.; Alonso, A.; Esteban, A.; Peris, P.; Cortés, A.; Castillo, J.A.; Larraga, V. IL12 p35 and p40 subunit genes administered as pPAL plasmid constructs do not improve protection of pPAL-LACK vaccine against canine leishmaniasis. PLoS ONE 2019, 14, e0212136. [Google Scholar] [CrossRef] [PubMed]
- Leifso, K.; Cohen-Freue, G.; Dogra, N.; Murray, A.; McMaster, W.R. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: The Leishmania genome is constitutively expressed. Mol. Biochem. Parasitol. 2007, 152, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Alcolea, P.J.A.; Alonso, A.; Larraga, V. Proteome Profiling of Leishmania Infantum Promastigotes. J. Eukaryot. Microbiol. 2011, 58, 352–358. [Google Scholar] [CrossRef]
- Rosenzweig, D.; Smith, D.; Myler, P.J.; Olafson, R.W.; Zilberstein, D. Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics 2008, 8, 1843–1850. [Google Scholar] [CrossRef]
- Alcolea, P.J.; Alonso, A.; Molina, R.; Jiménez, M.; Myler, P.J.; Larraga, V. Functional genomics in sand fly–derived Leishmania promastigotes. PLOS Negl. Trop. Dis. 2019, 13, e0007288. [Google Scholar] [CrossRef]
- Alcolea, P.J.A.; Alonso, A.; Domínguez, M.; Parro, V.; Jimenez, M.; Molina, R.; Larraga, V. Influence of the Microenvironment in the Transcriptome of Leishmania infantum Promastigotes: Sand Fly versus Culture. PLOS Neglected Trop. Dis. 2016, 10, e0004693. [Google Scholar] [CrossRef] [PubMed]
- Alcolea, P.J.; Alonso, A.; Larraga, V. Rationale for selection of developmentally regulated genes as vaccine candidates against Leishmania infantum infection. Vaccine 2016, 34, 5474–5478. [Google Scholar] [CrossRef] [PubMed]
- De Agüero, M.G.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- De Freitas, F.B.; Frouco, G.; Martins, C.; Ferreira, F. African swine fever virus encodes for an E2-ubiquitin conjugating enzyme that is mono- and di-ubiquitinated and required for viral replication cycle. Sci. Rep. 2018, 8, 3471. [Google Scholar] [CrossRef]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Dawson, T.M.; Dawson, V.L. Molecular Pathways of Neurodegeneration in Parkinson’s Disease. Science 2003, 302, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Hallengren, J.; Chen, P.-C.; Wilson, S.M. Neuronal Ubiquitin Homeostasis. Cell Biophys. 2013, 67, 67–73. [Google Scholar] [CrossRef]
- Gardner, R.G.; Nelson, Z.W.; Gottschling, D.E. Degradation-Mediated Protein Quality Control in the Nucleus. Cell 2005, 120, 803–815. [Google Scholar] [CrossRef]
- Muñoz, C.; Francisco, J.S.; Gutiérrez, B.; González, J. Role of the Ubiquitin-Proteasome Systems in the Biology and Virulence of Protozoan Parasites. BioMed Res. Int. 2015, 2015, 141526. [Google Scholar] [CrossRef]
