Knowledge of SARS-CoV-2 Epitopes and Population HLA Types Is Important in the Design of COVID-19 Vaccines
Abstract
:1. Introduction
1.1. SARS-CoV-2 Specific T Cell Epitopes
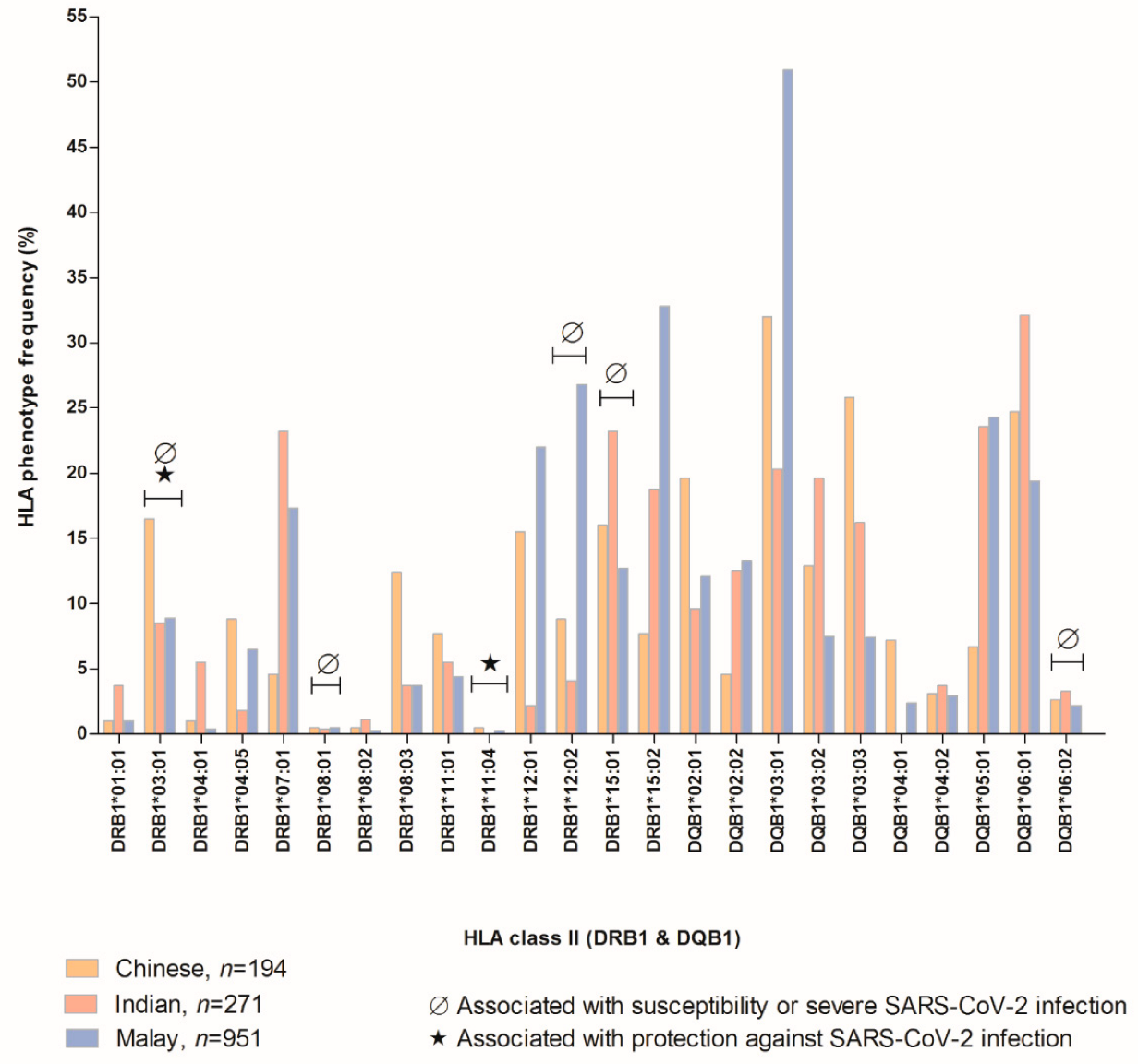
1.2. Susceptible, Severe, and Protective HLA to SARS-CoV-2 Virus Based on Malaysian Population Allele Frequencies
1.3. Host and Viral Factors in SARS-CoV-2 Infection
1.4. Current Vaccine Options for SARS-CoV-2 Virus
2. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization Coronavirus Disease (COVID-19) Dashboard. Available online: covid19.who.int (accessed on 12 October 2020).
