Evaluation of HSV-2 gE Binding to IgG-Fc and Application for Vaccine Development
Abstract
1. Introduction
2. Materials and Methods
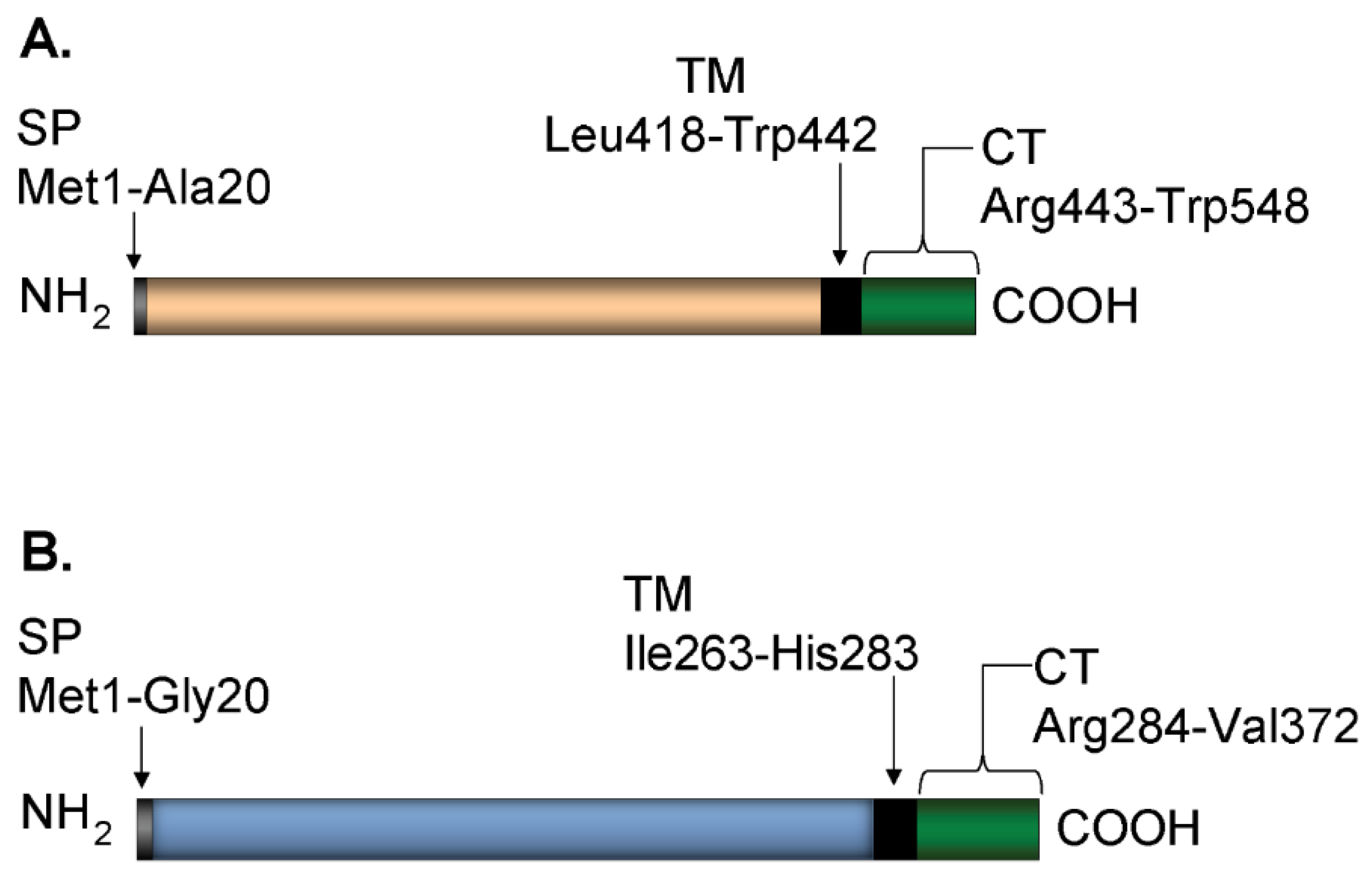
2.1. Plasmid Construction
2.2. Protein Expression
2.3. Purification of Soluble gE and gE/gI Heterodimers
2.4. SDS-PAGE and Western Blot
2.5. Surface Plasmon Resonance
2.6. Size Exclusion Chromatography/Multi-Angle Laser Light Scattering
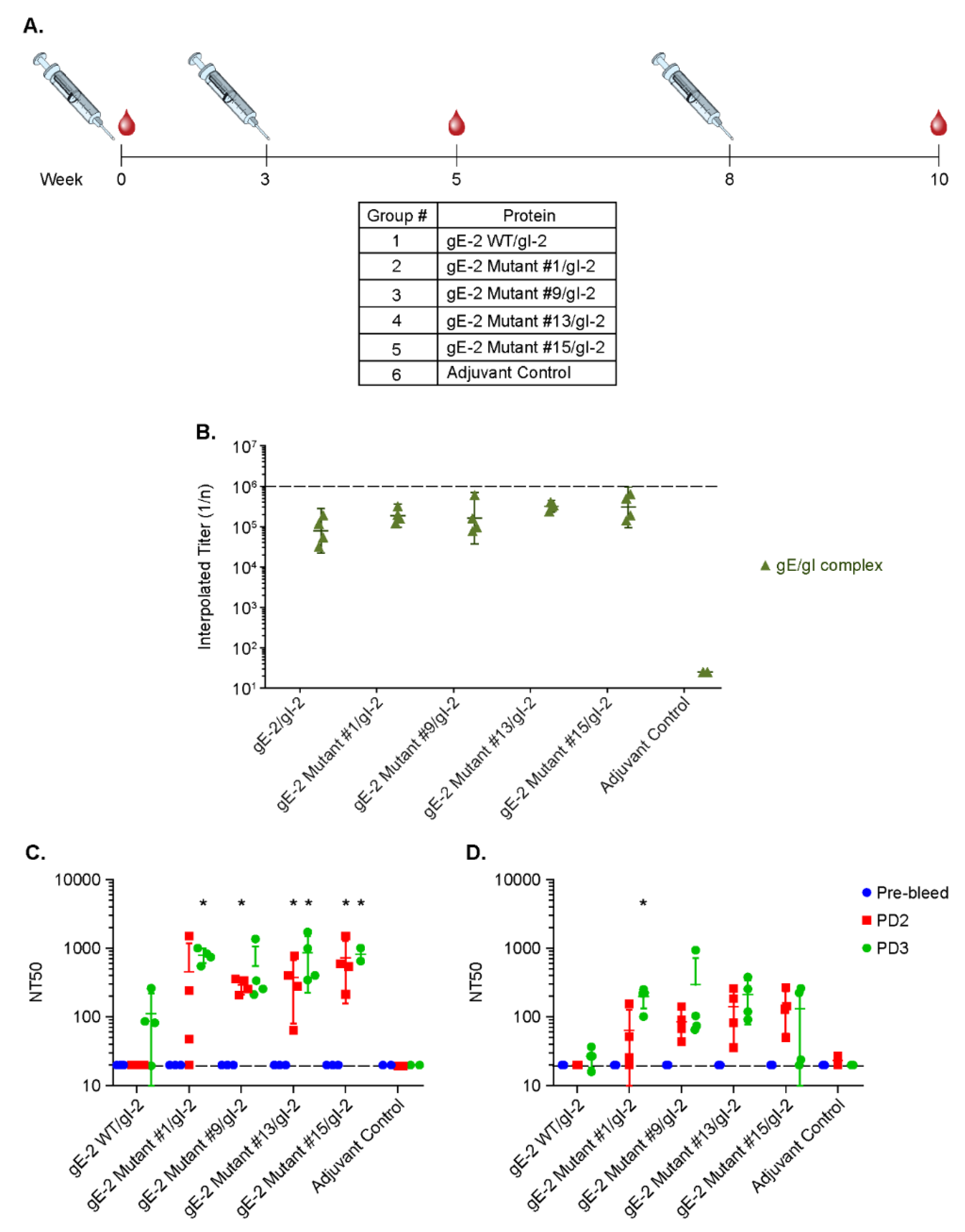
2.7. Rabbit Immunization Study
2.8. Antibody ELISA Assay
2.9. Neutralization Assay
3. Results
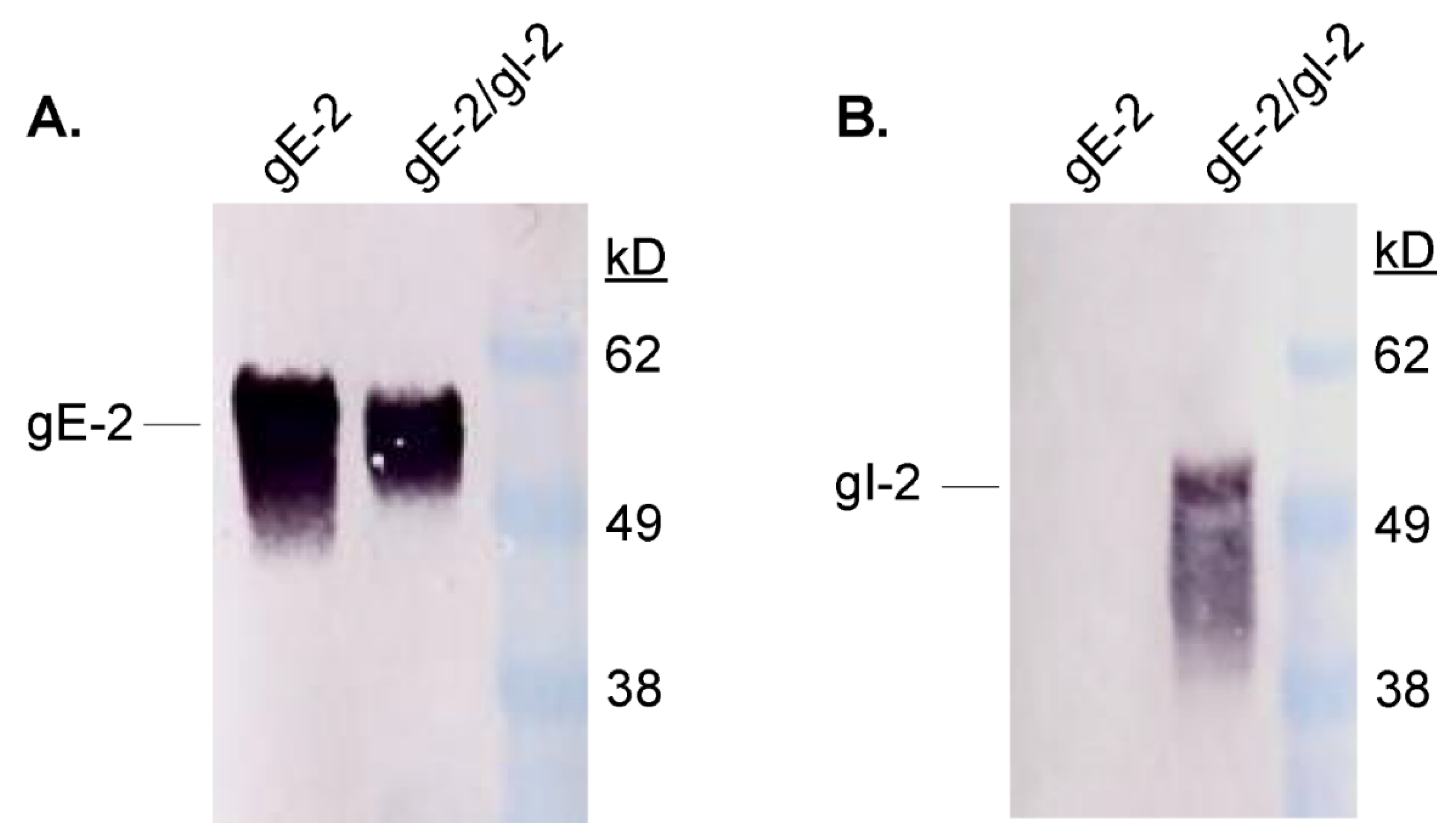
3.1. Soluble gE-2 Forms a Heterodimer with Soluble gI-2 When Co-Transfected into Mammalian Cells
3.2. gE-2 Requires gI-2 to Bind IgG Fc
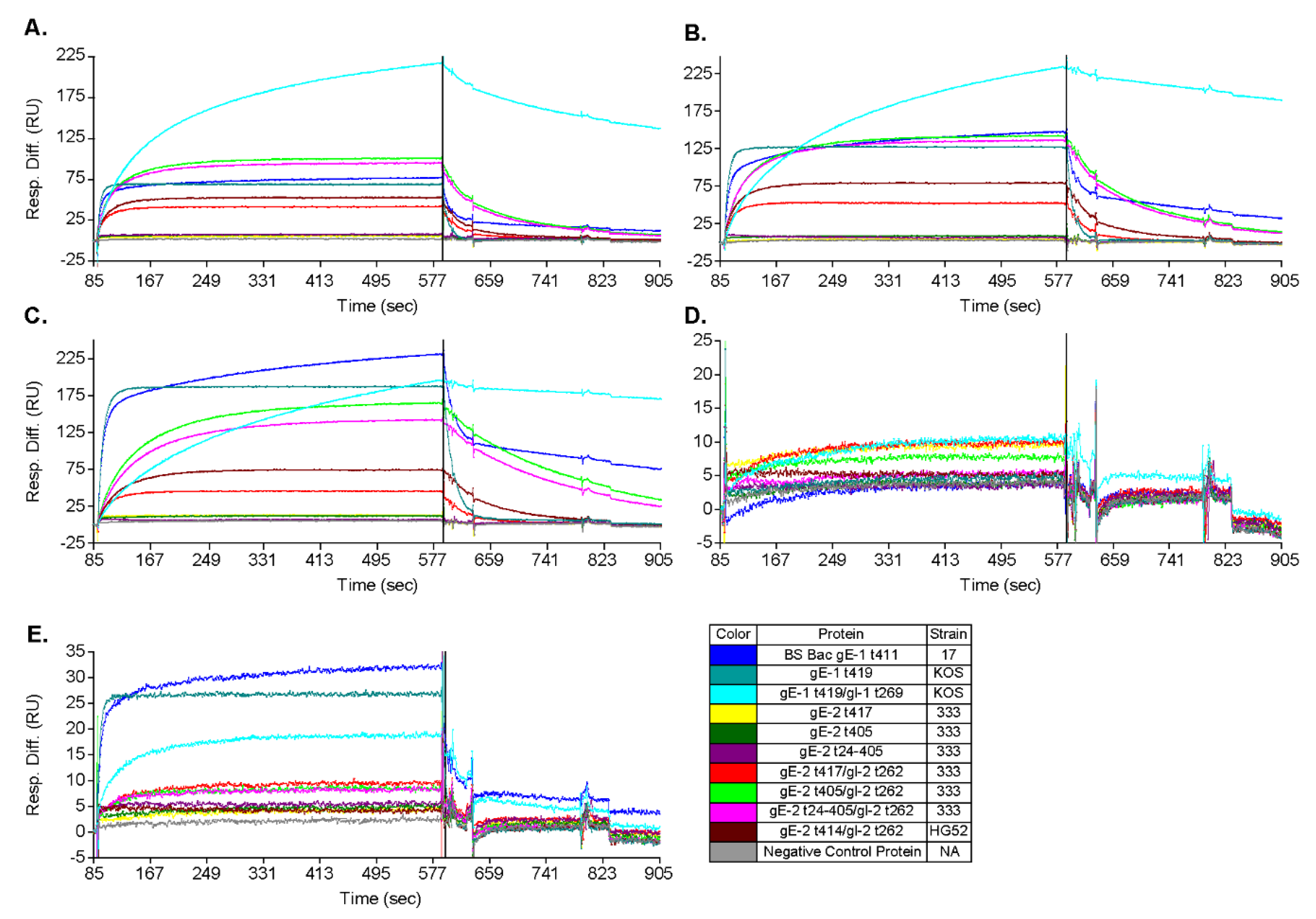
3.3. gE-2/gI-2 Heterodimers Appear to Have Different Association/Dissociation Profiles than gE-1/gI-1 Heterodimers
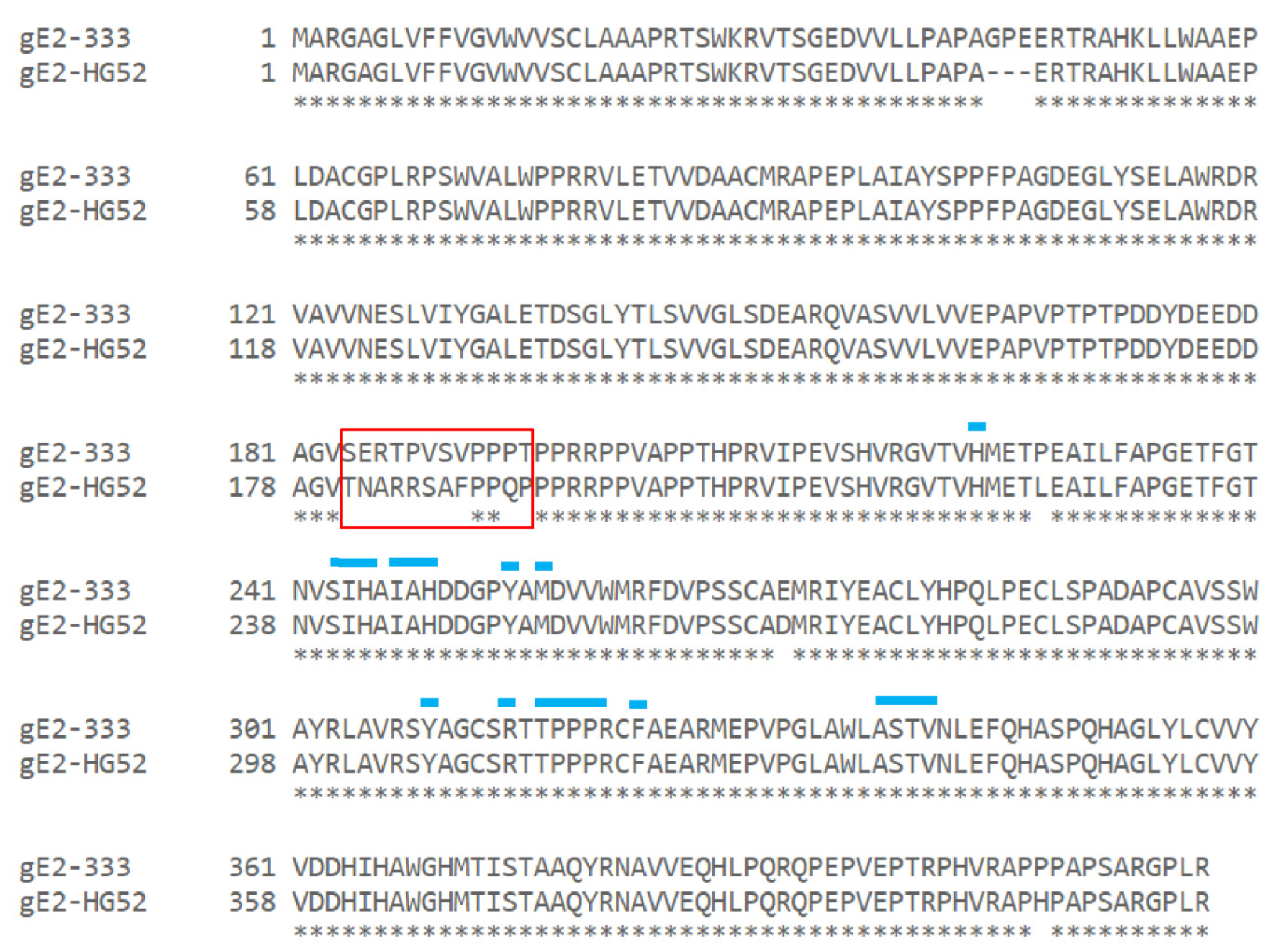
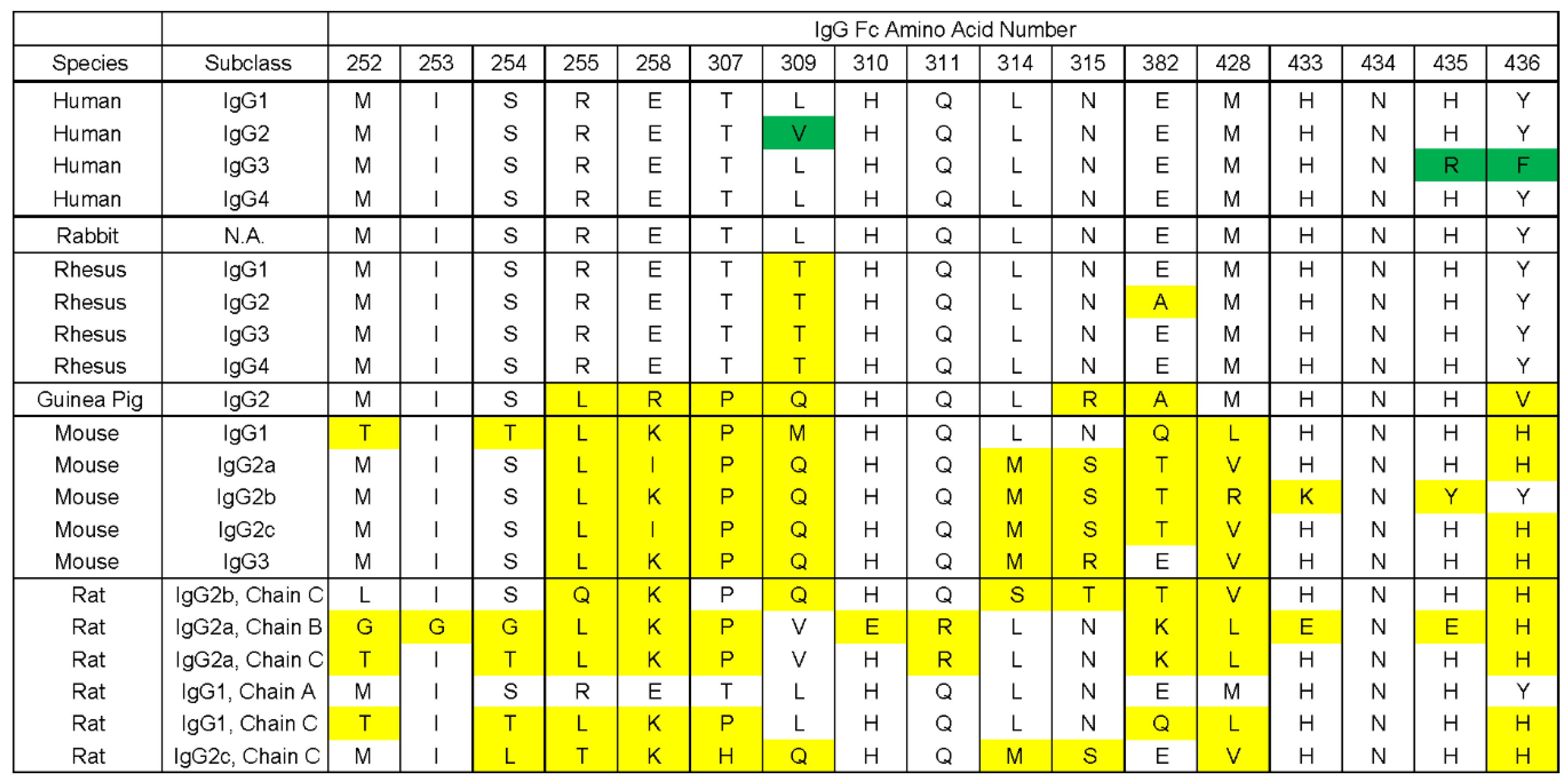
3.4. gE-2 Association with IgG Fc Is Modulated by the Length of Truncated gE, HSV Strain, and Species of IgG
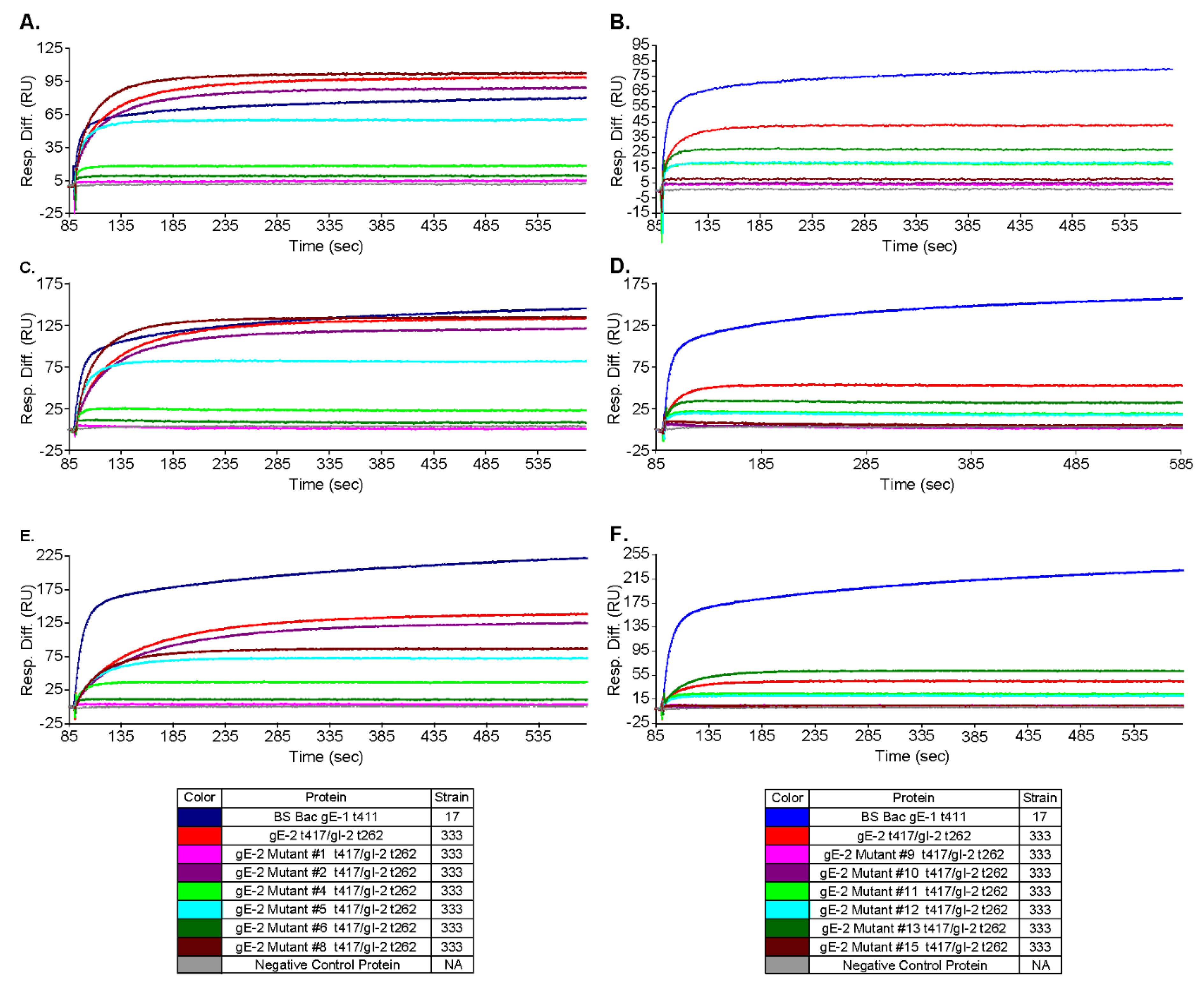
3.5. Mutations within the Putative gE-2:IgG Fc Interface Can Disrupt the Ability of gE-2 to Bind IgG Fc
3.6. Antibodies Elicited by gE-2/gI-2 Heterodimers Deficient in Binding IgG Fc Neutralize HSV-2 Virus Better than Those Elicited by Wildtype gE-2/gI-2 Heterodimers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnston, C.; Gottlieb, S.L.; Wald, A. Status of vaccine research and development of vaccines for herpes simplex virus. Vaccine 2016, 34, 2948–2952. [Google Scholar] [CrossRef] [PubMed]
- Baucke, R.B.; Spear, P.G. Membrane proteins specified by herpes simplex viruses. V. Identification of an Fc-binding glycoprotein. J. Virol. 1979, 32, 779–789. [Google Scholar] [CrossRef]
- Dingwell, K.S.; Johnson, D.C. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 1998, 72, 8933–8942. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zumbrun, E.E.; Huang, J.; Si, H.; Makaroun, L.; Friedman, H.M. Herpes simplex virus type 2 glycoprotein E is required for efficient virus spread from epithelial cells to neurons and for targeting viral proteins from the neuron cell body into axons. Virology 2010, 405, 269–279. [Google Scholar] [CrossRef][Green Version]
