Proinflammatory Innate Cytokines and Distinct Metabolomic Signatures Shape the T Cell Response in Active COVID-19
Abstract
1. Introduction
2. Materials and Methods
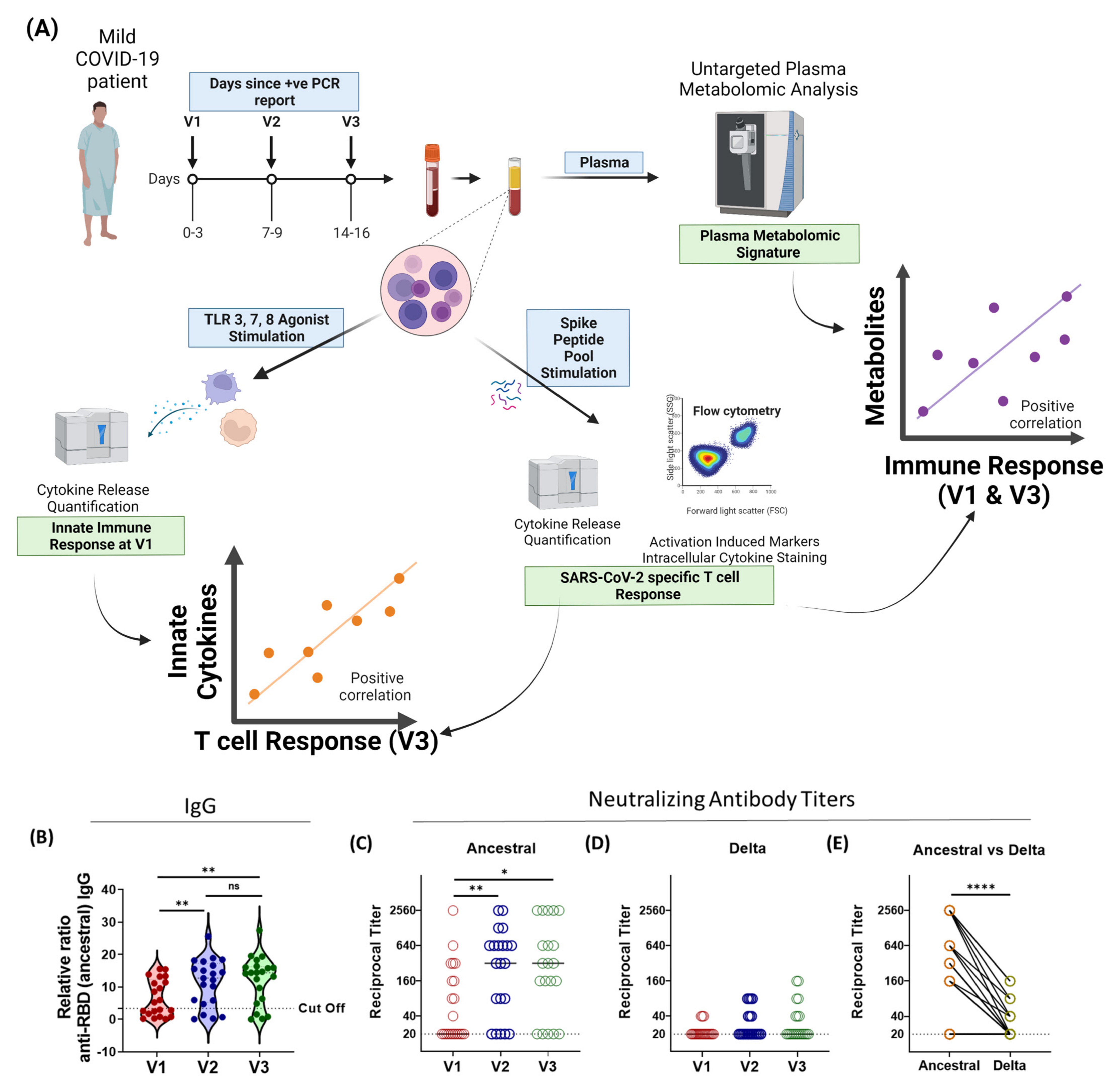
2.1. Study Plan
2.2. Human Ethics
2.3. Peripheral Blood Mononuclear Cells (PBMC) Isolation
2.4. THSTI In-House RBD IgG ELISA
2.5. Virus Neutralization Assay
2.6. Peptide Pool
2.7. SARS-CoV-2-Specific T Cell Response
2.8. Flow Cytometry
2.9. Metabolomic Analysis
2.10. In Vitro Stimulation of PBMCs with TLR Agonists
2.11. Analysis of Cytokine Secretion by Luminex
2.12. Data Representation and Statistical Analysis
3. Results
3.1. Humoral Immune Responses against SARS-CoV-2 Variants Is Broadly Inadequate in Active-COVID-19 Patients
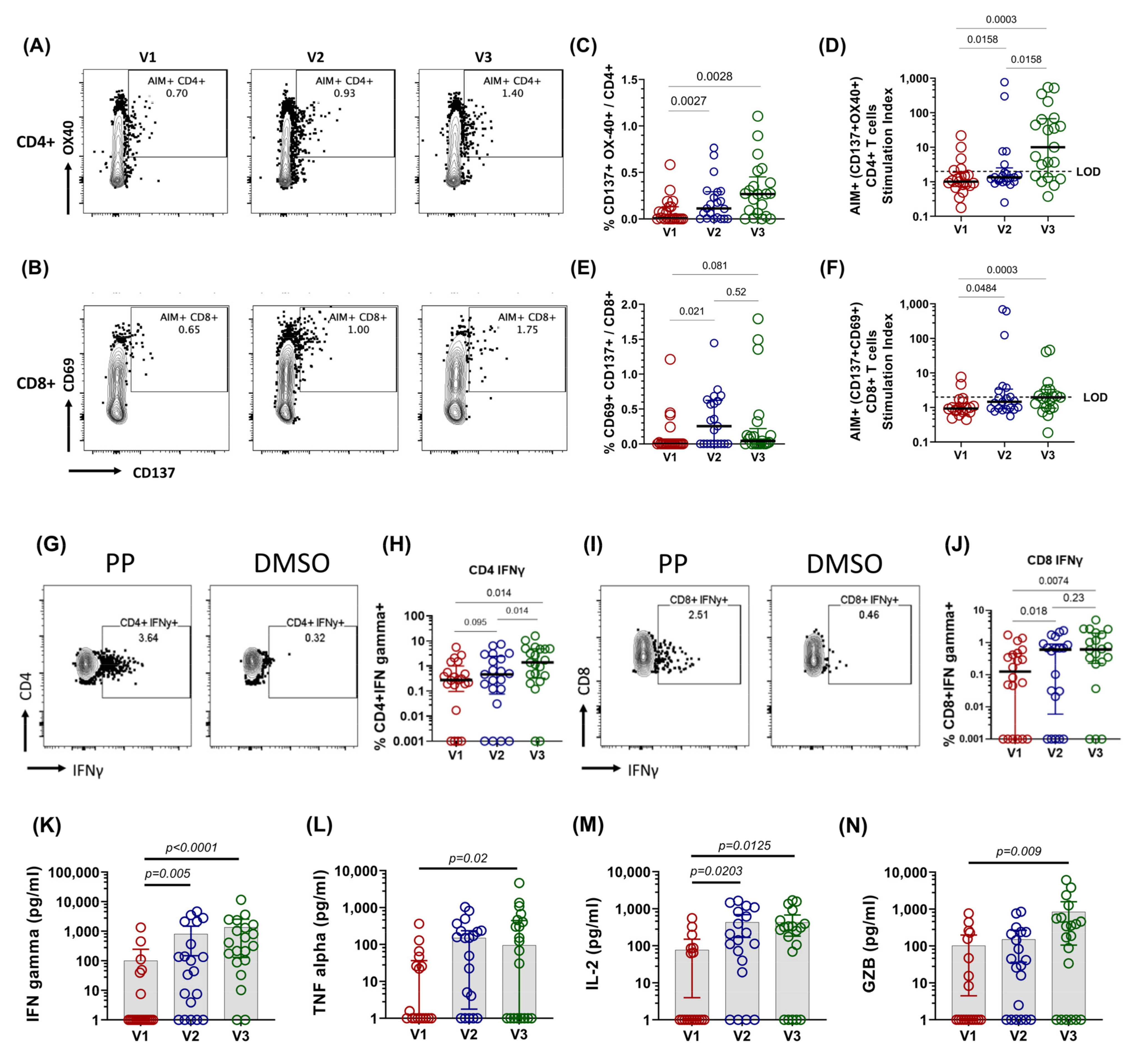
3.2. Spike-Specific T Cell Response in COVID-19 Patients
3.3. Cross-Reactive Spike-Specific T Cell Response against Delta and Omicron Variants Is Largely Preserved in Active COVID-19 Patients
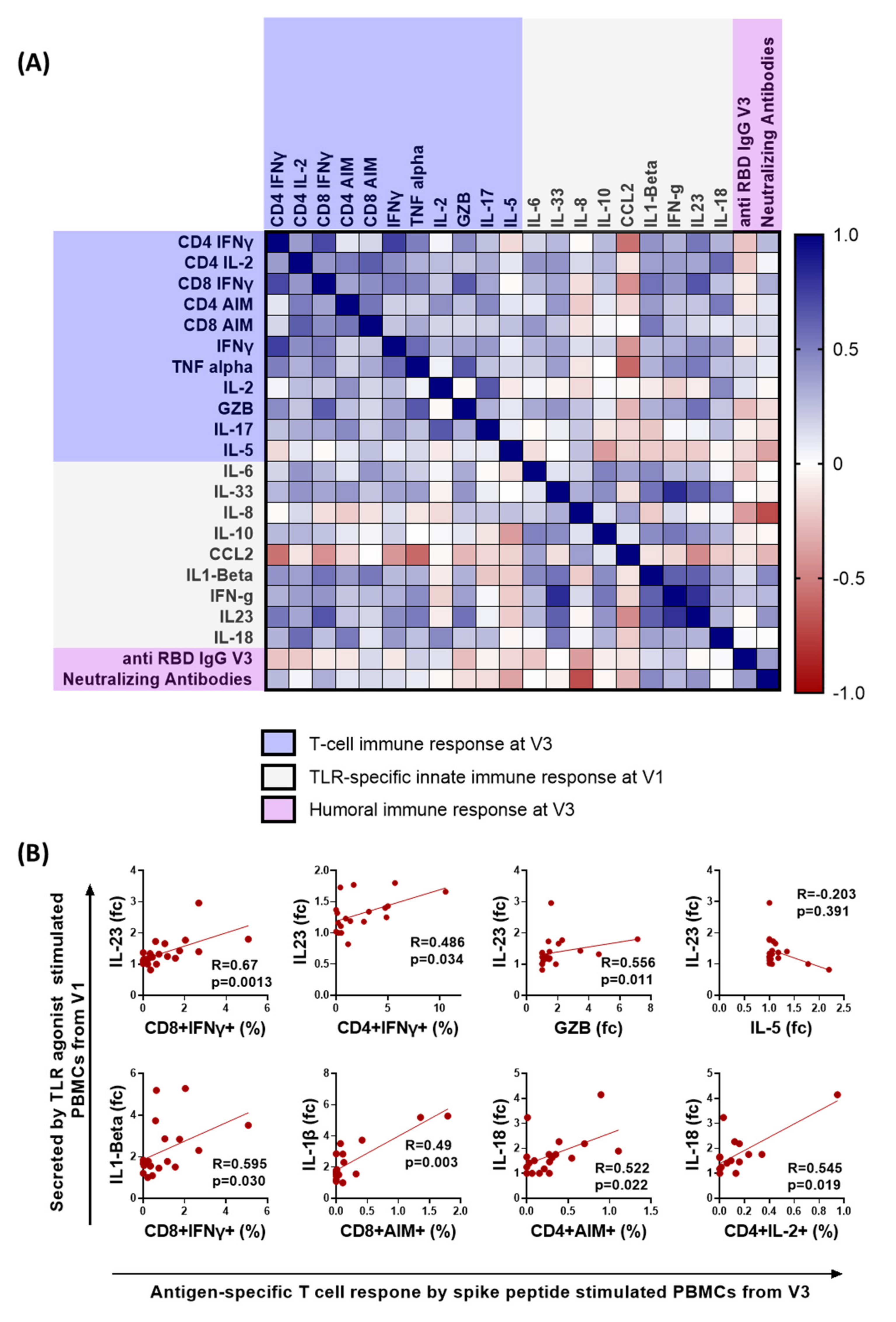
3.4. Early Proinflammatory Innate Cytokine Response Correlates with a Robust T Cell Response
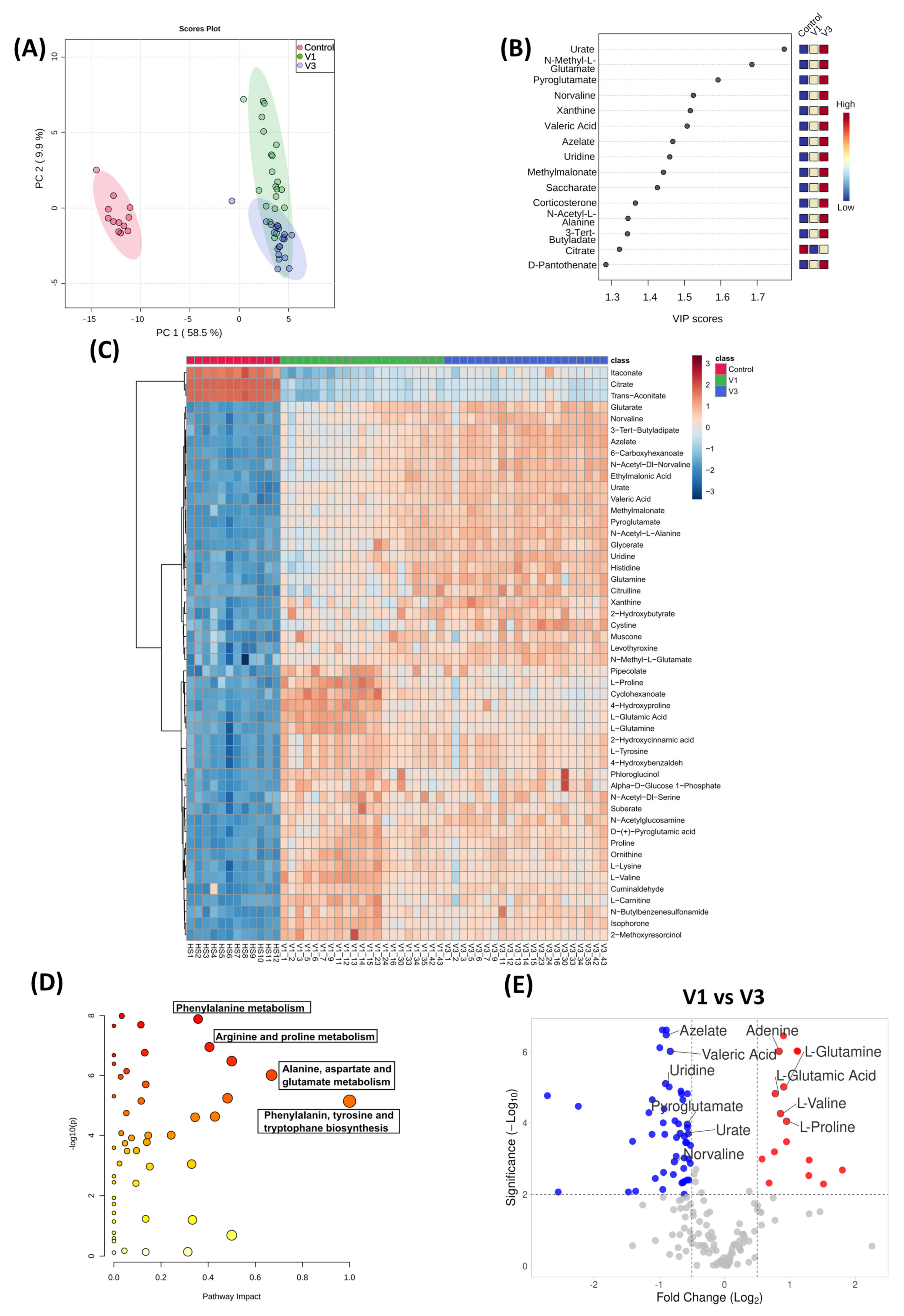
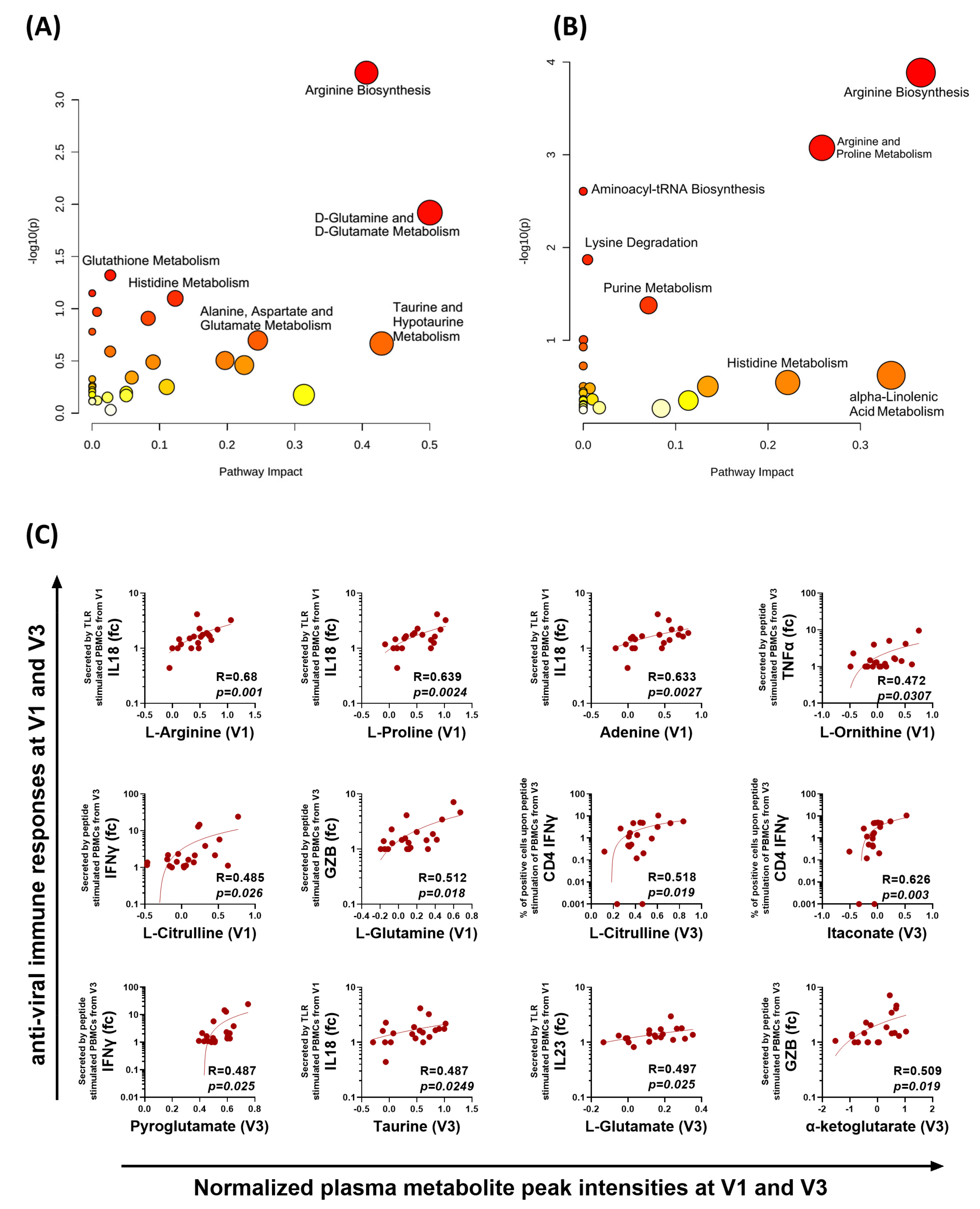
3.5. Distinct Metabolomic Alterations Characterize Acute COVID-19 Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stephens, D.S.; McElrath, M.J. COVID-19 and the Path to Immunity. JAMA 2020, 324, 1279–1281. [Google Scholar] [CrossRef] [PubMed]
- Sherina, N.; Piralla, A.; Du, L.; Wan, H.; Kumagai-Braesch, M.; Andréll, J.; Braesch-Andersen, S.; Cassaniti, I.; Percivalle, E.; Sarasini, A.; et al. Persistence of SARS-CoV-2-specific B and T cell responses in convalescent COVID-19 patients 6–8 months after the infection. Med 2021, 2, 281–295.e4. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Sidney, J.; Vita, R.; Peters, B.; Crotty, S.; Weiskopf, D.; Sette, A. SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe 2021, 29, 1076–1092. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cai, C.; Grifoni, A.; Müller, T.R.; Niessl, J.; Olofsson, A.; Humbert, M.; Hansson, L.; Österborg, A.; Bergman, P.; et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022, 28, 472–476. [Google Scholar] [CrossRef]
- Keeton, R.; Tincho, M.B.; Ngomti, A.; Baguma, R.; Benede, N.; Suzuki, A.; Khan, K.; Cele, S.; Bernstein, M.; Karim, F.; et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 2022, 603, 488–492. [Google Scholar] [CrossRef]
