Heterologous Systemic Prime–Intranasal Boosting Using a Spore SARS-CoV-2 Vaccine Confers Mucosal Immunity and Cross-Reactive Antibodies in Mice as well as Protection in Hamsters
Abstract
1. Introduction
2. Materials and Methods
2.1. General Methods
2.2. Construction of B. subtilis Spores Expressing SARS-CoV-2 Antigens
2.3. Immunogens
2.4. Prime Boost Vaccination Studies
2.5. Determination of Mucosal and Systemic Antibody Titers by Indirect ELISA
2.6. Cytokine Analysis
2.7. Hamster Challenge Study
2.8. Statistical Analysis
3. Results
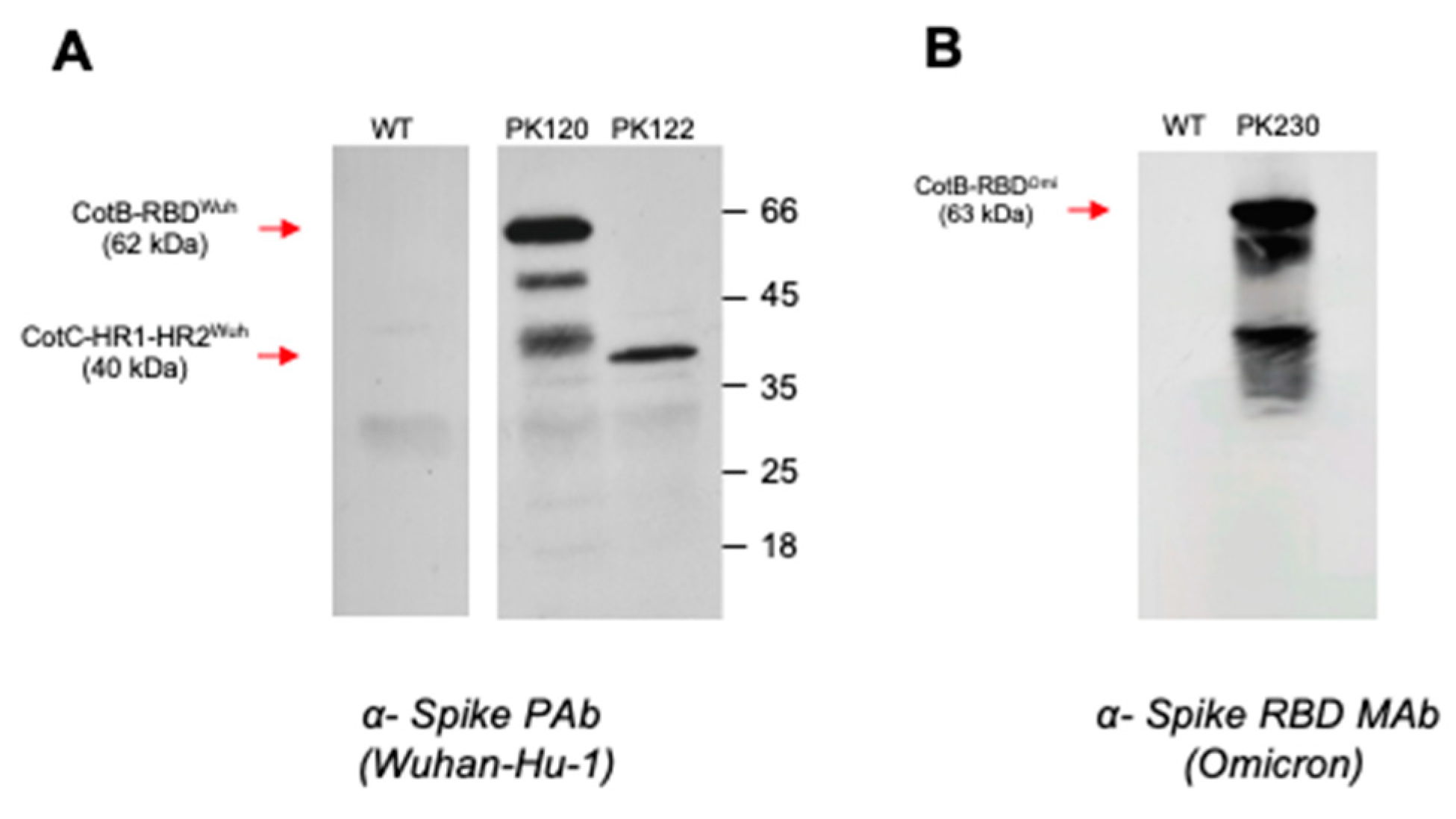