- Sangeeta, D.; Deepjyoti, K. Wuhan Coronavirus: A fast-emerging global threat. Int. J. Health Res. Med. Leg. Pract. 2020, 6, 79–82. [Google Scholar] [CrossRef]
- Heisbourg, F. From Wuhan to the world: How the pandemic will Reshape Geopolitics. Survival 2020, 62, 7–24. [Google Scholar] [CrossRef]
- Choi, E.M.; Chu, D.K.; Cheng, P.K.; Tsang, D.N.; Peiris, M.; Bausch, D.G.; Poon, L.L.; Watson-Jones, D. In-flight transmission of SARS-CoV-2. Emerg. Infect. Dis. 2020, 26, 2713. [Google Scholar] [CrossRef]
- Chen, L.; Zhong, L. Genomics functional analysis and drug screening of SARS-CoV-2. Genes Dis. 2020, 7, 542–550. [Google Scholar] [CrossRef]
- Lawton, G. The coronavirus evolves. New Sci. 2021, 249, 8–9. [Google Scholar] [CrossRef]
- Requena, D.; Médico, A.; Chacón, R.D.; Ramírez, M.; Marín-Sánchez, O. Identification of Novel Candidate Epitopes on SARS-CoV-2 Proteins for South America: A Review of HLA Frequencies by Country. Front. Immunol. 2020, 11, 2008. [Google Scholar] [CrossRef]
- Trowsdale, J. Genetic and functional relationships between MHC and NK receptor genes. Immunity 2001, 15, 363–374. [Google Scholar] [CrossRef]
- Shankarkumar, U. The human leukocyte antigen (HLA) system. Int. J. Hum. Genet. 2004, 4, 91–103. [Google Scholar] [CrossRef]
- Magner, W.J.; Kazim, A.L.; Stewart, C.; Romano, M.A.; Catalano, G.; Grande, C.; Keiser, N.; Santaniello, F.; Tomasi, T.B. Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J. Immunol. 2000, 165, 7017–7024. [Google Scholar] [CrossRef]
- De Wit, J.; Borghans, J.A.; Kesmir, C.; van Baarle, D. Role of HLA and KIR in Viral Infections. Front. Immunol. 2016, 7, 286. [Google Scholar] [CrossRef] [Green Version]
- Khanolkar, A.; Badovinac, V.P.; Harty, J.T. CD8 T cell memory development: CD4 T cell help is appreciated. Immunol. Res. 2007, 39, 94–104. [Google Scholar] [CrossRef]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lübke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021, 22, 74–85. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.; Hafezi, M.; Chia, A.; Chng, M.H.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Ferretti, A.P.; Kula, T.; Wang, Y.; Nguyen, D.M.; Weinheimer, A.; Dunlap, G.S.; Xu, Q.; Nabilsi, N.; Perullo, C.R.; Cristofaro, A.W.; et al. Unbiased screens show CD8+ T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity 2020, 53, 1095–1107. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Strålin, K.; Gorin, J.B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020, 183, 158–168. [Google Scholar] [CrossRef]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef]
- Crooke, S.N.; Ovsyannikova, I.G.; Kennedy, R.B.; Poland, G.A. Immunoinformatic identification of B cell and T cell epitopes in the SARS-CoV-2 proteome. Sci. Rep. 2020, 10, 1–5. [Google Scholar] [CrossRef]
- Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R.H.; Peters, B.; Sette, A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 2020, 27, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Yarmarkovich, M.; Warrington, J.M.; Farrel, A.; Maris, J.M. Identification of SARS-CoV-2 vaccine epitopes predicted to induce long-term population-scale immunity. Cell Rep. Med. 2020, 1, 100036. [Google Scholar] [CrossRef] [PubMed]
- Kiyotani, K.; Toyoshima, Y.; Nemoto, K.; Nakamura, Y. Bioinformatic prediction of potential T cell epitopes for SARS-CoV-2. J. Hum. Genet. 2020, 65, 569–575. [Google Scholar] [CrossRef]
- Keller, M.D.; Harris, K.M.; Jensen-Wachspress, M.A.; Kankate, V.V.; Lang, H.; Lazarski, C.A.; Durkee-Shock, J.; Lee, P.H.; Chaudhry, K.; Webber, K.; et al. SARS-CoV-2–specific T cells are rapidly expanded for therapeutic use and target conserved regions of the membrane protein. Blood 2020, 136, 2905–2917. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef]
- Lipsitch, M.; Grad, Y.H.; Sette, A.; Crotty, S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020, 20, 709–713. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Pre-existing immunity to SARS-CoV-2: The knowns and unknowns. Nat. Rev. Immunol. 2020, 20, 457–458. [Google Scholar] [CrossRef]