- Tarke, A.; Coelho, C.H.; Zhang, Z.; Dan, J.M.; Yu, E.D.; Methot, N.; Bloom, N.I.; Goodwin, B.; Phillips, E.; Mallal, S.; et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 2022, 185, 847–859.e11. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Schultze, J.L.; Aschenbrenner, A.C. COVID-19 and the human innate immune system. Cell 2021, 184, 1671–1692. [Google Scholar] [CrossRef]
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K.; et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020, 369, eabc8511. [Google Scholar] [CrossRef]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012.e9. [Google Scholar] [CrossRef]
- Dalal, R.; Sadhu, S.; Awasthi, A. Chapter 6—Role of Th17 cell in tissue inflammation and organ-specific autoimmunity. In Translational Autoimmunity; Rezaei, N., Ed.; Academic Press: Cambridge, MA, USA, 2022; Volume 1, pp. 93–121. [Google Scholar]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tak-Yin Tsang, O.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Capelle, C.M.; Ciré, S.; Domingues, O.; Ernens, I.; Hedin, F.; Fischer, A.; Snoeck, C.J.; Ammerlaan, W.; Konstantinou, M.; Grzyb, K.; et al. Combinatorial analysis reveals highly coordinated early-stage immune reactions that predict later antiviral immune responses in mild COVID-19 patients. Cell Rep. Med. 2022, 4, 100600. [Google Scholar] [CrossRef]
- Chandran, A.; Rosenheim, J.; Nageswaran, G.; Swadling, L.; Pollara, G.; Gupta, R.K.; Burton, A.R.; Guerra-Assunção, J.A.; Woolston, A.; Ronel, T.; et al. Rapid synchronous type 1 IFN and virus-specific T cell responses characterize first wave non-severe SARS-CoV-2 infections. Cell Rep. Med. 2022, 3, 100557. [Google Scholar] [CrossRef]
- Lee, J.W.; Su, Y.; Baloni, P.; Chen, D.; Pavlovitch-Bedzyk, A.J.; Yuan, D.; Duvvuri, V.R.; Ng, R.H.; Choi, J.; Xie, J.; et al. Integrated analysis of plasma and single immune cells uncovers metabolic changes in individuals with COVID-19. Nat. Biotechnol. 2022, 40, 110–120. [Google Scholar] [CrossRef]
- Rendeiro, A.F.; Vorkas, C.K.; Krumsiek, J.; Singh, H.K.; Kapadia, S.N.; Cappelli, L.V.; Cacciapuoti, M.T.; Inghirami, G.; Elemento, O.; Salvatore, M. Metabolic and Immune Markers for Precise Monitoring of COVID-19 Severity and Treatment. Front. Immunol. 2022, 12, 809937. [Google Scholar] [CrossRef]
- O’Carroll, S.M.; O’Neill, L.A.J. Targeting immunometabolism to treat COVID-19. Immunother. Adv. 2021, 1, ltab013. [Google Scholar] [CrossRef]