3.1. Construction of B. subtilis Spores Displaying SARS-CoV-2 Domains
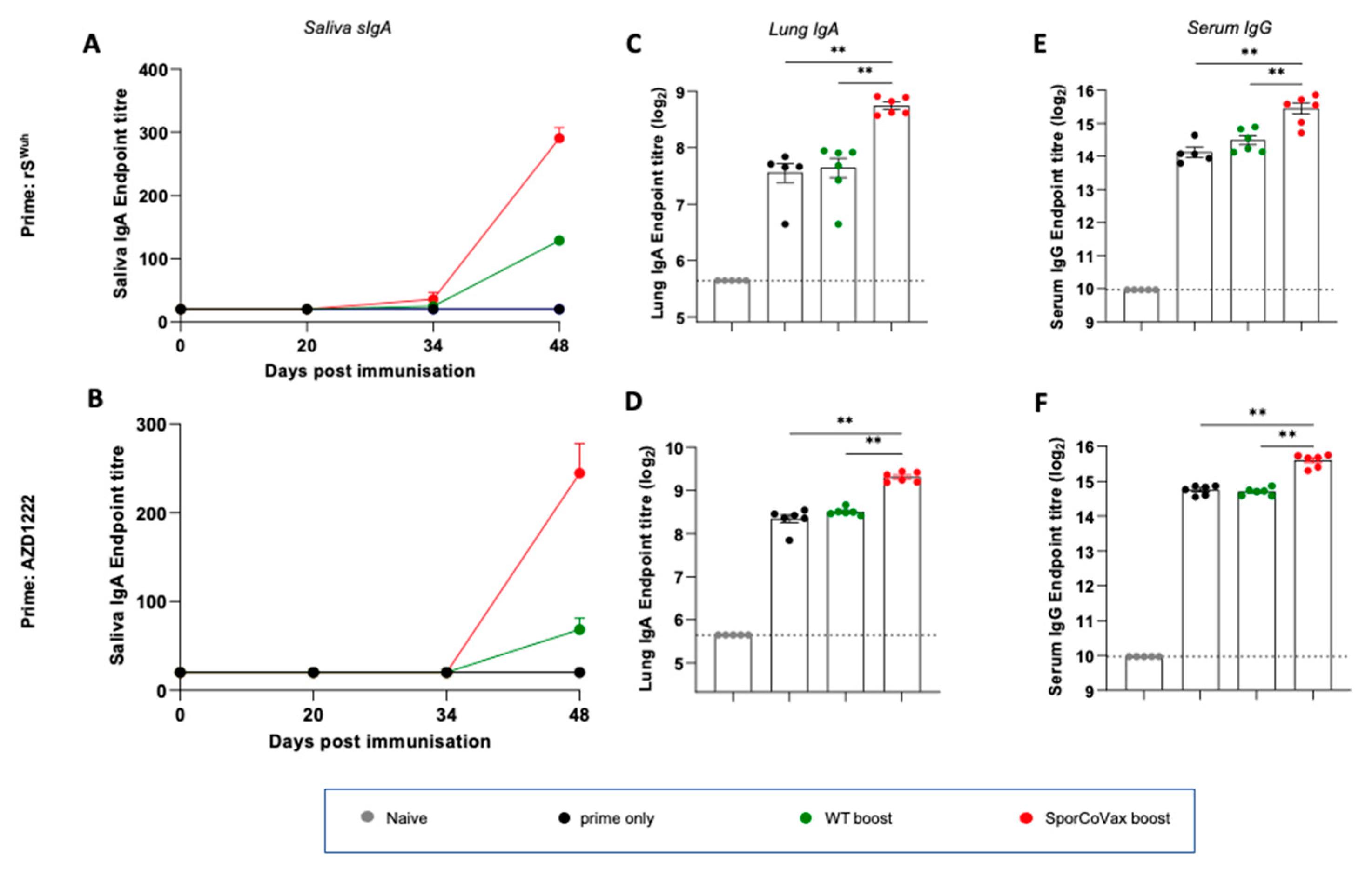
3.2. Intranasal Boosting with a Spore Vaccine following a Systemic Prime Evokes Mucosal Immune Responses
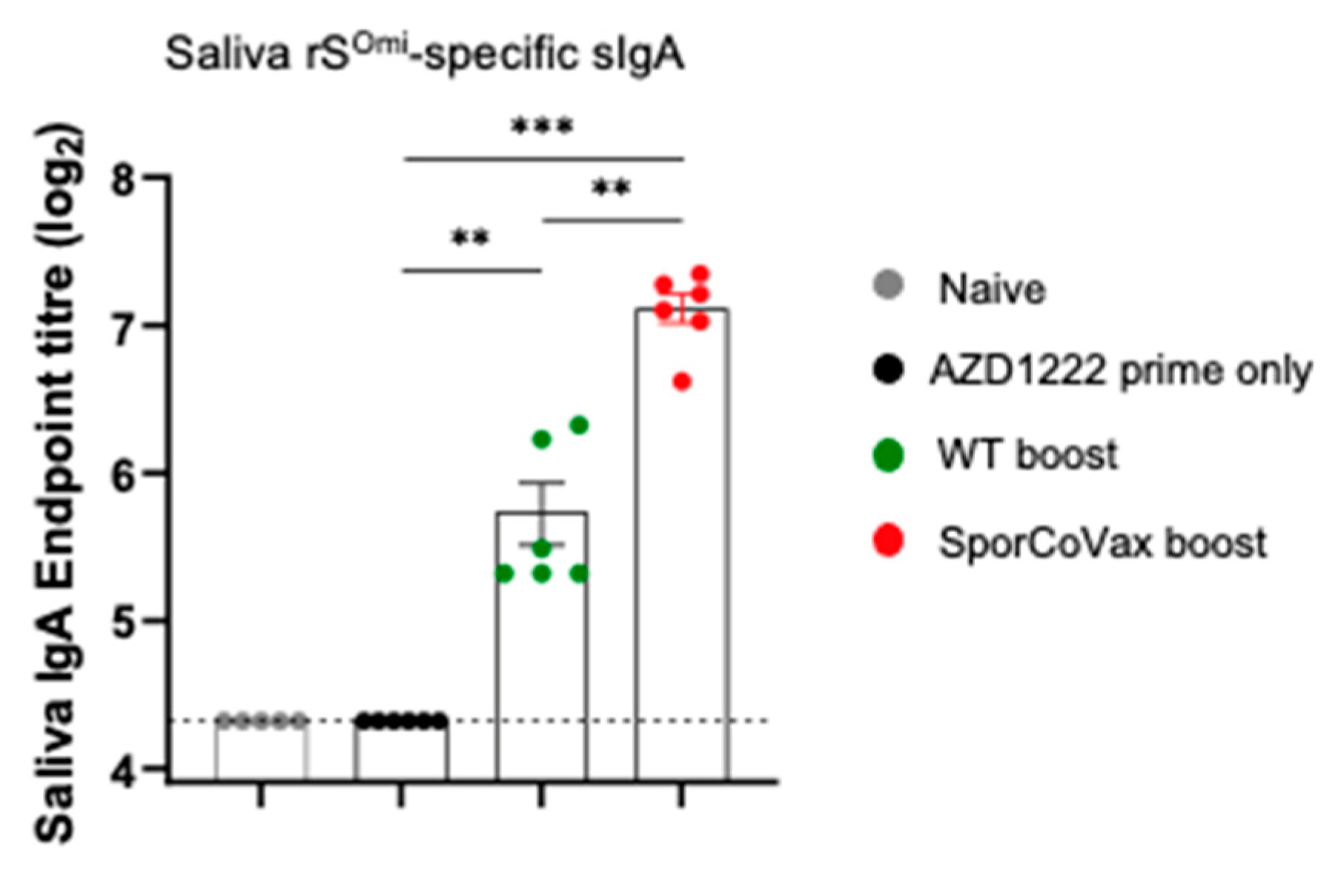
3.3. Boosting with Spores Evokes Cross-Reactive Antibodies
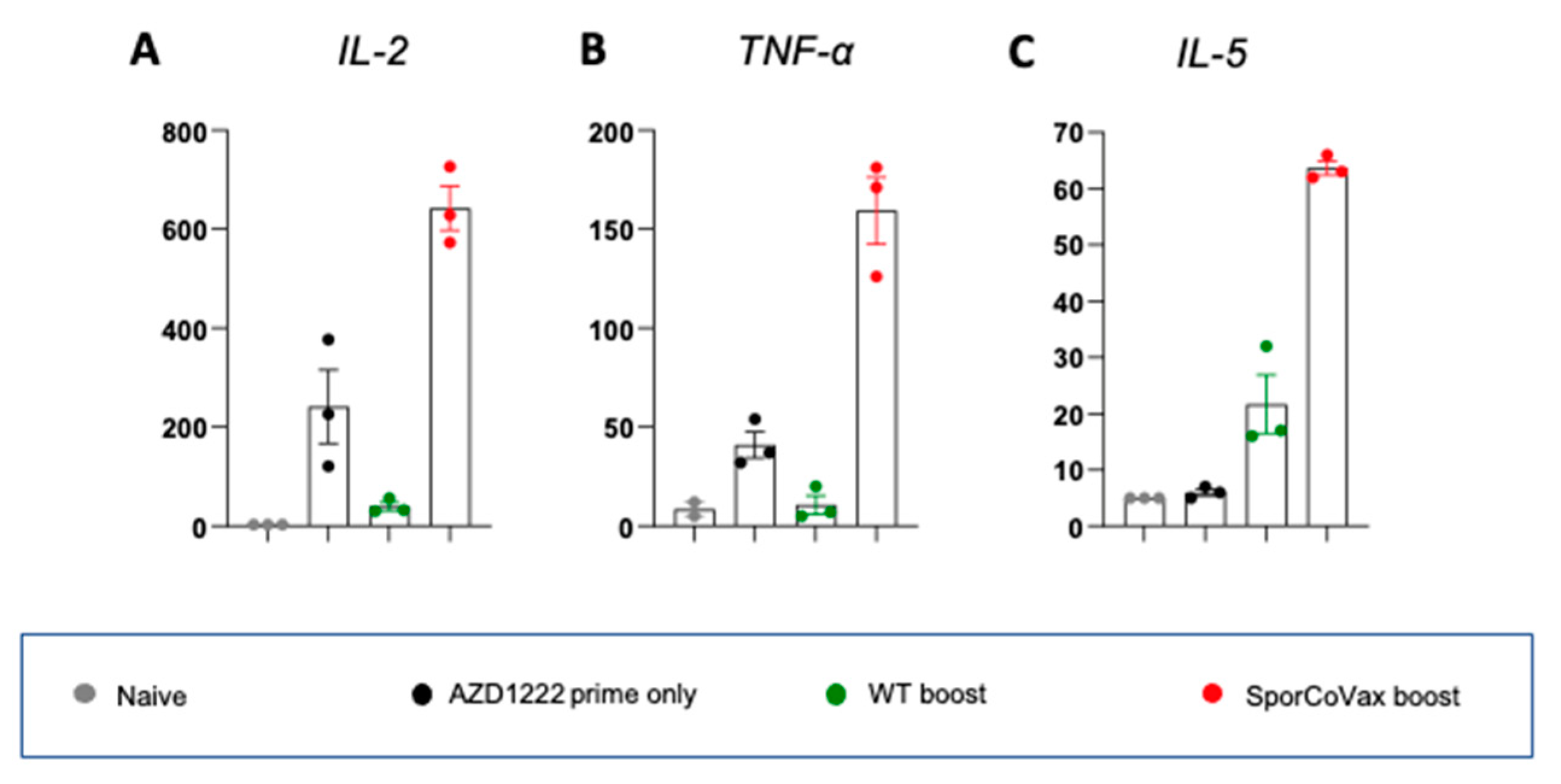
3.4. Intranasal Boosting with a Spore Vaccine Results in a Mixed T Cell Cytokine Profile
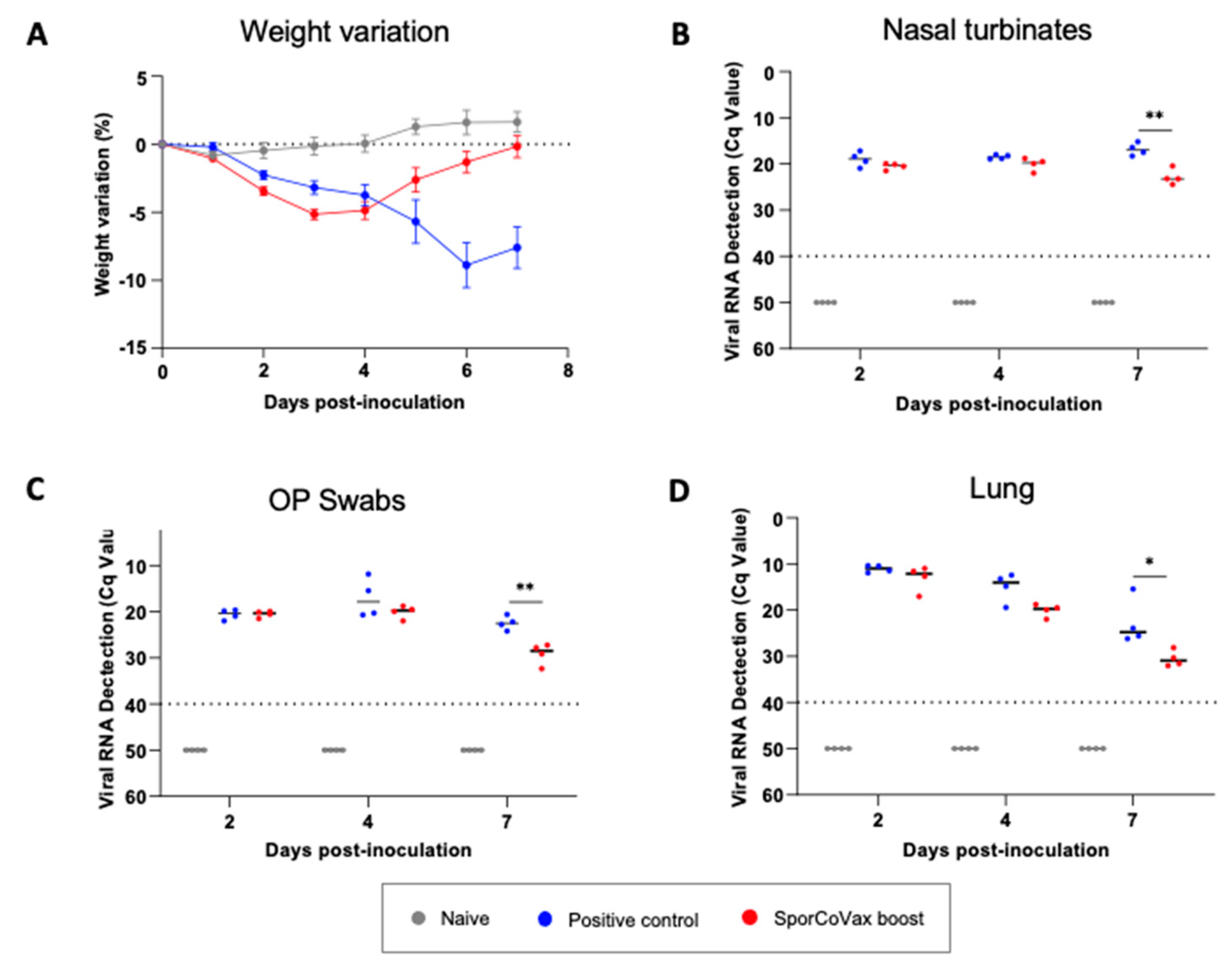
3.5. Intranasal Boosting with Spores Is Protective in the Hamster SARS-CoV-2 Model of Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mouro, V.; Fischer, A. Dealing