- Ng, K.W.; Faulkner, N.; Cornish, G.H.; Rosa, A.; Harvey, R.; Hussain, S.; Ulferts, R.; Earl, C.; Wrobel, A.G.; Benton, D.J.; et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 2020, 370, 1339–1343. [Google Scholar] [CrossRef]
- Gombar, S.; Bergquist, T.; Pejaver, V.; Hammarlund, N.E.; Murugesan, K.; Mooney, S.; Shah, N.; Pinsky, B.A.; Banaei, N. SARS-CoV-2 infection and COVID-19 severity in individuals with prior seasonal coronavirus infection. Diagn. Microbiol. Infect. Dis. 2021, 9, 115338. [Google Scholar] [CrossRef]
- Anderson, E.M.; Goodwin, E.C.; Verma, A.; Arevalo, C.P.; Bolton, M.J.; Weirick, M.E.; Gouma, S.; McAllister, C.M.; Christensen, S.R.; Weaver, J.; et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell 2021, 184, 1–7. [Google Scholar] [CrossRef]
- Too, C.L.; Tan, L.K.; Heselynn, H.; Nor-Shuhaila, S.; Eashwary, M.; Wahinuddin, S.; Lau, S.; Gun, S.C.; Mohd-Shahrir, M.S.; Ainon, M.M.; et al. HLA-A,-B,-C,-DRB1 and-DQB1 alleles and haplotypes in 194 Southeast Asia Chinese from Peninsular Malaysia. Hum. Immunol. 2019, 80, 906–907. [Google Scholar] [CrossRef] [PubMed]
- Nurul-Aain, A.F.; Tan, L.K.; Heselynn, H.; Nor-Shuhaila, S.; Eashwary, M.; Wahinuddin, S.; Lau, S.; Gun, S.C.; Mohd-Shahrir, M.S.; Ainon, M.M.; et al. HLA-A,-B,-C,-DRB1 and-DQB1 alleles and haplotypes in 271 Southeast Asia Indians from Peninsular Malaysia. Hum. Immunol. 2020, 81, 263–264. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.K.; Mohd-Farid, B.; Salsabil, S.; Heselynn, H.; Wahinuddin, S.; Lau, S.; Gun, S.C.; Nor-Suhaila, S.; Eashwary, M.; Mohd-Shahrir, M.S.; et al. HLA-A,-B,-C,-DRB1 and-DQB1 alleles and haplotypes in 951 Southeast Asia Malays from Peninsular Malaysia. Hum Immunol. 2016, 77, 818–819. [Google Scholar] [CrossRef] [PubMed]
- Pisanti, S.; Deelen, J.; Gallina, A.M.; Caputo, M.; Citro, M.; Abate, M.; Sacchi, N.; Vecchione, C.; Martinelli, R. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of COVID-19. J. Transl. Med. 2020, 18, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Littera, R.; Campagna, M.; Deidda, S.; Angioni, G.; Cipri, S.; Melis, M.; Firinu, D.; Santus, S.; Lai, A.; Porcella, R.; et al. Human Leukocyte Antigen Complex and Other Immunogenetic and Clinical Factors Influence Susceptibility or Protection to SARS-CoV-2 Infection and Severity of the Disease Course. The Sardinian Experience. Front. Immunol. 2020, 11, 3320. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.F.; Chen, K.H.; Chen, M.; Li, W.Y.; Chen, Y.J.; Tsao, C.H.; Yen, M.Y.; Huang, J.C.; Chen, Y.M. Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral. Immunol. 2011, 24, 421–426. [Google Scholar] [CrossRef]
- Tavasolian, F.; Rashidi, M.; Hatam, G.R.; Jeddi, M.; Hosseini, A.Z.; Mosawi, S.H.; Abdollahi, E.; Inman, R.D. HLA, Immune Response, and Susceptibility to COVID-19. Front. Immunol. 2021, 11, 3581. [Google Scholar] [CrossRef]
- Iturrieta-Zuazo, I.; Rita, C.G.; García-Soidán, A.; De Malet Pintos-Fonseca, A.; Alonso-Alarcón, N.; Pariente-Rodríguez, R.; Tejeda-Velarde, A.; Serrano-Villar, S.; Castañer-Alabau, J.L.; Nieto-Gañán, I. Possible role of HLA class-I genotype in SARS-CoV-2 infection and progression: A pilot study in a cohort of COVID-19 Spanish patients. Clin. Immunol. 2020, 219, 108572. [Google Scholar] [CrossRef]
- Sanchez-Mazas, A. HLA studies in the context of coronavirus outbreaks. Swiss. Med. Wkly. 2020, 150, w20248. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Du, H.; Zhang, J.; Li, Y.Y.; Qu, J.; Zhang, W.; Wang, Y.; Bao, S.; Li, Y.; et al. SARS-CoV-2 infection in children. N. Engl. J. Med. 2020, 382, 1663–1665. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, E.; Lipsitch, M.; Cevik, M. On the Effect of Age on the Transmission of SARS-CoV-2 in Households, Schools, and the Community. J. Infect. Dis. 2021, 223, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Ellington, S.; Strid, P.; Tong, V.T.; Woodworth, K.; Galang, R.R.; Zambrano, L.D.; Nahabedian, J.; Anderson, K.; Gilboa, S.M. Characteristics of women of reproductive age with laboratory-confirmed SARS-CoV-2 infection by pregnancy status—United States, January 22–June 7, 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 769. [Google Scholar] [CrossRef] [PubMed]