- Frank, I.; Friedman, H.M. A novel function of the herpes simplex virus type 1 Fc receptor: Participation in bipolar bridging of antiviral immunoglobulin G. J. Virol. 1989, 63, 4479–4488. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.C.; Feenstra, V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J. Virol. 1987, 61, 2208–2216. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.C.; Frame, M.C.; Ligas, M.W.; Cross, A.M.; Stow, N.D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 1988, 62, 1347–1354. [Google Scholar] [CrossRef]
- Dubin, G.; Socolof, E.; Frank, I.; Friedman, H.M. Herpes simplex virus type 1 Fc receptor protects infected cells from antibody-dependent cellular cytotoxicity. J. Virol. 1991, 65, 7046–7050. [Google Scholar] [CrossRef]
- Brideau, A.D.; Enquist, L.W.; Tirabassi, R.S. The role of virion membrane protein endocytosis in the herpesvirus life cycle. J. Clin. Virol. 2000, 17, 69–82. [Google Scholar] [CrossRef]
- McMillan, T.N.; Johnson, D.C. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 2001, 75, 1928–1940. [Google Scholar] [CrossRef]
- Johnson, D.C.; Webb, M.; Wisner, T.W.; Brunetti, C. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 2001, 75, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Polcicova, K.; Goldsmith, K.; Rainish, B.L.; Wisner, T.W.; Johnson, D.C. The extracellular domain of herpes simplex virus gE is indispensable for efficient cell-to-cell spread: Evidence for gE/gI receptors. J. Virol. 2005, 79, 11990–12001. [Google Scholar] [CrossRef] [PubMed]
- Sprague, E.R.; Martin, W.L.; Bjorkman, P.J. pH dependence and stoichiometry of binding to the Fc region of IgG by the herpes simplex virus Fc receptor gE-gI. J. Biol. Chem. 2004, 279, 14184–14193. [Google Scholar] [CrossRef] [PubMed]
- Sprague, E.R.; Wang, C.; Baker, D.; Bjorkman, P.J. Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging. PLoS Biol. 2006, 4, e148. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Dubin, G.; Basu, M.; Nguyen, V.; Friedman, H.M. Characterization of regions of herpes simplex virus type 1 glycoprotein E involved in binding the Fc domain of monomeric IgG and in forming a complex with glycoprotein I. J. Immunol. 1995, 154, 260–267. [Google Scholar]
- Dubin, G.; Frank, I.; Friedman, H.M. Herpes simplex virus type 1 encodes two Fc receptors which have different binding characteristics for monomeric immunoglobulin G (IgG) and IgG complexes. J. Virol. 1990, 64, 2725–2731. [Google Scholar] [CrossRef]