- Khwatenge, C.N.; Pate, M.; Miller, L.C.; Sang, Y. Immunometabolic Dysregulation at the Intersection of Obesity and COVID-19. Front. Immunol. 2021, 12, 732913. [Google Scholar] [CrossRef] [PubMed]
- Siska, P.J.; Decking, S.-M.; Babl, N.; Matos, C.; Bruss, C.; Singer, K.; Klitzke, J.; Schön, M.; Simeth, J.; Köstler, J.; et al. Metabolic imbalance of T cells in COVID-19 is hallmarked by basigin and mitigated by dexamethasone. J. Clin. Investig. 2021, 131, e148225. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, S.; Thiruvengadam, R.; Chattopadhyay, S.; Mehdi, F.; Kshetrapal, P.; Shrivastava, T.; Desiraju, B.K.; Batra, G.; Kang, G.; Bhatnagar, S. Comparative evaluation of SARS-CoV-2 IgG assays in India. J. Clin. Virol. 2020, 131, 104609. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, R.; Awasthi, A.; Medigeshi, G.; Bhattacharya, S.; Mani, S.; Sivasubbu, S.; Shrivastava, T.; Samal, S.; Rathna Murugesan, D.; Koundinya Desiraju, B.; et al. Effectiveness of ChAdOx1 nCoV-19 vaccine against SARS-CoV-2 infection during the delta (B.1.617.2) variant surge in India: A test-negative, case-control study and a mechanistic study of post-vaccination immune responses. Lancet Infect. Dis. 2021, 22, 473–482. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Gallart-Ayala, H.; Reinke, S.N.; Mathon, C.; Blankley, R.; Chaleckis, R.; Wheelock, C.E. Development of a Liquid Chromatography–High Resolution Mass Spectrometry Metabolomics Method with High Specificity for Metabolite Identification Using All Ion Fragmentation Acquisition. Anal. Chem. 2017, 89, 7933–7942. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, Y.; Sevak, J.K.; Kumar, S.; Kumar, N.; Gopinath, S.D. Metabolomic analysis of primary human skeletal muscle cells during myogenic progression. Sci. Rep. 2020, 10, 11824. [Google Scholar] [CrossRef]
- Sadhu, S.; Rizvi, Z.A.; Pandey, R.P.; Dalal, R.; Rathore, D.K.; Kumar, B.; Pandey, M.; Kumar, Y.; Goel, R.; Maiti, T.K.; et al. Gefitinib Results in Robust Host-Directed Immunity Against Salmonella Infection Through Proteo-Metabolomic Reprogramming. Front. Immunol. 2021, 12, 648710. [Google Scholar] [CrossRef]