- O’Driscoll, M.; Dos Santos, G.R.; Wang, L.; Cummings, D.A.; Azman, A.S.; Paireau, J.; Fontanet, A.; Cauchemez, S.; Salje, H. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature 2021, 590, 140–145. [Google Scholar] [CrossRef]
- Ayoub, H.H.; Chemaitelly, H.; Seedat, S.; Mumtaz, G.R.; Makhoul, M.; Abu-Raddad, L.J. Age could be driving variable SARS-CoV-2 epidemic trajectories worldwide. PLoS ONE 2020, 15, e0237959. [Google Scholar] [CrossRef]
- Wehl, G.; Laible, M.; Rauchenzauner, M. Co-infection of SARS CoV-2 and influenza A in a pediatric patient in Germany. klin padiatr. 2020, 232, 217–218. [Google Scholar] [CrossRef] [PubMed]
- Pinato, D.J.; Lee, A.J.; Biello, F.; Seguí, E.; Carbó, A.; Bruna, R.; Bower, M.; Rizzo, G.; Benafif, S.; Carmona, C.; et al. Presenting features and early mortality from SARS-CoV-2 infection in cancer patients during the initial stage of the COVID-19 pandemic in Europe. Cancers 2020, 12, 1841. [Google Scholar] [CrossRef]
- Eger, K.; Hashimoto, S.; Braunstahl, G.J.; Ten Brinke, A.; Patberg, K.W.; Beukert, A.; Smeenk, F.; Weersink, E.J.; Bel, E.H. Poor outcome of SARS-CoV-2 infection in patients with severe asthma on biologic therapy. Respir. Med. 2021, 177, 106287. [Google Scholar] [CrossRef]
- Maggiolo, F.; Zoboli, F.; Arosio, M.; Valenti, D.; Guarneri, D.; Sangiorgio, L.; Ripamonti, D.; Callegaro, A. SARS-CoV-2 infection in persons living with HIV: A single center prospective cohort. J. Med. Virol. 2021, 93, 1145–1149. [Google Scholar] [CrossRef]
- Yao, Q.; Wang, P.; Wang, X.; Qie, G.; Meng, M.; Tong, X.; Bai, X.; Ding, M.; Liu, W.; Liu, K.; et al. Retrospective study of risk factors for severe SARS-CoV-2 infections in hospitalized adult patients. Pol. Arch. Intern. Med. 2020, 130, 390–399. [Google Scholar] [CrossRef]
- Toraih, E.A.; Sedhom, J.A.; Dokunmu, T.M.; Hussein, M.H.; Ruiz, E.M.; Muthusamy, K.; Zerfaoui, M.; Kandil, E. Hidden in plain sight: The effects of BCG vaccination in the COVID-19 pandemic. J. Med. Virol. 2021, 93, 1950–1966. [Google Scholar] [CrossRef]
- Reche, P.A. Potential cross-reactive immunity to SARS-CoV-2 from common human pathogens and vaccines. Front. Immunol. 2020, 11, 2694. [Google Scholar] [CrossRef] [PubMed]
- Anbarasu, A.; Ramaiah, S.; Livingstone, P. Vaccine repurposing approach for preventing COVID 19: Can MMR vaccines reduce morbidity and mortality? Hum. Vaccin. Immunother. 2020, 16, 2217–2218. [Google Scholar] [CrossRef] [PubMed]
- Bonafè, M.; Prattichizzo, F.; Giuliani, A.; Storci, G.; Sabbatinelli, J.; Olivieri, F. Inflamm-aging: Why older men are the most susceptible to SARS-CoV-2 complicated outcomes. Cytokine Growth Factor Rev. 2020, 53, 33–37. [Google Scholar] [CrossRef]
- Vishvkarma, R.; Rajender, S. Could SARS-CoV-2 affect male fertility? Andrologia 2020, 52, e13712. [Google Scholar] [CrossRef] [PubMed]
- Conti, P.; Younes, A. Coronavirus COV-19/SARS-CoV-2 affects women less than men: Clinical response to viral infection. J. Biol. Regul. Homeost. Agents. 2020, 34, 339–343. [Google Scholar] [PubMed]
- Wu, C.; Yang, W.; Wu, X.; Zhang, T.; Zhao, Y.; Ren, W.; Xia, J. Clinical manifestation and laboratory characteristics of SARS-CoV-2 infection in pregnant women. Virol. Sin. 2020, 35, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Flannery, D.D.; Gouma, S.; Dhudasia, M.B.; Mukhopadhyay, S.; Pfeifer, M.R.; Woodford, E.C.; Gerber, J.S.; Arevalo, C.P.; Bolton, M.J.; Weirick, M.E.; et al. SARS-CoV-2 seroprevalence among parturient women in Philadelphia. Sc.i Immunol. 2020, 5, eabd5079. [Google Scholar] [CrossRef]
- Stringhini, S.; Wisniak, A.; Piumatti, G.; Azman, A.S.; Lauer, S.A.; Baysson, H.; De Ridder, D.; Petrovic, D.; Schrempft, S.; Marcus, K.; et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): A population-based study. Lancet 2020, 396, 313–319. [Google Scholar] [CrossRef]