- Dubin, G.; Basu, S.; Mallory, D.L.; Basu, M.; Tal-Singer, R.; Friedman, H.M. Characterization of domains of herpes simplex virus type 1 glycoprotein E involved in Fc binding activity for immunoglobulin G aggregates. J. Virol. 1994, 68, 2478–2485. [Google Scholar] [CrossRef]
- Rizvi, S.M.; Raghavan, M. An N-terminal domain of herpes simplex virus type Ig E is capable of forming stable complexes with gI. J. Virol. 2001, 75, 11897–11901. [Google Scholar] [CrossRef]
- Johansson, P.J.; Myhre, E.B.; Blomberg, J. Specificity of Fc receptors induced by herpes simplex virus type 1: Comparison of immunoglobulin G from different animal species. J. Virol. 1985, 56, 489–494. [Google Scholar] [CrossRef]
- Chapman, T.L.; You, I.; Joseph, I.M.; Bjorkman, P.J.; Morrison, S.L.; Raghavan, M. Characterization of the interaction between the herpes simplex virus type I Fc receptor and immunoglobulin G. J. Biol. Chem. 1999, 274, 6911–6919. [Google Scholar] [CrossRef]
- Wiger, D.; Michaelsen, T.E. Binding site and subclass specificity of the herpes simplex virus type 1-induced Fc receptor. Immunology 1985, 54, 565–572. [Google Scholar] [PubMed]
- Johansson, P.J.; Hallberg, T.; Oxelius, V.A.; Grubb, A.; Blomberg, J. Human immunoglobulin class and subclass specificity of Fc receptors induced by herpes simplex virus type 1. J. Virol. 1984, 50, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Stanberry, L.R.; Spruance, S.L.; Cunningham, A.L.; Bernstein, D.I.; Mindel, A.; Sacks, S.; Tyring, S.; Aoki, F.Y.; Slaoui, M.; Denis, M.; et al. Glycoprotein-D-adjuvant vaccine to prevent genital herpes. N. Engl. J. Med. 2002, 347, 1652–1661. [Google Scholar] [CrossRef] [PubMed]
- Belshe, R.B.; Leone, P.A.; Bernstein, D.I.; Wald, A.; Levin, M.J.; Stapleton, J.T.; Gorfinkel, I.; Morrow, R.L.; Ewell, M.G.; Stokes-Riner, A.; et al. Efficacy results of a trial of a herpes simplex vaccine. N. Engl. J. Med. 2012, 366, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Corey, L.; Langenberg, A.G.; Ashley, R.; Sekulovich, R.E.; Izu, A.E.; Douglas, J.M., Jr.; Handsfield, H.H.; Warren, T.; Marr, L.; Tyring, S.; et al. Recombinant glycoprotein vaccine for the prevention of genital HSV-2 infection: Two randomized controlled trials. Chiron HSV Vaccine Study Group. JAMA 1999, 282, 331–340. [Google Scholar] [CrossRef]