- Rizvi, Z.A.; Dalal, R.; Sadhu, S.; Kumar, Y.; Kumar, S.; Gupta, S.K.; Tripathy, M.R.; Rathore, D.K.; Awasthi, A. High-salt diet mediates interplay between NK cells and gut microbiota to induce potent tumor immunity. Sci. Adv. 2021, 7, eabg5016. [Google Scholar] [CrossRef] [PubMed]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Isho, B.; Abe, K.T.; Zuo, M.; Jamal, A.J.; Rathod, B.; Wang, J.H.; Li, Z.; Chao, G.; Rojas, O.L.; Bang, Y.M.; et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020, 5, eabb551. [Google Scholar] [CrossRef]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Kanneganti, T.-D. Innate immunity: The first line of defense against SARS-CoV-2. Nat. Immunol. 2022, 23, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Krieg, A.M.; Vollmer, J. Toll-like receptors 7, 8, and 9: Linking innate immunity to autoimmunity. Immunol. Rev. 2007, 220, 251–269. [Google Scholar] [CrossRef]
- Danlos, F.-X.; Grajeda-Iglesias, C.; Durand, S.; Sauvat, A.; Roumier, M.; Cantin, D.; Colomba, E.; Rohmer, J.; Pommeret, F.; Baciarello, G.; et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 2021, 12, 258. [Google Scholar] [CrossRef]
- Jia, H.; Liu, C.; Li, D.; Huang, Q.; Liu, D.; Zhang, Y.; Ye, C.; Zhou, D.; Wang, Y.; Tan, Y.; et al. Metabolomic analyses reveals new stage-specific features of the COVID-19. Eur. Respir. J. 2021, 59, 2100284. [Google Scholar] [CrossRef]
- Páez-Franco, J.C.; Torres-Ruiz, J.; Sosa-Hernández, V.A.; Cervantes-Díaz, R.; Romero-Ramírez, S.; Pérez-Fragoso, A.; Meza-Sánchez, D.E.; Germán-Acacio, J.M.; Maravillas-Montero, J.L.; Mejía-Domínguez, N.R.; et al. Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients. Sci. Rep. 2021, 11, 6350. [Google Scholar] [CrossRef]
- Sindelar, M.; Stancliffe, E.; Schwaiger-Haber, M.; Anbukumar, D.S.; Adkins-Travis, K.; Goss, C.W.; O’Halloran, J.A.; Mudd, P.A.; Liu, W.-C.; Albrecht, R.A.; et al. Longitudinal metabolomics of human plasma reveals prognostic markers of COVID-19 disease severity. Cell Rep. Med. 2021, 2, 100369. [Google Scholar] [CrossRef]
- Flor, L.S.; Friedman, J.; Spencer, C.N.; Cagney, J.; Arrieta, A.; Herbert, M.E.; Stein, C.; Mullany, E.C.; Hon, J.; Patwardhan, V.; et al. Quantifying the effects of the COVID-19 pandemic on gender equality on health, social, and economic indicators: A comprehensive review of data from March, 2020, to September, 2021. Lancet 2022, 399, 2381–2397. [Google Scholar] [CrossRef]
- Wilk, A.J.; Lee, M.J.; Wei, B.; Parks, B.; Pi, R.; Martínez-Colón, G.J.; Ranganath, T.; Zhao, N.Q.; Taylor, S.; Becker, W.; et al. Multi-omic profiling reveals widespread dysregulation of innate immunity and hematopoiesis in COVID-19. J. Exp. Med. 2021, 218, e20210582. [Google Scholar] [CrossRef]