- Bajema, K.L.; Wiegand, R.E.; Cuffe, K.; Patel, S.V.; Iachan, R.; Lim, T.; Lee, A.; Moyse, D.; Havers, F.P.; Harding, L.; et al. Estimated SARS-CoV-2 Seroprevalence in the US as of September 2020. JAMA Intern. Med. 2020, 181, 450–460. [Google Scholar] [CrossRef]
- Britton, T.; Ball, F.; Trapman, P. A mathematical model reveals the influence of population heterogeneity on herd immunity to SARS-CoV-2. Science 2020, 369, 846–849. [Google Scholar] [CrossRef]
- Arora, R.K.; Joseph, A.; Van Wyk, J.; Rocco, S.; Atmaja, A.; May, E.; Yan, T.; Bobrovitz, N.; Chevrier, J.; Cheng, M.P.; et al. SeroTracker: A global SARS-CoV-2 seroprevalence dashboard. Lancet Infect. Dis. 2020, 21, e75–e76. [Google Scholar] [CrossRef]
- Gudbjartsson, D.F.; Helgason, A.; Jonsson, H.; Magnusson, O.T.; Melsted, P.; Norddahl, G.L.; Saemundsdottir, J.; Sigurdsson, A.; Sulem, P.; Agustsdottir, A.B.; et al. Spread of SARS-CoV-2 in the Icelandic population. N. Engl. J. Med. 2020, 382, 2302–2315. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Baker, J.S.; Quan, W.; Shen, S.; Fekete, G.; Gu, Y. A preventive role of exercise across the coronavirus 2 (SARS-CoV-2) pandemic. Front. Physiol. 2020, 11, 572718. [Google Scholar] [CrossRef]
- Chesnut, W.M.; MacDonald, S.; Wambier, C.G. Could diet and exercise reduce risk of COVID-19 syndemic? Med. Hypotheses 2021, 148, 110502. [Google Scholar] [CrossRef] [PubMed]
- Khaleb, M.B.; Benajiba, N. The role of nutrition in strengthening immune system against newly emerging viral diseases: Case of SARS-CoV-2. N. Afr. J. Food Nutr. Res. 2020, 4, 280–284. [Google Scholar]
- Shah, V.K.; Firmal, P.; Alam, A.; Ganguly, D.; Chattopadhyay, S. Overview of immune response during SARS-CoV-2 infection: Lessons from the past. Front. Immunol. 2020, 11, 1949. [Google Scholar] [CrossRef] [PubMed]
- Jordan, R.E.; Adab, P. Who is most likely to be infected with SARS-CoV-2? Lancet Infect. Dis. 2020, 20, 995–996. [Google Scholar] [CrossRef]
- Barazzoni, R.; Bischoff, S.C.; Breda, J.; Wickramasinghe, K.; Krznaric, Z.; Nitzan, D.; Pirlich, M.; Singer, P. ESPEN expert statements and practical guidance for nutritional management of individuals with SARS-CoV-2 infection. Clin. Nutr. 2020, 39, 1631–1638. [Google Scholar] [CrossRef]
- Pironi, L.; Sasdelli, A.S.; Ravaioli, F.; Baracco, B.; Battaiola, C.; Bocedi, G.; Brodosi, L.; Leoni, L.; Mari, G.A.; Musio, A. Malnutrition and nutritional therapy in patients with SARS-CoV-2 disease. Clin. Nutr. 2021, 40, 1330–1337. [Google Scholar] [CrossRef]
- Jacob, M.C.; Feitosa, I.S.; Albuquerque, U.P. Animal-based food systems are unsafe: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) fosters the debate on meat consumption. Public Health Nutr. 2020, 23, 3250–3255. [Google Scholar] [CrossRef]
- Michie, S.; West, R.; Rogers, M.B.; Bonell, C.; Rubin, G.J.; Amlôt, R. Reducing SARS-CoV-2 transmission in the UK: A behavioural science approach to identifying options for increasing adherence to social distancing and shielding vulnerable people. Br. J. Health Psychol. 2020, 25, 945–956. [Google Scholar] [CrossRef] [PubMed]
- West, R.; Michie, S.; Rubin, G.J.; Amlôt, R. Applying principles of behaviour change to reduce SARS-CoV-2 transmission. Nat. Hum. Behav. 2020, 4, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Doung-Ngern, P.; Suphanchaimat, R.; Panjangampatthana, A.; Janekrongtham, C.; Ruampoom, D.; Daochaeng, N.; Eungkanit, N.; Pisitpayat, N.; Srisong, N.; Yasopa, O.; et al. Case-control study of use of personal protective measures and risk for SARS-CoV 2 infection, Thailand. Emerg. Infect. Dis. 2020, 26, 2607. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, F.; Pachetti, M.; Marini, B.; Ippodrino, R.; Ciccozzi, M.; Zella, D. SARS-CoV-2: March toward adaptation. J. Med. Virol. 2020, 92, 2274–2276. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082. [Google Scholar] [CrossRef]