- Hook, L.M.; Awasthi, S.; Dubin, J.; Flechtner, J.; Long, D.; Friedman, H.M. A trivalent gC2/gD2/gE2 vaccine for herpes simplex virus generates antibody responses that block immune evasion domains on gC2 better than natural infection. Vaccine 2019, 37, 664–669. [Google Scholar] [CrossRef]
- Awasthi, S.; Hook, L.M.; Pardi, N.; Wang, F.; Myles, A.; Cancro, M.P.; Cohen, G.H.; Weissman, D.; Friedman, H.M. Nucleoside-modified mRNA encoding HSV-2 glycoproteins C, D, and E prevents clinical and subclinical genital herpes. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Lubinski, J.M.; Shaw, C.E.; Barrett, S.M.; Cai, M.; Wang, F.; Betts, M.; Kingsley, S.; Distefano, D.J.; Balliet, J.W.; et al. Immunization with a vaccine combining herpes simplex virus 2 (HSV-2) glycoprotein C (gC) and gD subunits improves the protection of dorsal root ganglia in mice and reduces the frequency of recurrent vaginal shedding of HSV-2 DNA in guinea pigs compared to immunization with gD alone. J. Virol. 2011, 85, 10472–10486. [Google Scholar] [CrossRef] [PubMed]
- Egan, K.P.; Hook, L.M.; Naughton, A.; Pardi, N.; Awasthi, S.; Cohen, G.H.; Weissman, D.; Friedman, H.M. An HSV-2 nucleoside-modified mRNA genital herpes vaccine containing glycoproteins gC, gD, and gE protects mice against HSV-1 genital lesions and latent infection. PLoS Pathog. 2020, 16, e1008795. [Google Scholar] [CrossRef]
- Egan, K.; Hook, L.M.; Naughton, A.; Friedman, H.M.; Awasthi, S. Herpes simplex virus type 2 trivalent protein vaccine containing glycoproteins C, D and E protects guinea pigs against HSV-1 genital infection. Hum. Vaccin. Immunother. 2020, 16, 2109–2113. [Google Scholar] [CrossRef]
- Gonzalez-Quintela, A.; Alende, R.; Gude, F.; Campos, J.; Rey, J.; Meijide, L.M.; Fernandez-Merino, C.; Vidal, C. Serum levels of immunoglobulins (IgG, IgA, IgM) in a general adult population and their relationship with alcohol consumption, smoking and common metabolic abnormalities. Clin. Exp. Immunol. 2008, 151, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Kardar, G.A.; Shams, S.H.; Pourpak, Z.; Moin, M. Normal value of immunoglobulins IgA, IgG, and IgM in Iranian healthy adults, measured by nephelometry. J. Immunoass. Immunochem. 2003, 24, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Beernink, P.T.; Shaughnessy, J.; Braga, E.M.; Liu, Q.; Rice, P.A.; Ram, S.; Granoff, D.M. A meningococcal factor H binding protein mutant that eliminates factor H binding enhances protective antibody responses to vaccination. J. Immunol. 