- Stephenson, E.; Reynolds, G.; Botting, R.A.; Calero-Nieto, F.J.; Morgan, M.D.; Tuong, Z.K.; Bach, K.; Sungnak, W.; Worlock, K.B.; Yoshida, M.; et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021, 27, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Barh, D.; Tiwari, S.; Andrade, B.S.; Weener, M.E.; Góes-Neto, A.; Azevedo, V.; Ghosh, P.; Blum, K.; Ganguly, N.K. A novel multi-omics-based highly accurate prediction of symptoms, comorbid conditions, and possible long-term complications of COVID-19. Mol. Omics 2021, 17, 317–337. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021, 34, 108728. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Wang, Z.; Pan, H.; Jiang, B. Type I IFN deficiency: An immunological characteristic of severe COVID-19 patients. Signal Transduct. Target. Ther. 2020, 5, 198. [Google Scholar] [CrossRef]
- Thiruvengadam, R.; Binayke, A.; Awasthi, A. SARS-CoV-2 delta variant: A persistent threat to the effectiveness of vaccines. Lancet Infect. Dis. 2022, 22. [Google Scholar] [CrossRef]
- Nyberg, T.; Ferguson, N.M.; Nash, S.G.; Webster, H.H.; Flaxman, S.; Andrews, N.; Hinsley, W.; Bernal, J.L.; Kall, M.; Bhatt, S.; et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: A cohort study. Lancet 2022, 399, 1303–1312. [Google Scholar] [CrossRef]
- Liu, J.; Chandrashekar, A.; Sellers, D.; Barrett, J.; Jacob-Dolan, C.; Lifton, M.; McMahan, K.; Sciacca, M.; VanWyk, H.; Wu, C.; et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature 2022, 603, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Tennøe, S.; Gheorghe, M.; Stratford, R.; Clancy, T. The T cell epitope landscape of SARS-CoV-2 variants of concern. Vaccines 2022, 10, 1123. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Woo, H.G. Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies. Front. Immunol. 2022, 12, 830527. [Google Scholar] [CrossRef] [PubMed]
- Gil-Etayo, F.J.; Garcinuño, S.; Utrero-Rico, A.; Cabrera-Marante, O.; Arroyo-Sanchez, D.; Mancebo, E.; Pleguezuelo, D.E.; Rodríguez-Frías, E.; Allende, L.M.; Morales-Pérez, P. An Early Th1 Response Is a Key Factor for a Favorable COVID-19 Evolution. Biomedicines 2022, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Janssen, N.A.F.; Grondman, I.; de Nooijer, A.H.; Boahen, C.K.; Koeken, V.; Matzaraki, V.; Kumar, V.; He, X.; Kox, M.; Koenen, H.; et al. Dysregulated Innate and Adaptive Immune Responses Discriminate Disease Severity in COVID-19. J. Infect. Dis. 2021, 223, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Masood, K.I.; Yameen, M.; Ashraf, J.; Shahid, S.; Mahmood, S.F.; Nasir, A.; Nasir, N.; Jamil, B.; Ghanchi, N.K.; Khanum, I.; et al. Upregulated type I interferon responses in asymptomatic COVID-19 infection are associated with improved clinical outcome. Sci. Rep. 2021, 11, 22958. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef] [PubMed]
- Alavi Darazam, I.; Shokouhi, S.; Pourhoseingholi, M.A.; Naghibi Irvani, S.S.; Mokhtari, M.; Shabani, M.; Amirdosara, M.; Torabinavid, P.; Golmohammadi, M.; Hashemi, S.; et al. Role of interferon therapy in severe COVID-19: The COVIFERON randomized controlled trial. Sci. Rep. 2021, 11, 8059. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.-H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Nakanishi, K. Unique action of interleukin-18 on T cells and other immune cells. Front. Immunol. 2018, 9, 763. [Google Scholar] [CrossRef]
- Lee, Y.; Awasthi, A.; Yosef, N.; Quintana, F.J.; Xiao, S.; Peters, A.; Wu, C.; Kleinewietfeld, M.; Kunder, S.; Hafler, D.A.; et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 2012, 13, 991–999. [Google Scholar] [CrossRef]
- Rizvi, Z.A.; Dalal, R.; Sadhu, S.; Binayke, A.; Dandotiya, J.; Kumar, Y.; Shrivastava, T.; Gupta, S.K.; Aggarwal, S.; Tripathy, M.R.; et al. Golden Syrian hamster as a model to study cardiovascular complications associated with SARS-CoV-2 infection. eLife 2022, 11, e73522. [Google Scholar] [CrossRef]
- Pawlak, M.; DeTomaso, D.; Schnell, A.; Meyer zu Horste, G.; Lee, Y.; Nyman, J.; Dionne, D.; Regan, B.M.L.; Singh, V.; Delorey, T.; et al. Induction of a colitogenic phenotype in Th1-like cells depends on interleukin-23 receptor signaling. Immunity 2022, 55, 1663–1679. [Google Scholar] [CrossRef]
- DuPage, M.; Bluestone, J.A. Harnessing the plasticity of CD4+ T cells to treat immune-mediated disease. Nat. Rev. Immunol. 2016, 16, 149–163. [Google Scholar] [CrossRef]
- Lau, S.K.; Lau, C.C.; Chan, K.-H.; Li, C.P.; Chen, H.; Jin, D.-Y.; Chan, J.F.; Woo, P.C.; Yuen, K.-Y. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: Implications for pathogenesis and treatment. J. Gen. Virol. 2013, 94, 2679–2690. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef]
- Wculek, S.K.; Khouili, S.C.; Priego, E.; Heras-Murillo, I.; Sancho, D. Metabolic Control of Dendritic Cell Functions: Digesting Information. Front. Immunol. 2019, 10, 775. [Google Scholar] [CrossRef] [PubMed]