- Abdullahi, I.N.; Emeribe, A.U.; Mustapha, J.O.; Fasogbon, S.A.; Ofor, I.B.; Opeyemi, I.S.; Obi-George, C.; Sunday, A.O.; Nwofe, J. Exploring the genetics, ecology of SARS-CoV-2 and climatic factors as possible control strategies against COVID-19. Infez. Med. 2020, 28, 166–173. [Google Scholar]
- Ho, F.K.; Petermann-Rocha, F.; Gray, S.R.; Jani, B.D.; Katikireddi, S.V.; Niedzwiedz, C.L.; Foster, H.; Hastie, C.E.; Mackay, D.F.; Gill, J.M.; et al. Is older age associated with COVID-19 mortality in the absence of other risk factors? General population cohort study of 470,034 participants. PLoS ONE 2020, 15, e0241824. [Google Scholar] [CrossRef]
- D’ascanio, M.; Innammorato, M.; Pasquariello, L.; Pizzirusso, D.; Guerrieri, G.; Castelli, S.; Pezzuto, A.; Anibaldi, P.; Marcolongo, A.; Mancini, R.; et al. Age is not the only risk factor in COVID-19: The role of comorbidities and of long staying in residential care homes. BMC Geriatr. 2021, 21, 63. [Google Scholar] [CrossRef]
- Noor, F.M.; Islam, M.M. Prevalence and associated risk factors of mortality among COVID-19 patients: A meta-analysis. J. Community Health 2020, 45, 1270–1282. [Google Scholar] [CrossRef]
- Biswas, M.; Rahaman, S.; Biswas, T.K.; Haque, Z.; Ibrahim, B. Association of sex, age, and comorbidities with mortality in COVID-19 patients: A systematic review and meta-analysis. Intervirology 2020, 64, 1–2. [Google Scholar] [CrossRef]
- Wu, C.; Chen, X.; Cai, Y.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; Zhang, Y.; et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Onder, G.; Rezza, G.; Brusaferro, S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA 2020, 323, 1775–1776. [Google Scholar] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Grasselli, G.; Zangrillo, A.; Zanella, A.; Antonelli, M.; Cabrini, L.; Castelli, A.; Cereda, D.; Coluccello, A.; Foti, G.; Fumagalli, R.; et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA 2020, 323, 1574–1581. [Google Scholar] [CrossRef]
- Pagani, G.; Conti, F.; Giacomelli, A.; Bernacchia, D.; Rondanin, R.; Prina, A.; Scolari, V.; Gandolfi, C.E.; Castaldi, S.; Marano, G.; et al. Seroprevalence of SARS-CoV-2 significantly varies with age: Preliminary results from a mass population screening. J. Infect. 2020, 81, e10. [Google Scholar] [CrossRef]
- Romagnoli, S.; Peris, A.; De Gaudio, A.R.; Geppetti, P. SARS-CoV-2 and COVID-19: From the bench to the bedside. Physiol. Rev. 2020, 100, 1455–1466. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Bosso, M.; Thanaraj, T.A.; Abu-Farha, M.; Alanbaei, M.; Abubaker, J.; Al-Mulla, F. The Two Faces of ACE2: The Role of ACE2 Receptor and Its Polymorphisms in Hypertension and COVID-19. Mol. Ther. Methods Clin. Dev. 2020, 18, 321–327. [Google Scholar] [CrossRef]
- Pal, R.; Bhansali, A. COVID-19, diabetes mellitus and ACE2: The conundrum. Diabetes Res. Clin. Pract. 2020, 162, 108132. [Google Scholar] [CrossRef]
- Wicik, Z.; Eyileten, C.; Jakubik, D.; Simões, S.N.; Martins, D.C.; Pavão, R.; Siller-Matula, J.M.; Postula, M. ACE2 interaction networks in COVID-19: A physiological framework for prediction of outcome in patients with cardiovascular risk factors. J. Clin. Med. 2020, 9, 3743. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.M.; Niikura, M.; Yang, C.W.; Sin, D.D. COVID-19 and COPD. Eur. Respir. J. 2020, 56, 2002108. [Google Scholar] [CrossRef] [PubMed]
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D.; et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol. 2020, 251, 228–248. [Google Scholar] [CrossRef] [PubMed]
- Rathnasinghe, R.; Jangra, S.; Cupic, A.; Martínez-Romero, C.; Mulder, L.C.; Kehrer, T.; Yildiz, S.; Choi, A.; Mena, I.; De Vrieze, J.; et al. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv 2021, 9592. [Google Scholar] [CrossRef]
- Devaux, C.A.; Rolain, J.M.; Raoult. D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020, 53, 425. [Google Scholar] [CrossRef]
- Wijnant, S.R.; Jacobs, M.; Van Eeckhoutte, H.P.; Lapauw, B.; Joos, G.F.; Bracke, K.R.; Brusselle, G.G. Expression of ACE2, the SARS-CoV-2 receptor, in lung tissue of patients with type 2 diabetes. Diabetes 2020, 69, 2691–2699. [Google Scholar] [CrossRef]