2011, 186, 3606–3614. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Arakawa, T.; Philo, J.S. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal. Biochem. 1996, 240, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, B.S.; Kerwin, B.A.; Chang, B.S.; Philo, J.S. Online size-exclusion high-performance liquid chromatography light scattering and differential refractometry methods to determine degree of polymer conjugation to proteins and protein-protein or protein-ligand association states. Anal. Biochem. 2001, 299, 136–146. [Google Scholar] [CrossRef]
- National Research Council Committee for the Update of the Guide for the Care Use of Laboratory Animals. The National Academies Collection: Reports Funded by National Institutes of Health; National Academies Press (US): Washington, DC, USA, 2011. [CrossRef]
- Zielinska, E.; Liu, D.; Wu, H.Y.; Quiroz, J.; Rappaport, R.; Yang, D.P. Development of an improved microneutralization assay for respiratory syncytial virus by automated plaque counting using imaging analysis. Virol. J. 2005, 2, 84. [Google Scholar] [CrossRef]
- Caulfield, M.J.; Shaffer, D. A computer program for the evaluation of ELISA data obtained using an automated microtiter plate absorbance reader. J. Immunol. Methods 1984, 74, 205–215. [Google Scholar] [CrossRef]
- Akhrameyeva, N.V.; Zhang, P.; Sugiyama, N.; Behar, S.M.; Yao, F. Development of a glycoprotein D-expressing dominant-negative and replication-defective herpes simplex virus 2 (HSV-2) recombinant viral vaccine against HSV-2 infection in mice. J. Virol. 2011, 85, 5036–5047. [Google Scholar] [CrossRef]
- Awasthi, S.; Huang, J.; Shaw, C.; Friedman, H.M. Blocking herpes simplex virus 2 glycoprotein E immune evasion as an approach to enhance efficacy of a trivalent subunit antigen vaccine for genital herpes. J. Virol. 2014, 88, 8421–8432. [Google Scholar] [CrossRef]
- DeLano, W.L.; Ultsch, M.H.; de Vos, A.M.; Wells, J.A. Convergent solutions to binding at a protein-protein interface. Science 2000, 287, 1279–1283. [Google Scholar] [CrossRef]
- Burton, D.R. Immunoglobulin G: Functional sites. Mol. Immunol. 1985, 22, 161–206. [Google Scholar] [CrossRef]