with a mucosal viral pandemic: Lessons from COVID-19 vaccines. Mucosal Immunol. 2022, 15, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Brosh-Nissimov, T.; Orenbuch-Harroch, E.; Chowers, M.; Elbaz, M.; Nesher, L.; Stein, M.; Maor, Y.; Cohen, R.; Hussein, K.; Weinberger, M.; et al. BNT162b2 vaccine breakthrough: Clinical characteristics of 152 fully vaccinated hospitalized COVID-19 patients in Israel. Clin. Microbiol. Infect. 2021, 27, 1652–1657. [Google Scholar] [CrossRef] [PubMed]
- Farinholt, T.; Doddapaneni, H.; Qin, X.; Menon, V.; Meng, Q.; Metcalf, G.; Chao, H.; Gingras, M.C.; Avadhanula, V.; Farinholt, P.; et al. Transmission event of SARS-CoV-2 delta variant reveals multiple vaccine breakthrough infections. BMC Med. 2021, 19, 255. [Google Scholar] [CrossRef] [PubMed]
- Okuya, K.; Yoshida, R.; Manzoor, R.; Saito, S.; Suzuki, T.; Sasaki, M.; Saito, T.; Kida, Y.; Mori-Kajihara, A.; Kondoh, T.; et al. Potential Role of Nonneutralizing IgA Antibodies in Cross-Protective Immunity against Influenza A Viruses of Multiple Hemagglutinin Subtypes. J. Virol. 2020, 94, e00408-20. [Google Scholar] [CrossRef] [PubMed]
- Mantis, N.J.; Rol, N.; Corthesy, B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef]