- Bhandari, D.; Guha, K.; Bhaduri, N.; Saha, P. Ubiquitination of mRNA cycling sequence binding protein from Leishmania donovani (LdCSBP) modulates the RNA endonuclease activity of its Smr domain. FEBS Lett. 2011, 585, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Kazemirad, E.; Mohebali, M.; Khadem-Erfan, M.B.; Hajjaran, H.; Hadighi, R.; Khamesipour, A.; Rezaie, S.; Saffari, M.; Raoofian, R.; Heidari, M. Overexpression of Ubiquitin and Amino Acid Permease Genes in Association with Antimony Resistance in Leishmania tropica Field Isolates. Korean J. Parasitol. 2013, 51, 413–419. [Google Scholar] [CrossRef]
- González-Aseguinolaza, G.; Almazan, F.; Rodríguez, J.; Marquet, A.; Larraga, V. Cloning of the gp63 surface protease of Leishmania infantum: Differential post-translational modifications correlated with different infective forms. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 1997, 1361, 92–102. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hannaert, V.; Opperdoes, F.R.; Michels, P.A. Comparison and Evolutionary Analysis of the Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Different Kinetoplastida. J. Mol. Evol. 1998, 47, 728–738. [Google Scholar] [CrossRef]
- Alzate, J.F.; Alvarez-Barrientos, A.; Gonzalez, V.M.; Jimenez-Ruiz, A. Heat-induced programmed cell death in Leishmania infantum is reverted by Bcl-X(L) expression. Apoptosis 2006, 11, 161–171. [Google Scholar] [CrossRef]
- Alcolea, P.J.; Alonso, A.; García-Tabares, F.; Mena, M.D.C.; Ciordia, S.; Larraga, V. An Insight into the Constitutive Proteome Throughout Leishmania donovani Promastigote Growth and Differentiation. Int. Microbiol. 2018, 22, 143–154. [Google Scholar] [CrossRef]
- Cristobo, I.; Larriba, M.J.; Ríos, V.D.L.; García, F.; Muñoz, A.; Casal, J.I. Proteomic analysis of 1α,25-Dihydroxyvitamin D3 action on human colon cancer cells reveals a link to splicing regulation. J. Proteom. 2011, 75, 384–397. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Buffet, P.A.; Sulahian, A.; Garin, Y.J.; Nassar, N.; Derouin, F. Culture microtitration: A sensitive method for quantifying Leishmania infantum in tissues of infected mice. Antimicrob. Agents Chemother. 1995, 39, 2167–2168. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sheng, Y.; Hong, J.H.; Doherty, R.; Srikumar, T.; Shloush, J.; Avvakumov, G.V.; Walker, J.R.; Xue, S.; Neculai, D.; Wan, J.W.; et al. A Human Ubiquitin Conjugating Enzyme (E2)-HECT E3 Ligase Structure-function Screen. Mol. Cell. Proteom. 2012, 11, 329–341. [Google Scholar] [CrossRef]
- Merkley, N.; Shaw, G.S. Solution Structure of the Flexible Class II Ubiquitin-conjugating Enzyme Ubc1 Provides Insights for Polyubiquitin Chain Assembly. J. Biol. Chem. 2004, 279, 47139–47147. [Google Scholar] [CrossRef] [PubMed]
- Besteiro, S.; Williams, R.; Morrison, L.S.; Coombs, G.H.; Mottram, J. Endosome Sorting and Autophagy Are Essential for Differentiation and Virulence of Leishmania major. J. Biol. Chem. 2006, 281, 11384–11396. [Google Scholar] [CrossRef] [PubMed]