- Taneera, J.; El-Huneidi, W.; Hamad, M.; Mohammed, A.K.; Elaraby, E.; Hachim, M.Y. Expression profile of SARS-CoV-2 host receptors in human pancreatic islets revealed upregulation of ACE2 in diabetic donors. Biology 2020, 9, 215. [Google Scholar] [CrossRef]
- Soldo, J.; Heni, M.; Königsrainer, A.; Häring, H.U.; Birkenfeld, A.L.; Peter, A. Increased Hepatic ACE2 Expression in NAFL and Diabetes—A Risk for COVID-19 Patients? Diabetes Care 2020, 43, e134–e136. [Google Scholar] [CrossRef]
- Higham, A.; Singh, D. Increased ACE2 expression in bronchial epithelium of COPD patients who are overweight. Obesity 2020, 28, 1586–1589. [Google Scholar] [CrossRef]
- Sharif-Askari, N.S.; Sharif-Askari, F.S.; Alabed, M.; Temsah, M.H.; Al Heialy, S.; Hamid, Q.; Halwani, R. Airways expression of SARS-CoV-2 receptor, ACE2, and TMPRSS2 is lower in children than adults and increases with smoking and COPD. Mol. Ther. Methods Clin. Dev. 2020, 18, 1–6. [Google Scholar] [CrossRef]
- Eapen, M.S.; Lu, W.; Hackett, T.L.; Singhera, G.K.; Thompson, I.E.; McAlinden, K.D.; Hardikar, A.; Weber, H.C.; Haug, G.; Wark, P.A.; et al. Dysregulation of endocytic machinery and ACE2 in small airways of smokers and COPD patients can augment their susceptibility to SARS-CoV-2 (COVID-19) infections. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 320, L158–L163. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.; Van Eeckhoutte, H.P.; Wijnant, S.R.; Janssens, W.; Joos, G.F.; Brusselle, G.G.; Bracke, K.R. Increased expression of ACE2, the SARS-CoV-2 entry receptor, in alveolar and bronchial epithelium of smokers and COPD subjects. Eur. Respir. J. 2020, 56, 2002378. [Google Scholar] [CrossRef] [PubMed]
- Riccò, M.; Gualerzi, G.; Ranzieri, S.; Bragazzi, N.L. Stop playing with data: There is no sound evidence that Bacille Calmette-Guérin may avoid SARS-CoV-2 infection (for now). Acta Biomed. 2020, 91, 207. [Google Scholar]
- Agerer, B.; Koblischke, M.; Gudipati, V.; Montaño-Gutierrez, L.F.; Smyth, M.; Popa, A.; Genger, J.W.; Endler, L.; Florian, D.M.; Mühlgrabner, V.; et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8+ T cell responses. Sci. Immunol. 2021, 6, eabg6461. [Google Scholar] [CrossRef] [PubMed]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the effects of SARS-CoV-2 Spike mutation D614G on transmissibility and pathogenicity. Cell 2021, 184, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Liu, Y.; Liu, J.; Zhang, X.; Zou, J.; Fontes-Garfias, C.R.; Xia, H.; Swanson, K.A.; Cutler, M.; Cooper, D.; et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021, 27, 620–621. [Google Scholar] [CrossRef]
- World Health Organization. R&D Blueprint and COVID-19. Draft Landscape and Tracker of COVID-19 Candidate Vaccines. Available online: www.who.int/teams/blueprint/covid-19 (accessed on 2 February 2021).
- Chakraborty, C.; Agoramoorthy, G. India’s cost-effective COVID-19 vaccine development initiatives. Vaccine 2020, 38, 7883. [Google Scholar] [CrossRef]
- Gupta, J.; Gupta, R.; Vanshita. Microneedle Technology: An Insight into Recent Advancements and Future Trends in Drug and Vaccine Delivery. Assay Drug Dev Technol. 2020, 19. [Google Scholar] [CrossRef]
- World Health Organization. Coronavirus Disease (COVID-19) Pandemic-Emergency Use Listing Procedure (EUL). Status of COVID-19 Vaccines within WHO EUL/PQ evaluation process. Available online: extranet.who.int (accessed on 8 October 2020).
- National COVID-19 Immunisation Programme. The Special Committee for Ensuring Access to COVID-19 Vaccine Supply (JKJAV) 2021. Available online: www.covidnow.moh.my (accessed on 13 September 2021).
- European Medicines Agency-Pharmacovigilance Risk Assessment Committee. COVID-19 Vaccine AstraZeneca: Benefits still outweigh the risks despite possible link to rare blood clots with low blood platelets. 2021. Available online: www.ema.europa.eu (accessed on 19 March 2021).