- Ansone, L.; Briviba, M.; Silamikelis, I.; Terentjeva, A.; Perkons, I.; Birzniece, L.; Rovite, V.; Rozentale, B.; Viksna, L.; Kolesova, O.; et al. Amino Acid Metabolism is Significantly Altered at the Time of Admission in Hospital for Severe COVID-19 Patients: Findings from Longitudinal Targeted Metabolomics Analysis. Microbiol. Spectr. 2021, 9, e00338-00321. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.S.; Ansari, M.; Wagner, K.I.; Jarosch, S.; Huang, Y.; Mayr, C.H.; Strunz, M.; Lang, N.J.; D’Ippolito, E.; Hammel, M.; et al. Single-cell RNA sequencing reveals ex vivo signatures of SARS-CoV-2-reactive T cells through ‘reverse phenotyping’. Nat. Commun. 2021, 12, 4515. [Google Scholar] [CrossRef]
- Meckiff, B.J.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Kusnadi, A.; Simon, H.; Eschweiler, S.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; et al. Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4(+) T Cells in COVID-19. Cell 2020, 183, 1340–1353. [Google Scholar] [CrossRef]
- Bronte, V.; Zanovello, P. Regulation of immune responses by L-arginine metabolism. Nat. Rev. Immunol. 2005, 5, 641–654. [Google Scholar] [CrossRef]
- Kishton Rigel, J.; Sukumar, M.; Restifo Nicholas, P. Arginine Arms T Cells to Thrive and Survive. Cell Metab. 2016, 24, 647–648. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Roszik, J.; Grimm, E.A.; Ekmekcioglu, S. Impact of l-arginine metabolism on immune response and anticancer immunotherapy. Front. Oncol. 2018, 8, 67. [Google Scholar] [CrossRef]
- Sacchi, A.; Grassi, G.; Notari, S.; Gili, S.; Bordoni, V.; Tartaglia, E.; Casetti, R.; Cimini, E.; Mariotti, D.; Garotto, G. Expansion of myeloid derived suppressor cells contributes to platelet activation by L-arginine deprivation during SARS-CoV-2 infection. Cells 2021, 10, 2111. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, G.; Coppola, A.; Izzo, R.; Annunziata, A.; Bernardo, M.; Lombardi, A.; Trimarco, V.; Santulli, G.; Trimarco, B. Effects of adding L-arginine orally to standard therapy in patients with COVID-19: A randomized, double-blind, placebo-controlled, parallel-group trial. Results of the first interim analysis. eClinicalMedicine 2021, 40, 101125. [Google Scholar] [CrossRef] [PubMed]
- Grimes, J.M.; Khan, S.; Badeaux, M.; Rao, R.M.; Rowlinson, S.W.; Carvajal, R.D. Arginine depletion as a therapeutic approach for patients with COVID-19. Int. J. Infect. Dis. 2021, 102, 566–570. [Google Scholar] [CrossRef]