- Johansson, P.J.; Ota, T.; Tsuchiya, N.; Malone, C.C.; Williams, R.C., Jr. Studies of protein A and herpes simplex virus-1 induced Fc gamma-binding specificities. Different binding patterns for IgG3 from Caucasian and Oriental subjects. Immunology 1994, 83, 631–638. [Google Scholar] [PubMed]
- Jazayeri, M.H.; Pourfathollah, A.A.; Rasaee, M.J.; Porpak, Z.; Jafari, M.E. The concentration of total serum IgG and IgM in sera of healthy individuals varies at different age intervals. Biomed. Aging Pathol. 2013, 3, 241–245. [Google Scholar] [CrossRef]

| Construct | Amino Acid Number (Based on gE-2 Sequence) | Mutation Type | Obtained Protein Expression (Y/N) | ||||
|---|---|---|---|---|---|---|---|
| 337 | 338 | 339 | 340 | ||||
| Putative Surface-exposed Loop Mutations | gE-2t 417 WT | A | S | T | V | N.A. | Y |
| gE-2t 417 Mutant # 1 | AARAA | S | T | V | Insertion | Y | |
| gE-2t 417 Mutant # 2 | A | G | T | V | Substitution | Y | |
| gE-2t 417 Mutant # 3 | A | - | T | V | Deletion | N | |
| gE-2t 417 Mutant # 4 | A | S | G | V | Substitution | Y | |
| gE-2t 417 Mutant # 5 | A | S | - | V | Deletion | Y | |
| gE-2t 417 Mutant # 6 | A | G | G | V | Substitution | Y | |
| gE-2t 417 Mutant # 7 | A | - | - | V | Deletion | N | |
| gE-2t 417 Mutant # 8 | G | G | G | V | Substitution | Y | |
| gE-2t 417 Mutant # 9 | A | G | G | G | Substitution | Y | |
| 245 | 317 | 319 | |||||
| Putative Proximal IgG Fc Binding Interface Mutations | gE-2t 417 WT | H | P | P | N.A. | Y | |
| gE-2t 417 Mutant # 10 | G | P | P | Substitution | Y | ||
| gE-2t 417 Mutant # 11 | H | G | P | Substitution | Y | ||
| gE-2t 417 Mutant # 12 | H | P | G | Substitution | Y | ||
| gE-2t 417 Mutant # 13 | G | G | P | Substitution | Y | ||
| gE-2t 417 Mutant # 14 | G | P | G | Substitution | N | ||
| gE-2t 417 Mutant # 15 | H | G | G | Substitution | Y | ||
| Line # | Protein | Strain | Theoretical Molecular Weight (Da) | Apparent Molecular Weight (Da) | Apparent Molecular Weight Carbohydrate (Da) | Hydrodynamic Radius (nm) |
|---|---|---|---|---|---|---|
| 1 | Bac gE-1 t411 | 17 | 42,817 | 53,390 | N.A. | 5.556 |
| 2 | gE-1 t419 | KOS | 44,273 | 44,462 | 6847 | 4.512 |
| 3 | gE-2 t417 | 333 | 44,795 | 49,907 | 17,717 | 5.622 |
| 4 | gE-2 t-405 | 333 | 43,597 | 42,428 | 15,667 | 5.011 |
| 5 | gE-1 t419/gI-1 t269 | KOS | 72,408 | 70,073 | 28,146 | 5.957 |
| 6 | gE-2 t417/gI-2 t262 | 333 | 72,169 | 69,905 | 26,006 | 5.446 |
| 7 | gE-2 t414/gI-2 t262 | HG52 | 72,012 | 69,167 | 28,374 | 5.526 |