- Goos, C.; DeJung, M.; Wehman, A.M.; M-Natus, E.; Schmidt, J.; Sunter, J.; Engstler, M.; Butter, F.; Kramer, S. Trypanosomes can initiate nuclear export co-transcriptionally. Nucleic Acids Res. 2018, 47, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Madhubala, R. Ubiquitin Conjugation of Open Reading Frame F DNA Vaccine Leads to Enhanced Cell-Mediated Immune Response and Induces Protection against Both Antimony-Susceptible and -Resistant Strains of Leishmania donovani. J. Immunol. 2009, 183, 7719–7731. [Google Scholar] [CrossRef]
- Heidari, S.; Hajjaran, H.; Kazemi, B.; Gharechahi, J.; Mohebali, M.; Ranjbar, M.M.; Akhoundi, B.; Azarian, B.; Mirshahvaladi, S.; Raoofian, R. Identification of immunodominant proteins of Leishmania infantum by immunoproteomics to evaluate a recombinant multi-epitope designed antigen for serodiagnosis of human visceral leishmaniasis. Exp. Parasitol. 2021, 222, 108065. [Google Scholar] [CrossRef]
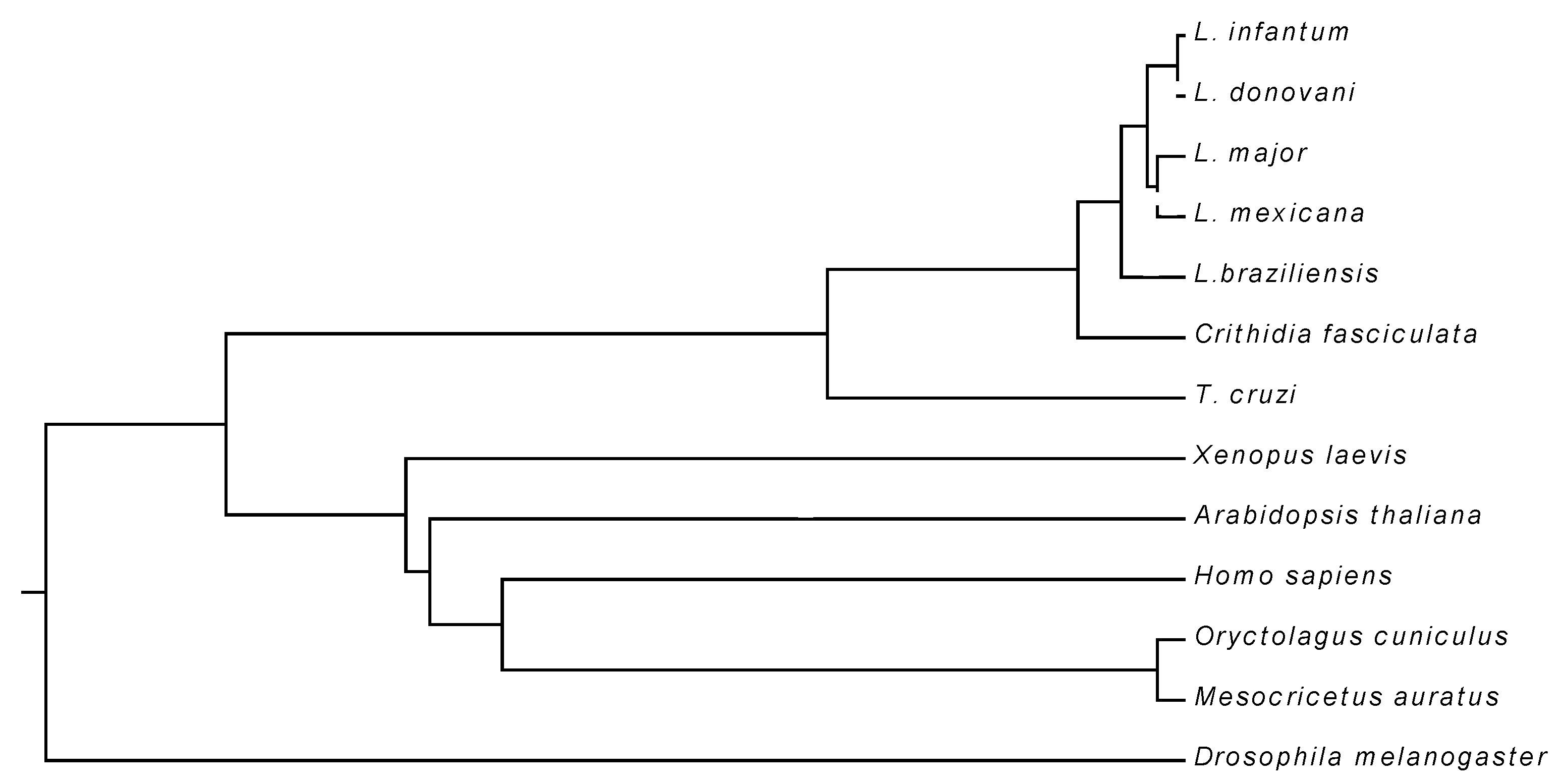


| Species | % Identity |
|---|---|
| Leishmania donovani | 100 |
| Leishmania major Leishmania mexicana Leishmania braziliensis Trypanosoma cruzi Oryctolagus cuniculus Homo sapiens Xenopus laevis Arabidopsis thaliana Drosophila melanogaster Mesocricetus auratus | 100 91.70 80.50 48.98 38.09 31.30 31.30 30.82 27.14 20.17 |
| BAND | Gene | Description | Score | Coverage |
|---|---|---|---|---|
| 1 | LinJ.33.2910 | Ubiquitin-conjugating enzyme E2 | 1235 | 91.70 |
| 2 | LinJ.33.2910 | Ubiquitin-conjugating enzyme E2 | 487 | 70.54 |
| Group | Sample Nº | Spleen Weight (g) | MG Dilution Limit | Amas/g Spleen |
|---|---|---|---|---|
| LiUBC1 | 1 2 3 4 5 6 7 8 | 0.1123 0.1099 0.0844 0.1236 0.1025 0.0844 0.0925 0.1299 | - - - - - - - - | - - - - - - - - |
| Control | 9 10 11 12 13 14 | 0.1371 0.0972 0.1161 0.1171 0.0752 0.1738 | - 16 - - 32 256 | - 2469 - - 6382 22094 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larraga, J.; Alcolea, P.J.; Alonso, A.; Martins, L.T.C.; Moreno, I.; Domínguez, M.; Larraga, V. Leishmania infantum UBC1 in Metacyclic Promastigotes from Phlebotomus perniciosus, a Vaccine Candidate for Zoonotic Visceral Leishmaniasis. Vaccines 2022, 10, 231. https://doi.org/10.3390/vaccines10020231
Larraga J, Alcolea PJ, Alonso A, Martins LTC, Moreno I, Domínguez M, Larraga V. Leishmania infantum UBC1 in Metacyclic Promastigotes from Phlebotomus perniciosus, a Vaccine Candidate for Zoonotic Visceral Leishmaniasis. Vaccines. 2022; 10(2):231. https://doi.org/10.3390/vaccines10020231
Chicago/Turabian StyleLarraga, Jaime, Pedro J. Alcolea, Ana Alonso, Luis T. C. Martins, Inmaculada Moreno, Mercedes Domínguez, and Vicente Larraga. 2022. "Leishmania infantum UBC1 in Metacyclic Promastigotes from Phlebotomus perniciosus, a Vaccine Candidate for Zoonotic Visceral Leishmaniasis" Vaccines 10, no. 2: 231. https://doi.org/10.3390/vaccines10020231
APA StyleLarraga, J., Alcolea, P. J., Alonso, A., Martins, L. T. C., Moreno, I., Domínguez, M., & Larraga, V. (2022). Leishmania infantum UBC1 in Metacyclic Promastigotes from Phlebotomus perniciosus, a Vaccine Candidate for Zoonotic Visceral Leishmaniasis. Vaccines, 10(2), 231. https://doi.org/10.3390/vaccines10020231