- Buck, M.D.; O’Sullivan, D.; Pearce, E.L. T cell metabolism drives immunity. J. Exp. Med. 2015, 212, 1345–1360. [Google Scholar] [CrossRef]
- Swain, A.; Bambouskova, M.; Kim, H.; Andhey, P.S.; Duncan, D.; Auclair, K.; Chubukov, V.; Simons, D.M.; Roddy, T.P.; Stewart, K.M.; et al. Comparative evaluation of itaconate and its derivatives reveals divergent inflammasome and type I interferon regulation in macrophages. Nat. Metab. 2020, 2, 594–602. [Google Scholar] [CrossRef]
- Mills, E.L.; Ryan, D.G.; Prag, H.A.; Dikovskaya, D.; Menon, D.; Zaslona, Z.; Jedrychowski, M.P.; Costa, A.S.H.; Higgins, M.; Hams, E.; et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 2018, 556, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef]
- Ye, L.; Park, J.J.; Peng, L.; Yang, Q.; Chow, R.D.; Dong, M.B.; Lam, S.Z.; Guo, J.; Tang, E.; Zhang, Y.; et al. A genome-scale gain-of-function CRISPR screen in CD8 T cells identifies proline metabolism as a means to enhance CAR-T therapy. Cell Metab 2022, 34, 595–614.e514. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Binayke, A.; Zaheer, A.; Dandotiya, J.; Gupta, S.K.; Mani, S.; Tripathy, M.R.; Madan, U.; Shrivastava, T.; Kumar, Y.; Pandey, A.K.; et al. Proinflammatory Innate Cytokines and Distinct Metabolomic Signatures Shape the T Cell Response in Active COVID-19. Vaccines 2022, 10, 1762. https://doi.org/10.3390/vaccines10101762
Binayke A, Zaheer A, Dandotiya J, Gupta SK, Mani S, Tripathy MR, Madan U, Shrivastava T, Kumar Y, Pandey AK, et al. Proinflammatory Innate Cytokines and Distinct Metabolomic Signatures Shape the T Cell Response in Active COVID-19. Vaccines. 2022; 10(10):1762. https://doi.org/10.3390/vaccines10101762
Chicago/Turabian StyleBinayke, Akshay, Aymaan Zaheer, Jyotsna Dandotiya, Sonu Kumar Gupta, Shailendra Mani, Manas Ranjan Tripathy, Upasna Madan, Tripti Shrivastava, Yashwant Kumar, Anil Kumar Pandey, and et al. 2022. "Proinflammatory Innate Cytokines and Distinct Metabolomic Signatures Shape the T Cell Response in Active COVID-19" Vaccines 10, no. 10: 1762. https://doi.org/10.3390/vaccines10101762
APA StyleBinayke, A., Zaheer, A., Dandotiya, J., Gupta, S. K., Mani, S., Tripathy, M. R., Madan, U., Shrivastava, T., Kumar, Y., Pandey, A. K., Rathore, D. K., & Awasthi, A. (2022). Proinflammatory Innate Cytokines and Distinct Metabolomic Signatures Shape the T Cell Response in Active COVID-19. Vaccines, 10(10), 1762. https://doi.org/10.3390/vaccines10101762