| 8 | gE-2 t405/gI-2 t262 | 333 | 70,971 | 67,927 | 21,985 | 5.087 |
| 9 | gE-2 t24-405/gI-2 t262 | 333 | 70,008 | 66,664 | 25,057 | 5.149 |
| 10 | gE-2 t417 Mutant #1/gI-2 t262 | 333 | 72,538 | 68,290 | 28,414 | 5.427 |
| 11 | gE-2 t417 Mutant #2/gI-2 t262 | 333 | 72,139 | 68,020 | 27,977 | 5.396 |
| 12 | gE-2 t417 Mutant #4/gI-2 t262 | 333 | 72,124 | 69,350 | 26,852 | 5.474 |
| 13 | gE-2 t417 Mutant #5/gI-2 t262 | 333 | 72,067 | 69,582 | 28,452 | 5.398 |
| 14 | gE-2 t417 Mutant #6/gI-2 t262 | 333 | 72,094 | 68,749 | 26,822 | 5.33 |
| 15 | gE-2 t417 Mutant #8/gI-2 t262 | 333 | 72,080 | 69,444 | 25,675 | 5.222 |
| 16 | gE-2 t417 Mutant #9/gI-2 t262 | 333 | 72,052 | 69,105 | 26,540 | 5.236 |
| 17 | gE-2 t417 Mutant #10/gI-2 t262 | 333 | 72,088 | 69,563 | 26,205 | 5.312 |
| 18 | gE-2 t417 Mutant #11/gI-2 t262 | 333 | 72,128 | 70,121 | 26,437 | 5.318 |
| 19 | gE-2 t417 Mutant #12/gI-2 t262 | 333 | 72,128 | 69,323 | 27,000 | 5.309 |
| 20 | gE-2 t417 Mutant #13/gI-2 t262 | 333 | 72,048 | 70,327 | 28,454 | 5.395 |
| 21 | gE-2 t417 Mutant #15/gI-2 t262 | 333 | 72,088 | 69,574 | 26,905 | 5.336 |
| Line # | Protein | Virus Strain | Expression System | RU Post Association (RU Post Dissociation) | ||||
|---|---|---|---|---|---|---|---|---|
| Human IgG Fc | Human IgG | Rabbit IgG | Mouse IgG | Guinea Pig IgG | ||||
| 1 | Bac gE-1 t411 | 17 | Insect | 77 (12) | 147 (32) | 231 (75) | 4 (−3.1) | 32 (3.3) |
| 2 | gE-1 t419 | KOS | Mam. | 69 (1.2) | 127 (−1.9) | 188 (0.6) | 4 (−4.0) | 27 (−2.4) |
| 3 | gE-1 t419/gI-1 t269 | KOS | Mam. | 216 (137) | 232 (189) | 195 (170) | 11 (−2.3) | 19 (0.8) |
| 4 | gE-2 t417 | 333 | Mam. | 5 (0.2) | 4 (−2.7) | 11 (−2.7) | 10 (−2.7) | 5 (−0.7) |
| 5 | gE-2 t405 | 2.12 | Mam. | 5 (0.2) | 8 (−1.7) | 6 (−1.8) | 4 (−4.0) | 5 (−1.4) |
| 6 | gE-2 t24-405 | 333 | Mam. | 8 (1.2) | 7 (−2.2) | 6 (−1.2) | 4 (−3.8) | 5 (−0.03) |
| 7 | gE-2 t417/gI-2 t262 | 333 | Mam. | 41 (0.8) | 52 (−2.1) | 46 (−1.5) | 10 (−2.0) | 9 (−0.07) |
| 8 | gE-2 t405/gI-2 t262 | 333 | Mam. | 100 (7.7) | 142 (14) | 165 (34) | 8 (−3.5) | 9 (−1.5) |
| 9 | gE-2 t24-405/gI-2 t262 | 333 | Mam. | 95 (6.6) | 136 (12) | 142 (25) | 6 (−4.2) | 8 (−1.9) |
| 10 | gE-2 t414/gI-2 t262 | HG52 | Mam. | 53 (0.6) | 79 (−0.4) | 75 (−0.6) | 5 (−3.8) | 4 (−2.1) |
| 11 | Negative Control Protein | N.A. | Mam. | 4 (−1.3) | 2 (−2.2) | 11 (−2.8) | 4 (−3.5) | 2 (−1.8) |