- Teixeira, S.K.; Pereira, A.C.; Krieger, J.E. Genetics of resistant hypertension: The missing heritability and opportunities. Curr. Hypertens Rep. 2018, 20, 48. [Google Scholar] [CrossRef]
- Rossi, G.P.; Ceolotto, G.; Caroccia, B.; Lenzini, L. Genetic screening in arterial hypertension. Na.t Rev. Endocrinol. 2017, 13, 289–298. [Google Scholar] [CrossRef]
- Ciccarelli, M.; Finelli, R.; Rivera, N.; Santulli, G.; Izzo, R.; De Luca, N.; Rozza, F.; Ceccarelli, M.; Pagnotta, S.; Uliano, F.; et al. The possible role of chromosome X variability in hypertensive familiarity. J. Hum. Hypertens. 2017, 31, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Kujovich, J. Factor V Leiden thrombophilia. Genet. Med. 2011, 13, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| No. | Protein(s) and Their (Respective Numbers of Epitopes) | Subset | Ref. |
|---|---|---|---|
| 1 | N (4) non-RBD-S (90), RBD-S (23), E (2), ORF3a (7), ORF7a (3), ORF6 (7), ORF8 (4), nsp1 (1), nsp2 (7), nsp3 (11), nsp4 (10), nsp5 (4), nsp6 (9), nsp8 (2), nsp10 (2), nsp12 (9), nsp13 (4), nsp14 (4), nsp15 (1), nsp16 (2) | CD4 | [19] |
| 2 | S (29), N (15), M (27), ORF7a (1) | CD4 | [20] |
| 3 | S (11), N (21), ORF3a (4) | CD8 | [20] |
| 4 | ORF9+N (50), ORF5+M (11), ORF2+S (3), ORF3 (4), ORF4+E (1), ORF6 (1), ORF8 (6) | CD4 | [13] |
| 5 | ORF1 (4), ORF2+S (2), ORF9+N (5), ORF5+M (1) | CD8 | [13] |
| 6 | S (1), ORF1ab (4) | CD8 | [21] |
| 7 | M (2), S (5), ORF1ab (24), ORF3 (1), ORF6 (1), ORF7 (2), ORF8 (1) | CD4 | [21] |
| 8 | nsp1 (2), nsp2 (14), PLpro (34), nsp4 (22), 3CL (6), nsp6 (18), nsp7 (4), nsp8 (4), nsp9 (2), nsp10 (2), RdRpol (19), Hel (14), nsp14 (19), nsp15 (4), nsp16 (9), S (20), ORF3a (10), E (8), M (8), ORF6 (6), ORF7a (4), ORF8 (3), N (7), ORF10 (1) | CD4 | [22] |
| 9 | nsp1 (13), nsp2 (40), PLpro (128), nsp4 (40), 3CL (15), nsp6 (17), nsp7 (6), nsp8 (17), nsp9 (13), nsp10 (7), RdRpol (68), Hel (38), nsp14 (33), nsp15 (25), nsp16 (16), S (86), ORF3a (20), E (2), M (15), ORF6 (2), ORF7a (8), ORF8 (3), N (13), ORF10 (3) | CD8 | [22] |
| 10 | N (8), nsp7 (1) | CD4 | [15] |
| 11 | N (3) | CD8 | [15] |
| 12 | ORF1ab (40), M (3), S (6), N (2), ORF3a (3), ORF7a (1) | CD8 | [23] |
| 13 | ORF1ab (40), M (3), S (6), N (2), ORF3a (3), ORF7a (1) | CD4 | [23] |
| 14 | ORF1ab (1478), S (248), ORF3a (69), E (18), M (72), ORF6 (8), ORF7a (26) ORF8 (22), N (60), ORF10 (12) | CD8 | [24] |
| 15 | ORF1ab (1002), S (154), ORF3a (74), E (11), M (57), ORF6 (28), ORF7a (16) ORF8 (18), N (32), ORF10 (7) | CD4 | [24] |
| 16 | M (5), N (2), S (5) | CD4 | [25] |
| 17 | N (2) | CD8 | [25] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lani, R.; Senin, N.A.; AbuBakar, S.; Hassandarvish, P. Knowledge of SARS-CoV-2 Epitopes and Population HLA Types Is Important in the Design of COVID-19 Vaccines. Vaccines 2022, 10, 1606. https://doi.org/10.3390/vaccines10101606
Lani R, Senin NA, AbuBakar S, Hassandarvish P. Knowledge of SARS-CoV-2 Epitopes and Population HLA Types Is Important in the Design of COVID-19 Vaccines. Vaccines. 2022; 10(10):1606. https://doi.org/10.3390/vaccines10101606
Chicago/Turabian StyleLani, Rafidah, Nurul Aqidah Senin, Sazaly AbuBakar, and Pouya Hassandarvish. 2022. "Knowledge of SARS-CoV-2 Epitopes and Population HLA Types Is Important in the Design of COVID-19 Vaccines" Vaccines 10, no. 10: 1606. https://doi.org/10.3390/vaccines10101606
APA StyleLani, R., Senin, N. A., AbuBakar, S., & Hassandarvish, P. (2022). Knowledge of SARS-CoV-2 Epitopes and Population HLA Types Is Important in the Design of COVID-19 Vaccines. Vaccines, 10(10), 1606. https://doi.org/10.3390/vaccines10101606