| Protein | Virus Strain | Expression System | RU Post Association | |||
|---|---|---|---|---|---|---|
| Human IgG Fc | Human IgG | Rabbit IgG | ||||
| Run 1 | gE-1 t411 | 17 | Insect | 80 | 145 | 221 |
| gE-2t 417/gI-2 t262 | 333 | Mam. | 98 | 133 | 138 | |
| gE-2 t417 Mutant # 1/ gI-2 t262 | 333 | Mam. | 5 | 1 | 3 | |
| gE-2 t417 Mutant # 2/ gI-2 t262 | 333 | Mam. | 89 | 121 | 125 | |
| gE-2 t417 Mutant # 4/gI-2 t262 | 333 | Mam. | 18 | 23 | 37 | |
| gE-2 t417 Mutant # 5/gI-2 t262 | 333 | Mam. | 60 | 82 | 72 | |
| gE-2 t417 Mutant # 6/gI-2 t262 | 333 | Mam. | 9 | 8 | 11 | |
| gE-2 t417 Mutant # 8/gI-2 t262 | 333 | Mam. | 102 | 134 | 87 | |
| Negative Control Protein | N.A. | Mam. | 2 | 4 | 1 | |
| Run 2 | gE-1 t411 | 17 | Insect | 80 | 157 | 228 |
| gE-2t 417/gI-2t 262 | 333 | Mam. | 43 | 53 | 45 | |
| gE-2 t417 Mutant # 9/gI-2 t262 | 333 | Mam. | 4 | 2 | 4 | |
| gE-2 t417 Mutant # 10/gI-2 t262 | 333 | Mam. | 18 | 19 | 23 | |
| gE-2 t417 Mutant # 11/gI-2 t262 | 333 | Mam. | 19 | 18 | 21 | |
| gE-2 t417 Mutant # 12/gI-2 t262 | 333 | Mam. | 27 | 32 | 62 | |
| gE-2 t417 Mutant # 13/gI-2 t262 | 333 | Mam. | 5 | 3 | 5 | |
| gE-2 t417 Mutant # 15/gI-2 t262 | 333 | Mam. | 8 | 5 | 4 | |
| Negative Control Protein | N.A. | Mam. | 1 | 3 | 2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galli, J.D.; Horton, M.; Durr, E.; Heidecker, G.J.; Freed, D.; Fridman, A.; Wang, D.; Zhang, L. Evaluation of HSV-2 gE Binding to IgG-Fc and Application for Vaccine Development. Vaccines 2022, 10, 184. https://doi.org/10.3390/vaccines10020184
Galli JD, Horton M, Durr E, Heidecker GJ, Freed D, Fridman A, Wang D, Zhang L. Evaluation of HSV-2 gE Binding to IgG-Fc and Application for Vaccine Development. Vaccines. 2022; 10(2):184. https://doi.org/10.3390/vaccines10020184
Chicago/Turabian StyleGalli, Jennifer D., Melanie Horton, Eberhard Durr, Gwendolyn J. Heidecker, Daniel Freed, Arthur Fridman, Dai Wang, and Lan Zhang. 2022. "Evaluation of HSV-2 gE Binding to IgG-Fc and Application for Vaccine Development" Vaccines 10, no. 2: 184. https://doi.org/10.3390/vaccines10020184
APA StyleGalli, J. D., Horton, M., Durr, E., Heidecker, G. J., Freed, D., Fridman, A., Wang, D., & Zhang, L. (2022). Evaluation of HSV-2 gE Binding to IgG-Fc and Application for Vaccine Development. Vaccines, 10(2), 184. https://doi.org/10.3390/vaccines10020184