- Moldoveanu, Z.; Clements, M.L.; Prince, S.J.; Murphy, B.R.; Mestecky, J. Human immune responses to influenza virus vaccines administered by systemic or mucosal routes. Vaccine 1995, 13, 1006–1012. [Google Scholar] [CrossRef]
- Alu, A.; Chen, L.; Lei, H.; Wei, Y.; Tian, X.; Wei, X. Intranasal COVID-19 vaccines: From bench to bed. EBioMedicine 2022, 76, 103841. [Google Scholar] [CrossRef]
- Tiboni, M.; Casettari, L.; Illum, L. Nasal vaccination against SARS-CoV-2: Synergistic or alternative to intramuscular vaccines? Int. J. Pharm. 2021, 603, 120686. [Google Scholar] [CrossRef]
- Jiang, W.; Shi, L.; Cai, L.; Wang, X.; Li, J.; Li, H.; Liang, J.; Gu, Q.; Ji, G.; Li, J.; et al. A two-adjuvant multiantigen candidate vaccine induces superior protective immune responses against SARS-CoV-2 challenge. Cell Rep. 2021, 37, 110112. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Zhang, Z.; Wu, J.; Zhang, J.; Hu, H.; Zhu, T.; Zhang, J.; Luo, L.; Fan, P.; et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type-5 vector-based COVID-19 vaccine (Ad5-nCoV) in adults: Preliminary report of an open-label and randomised phase 1 clinical trial. Lancet Infect. Dis. 2021, 21, 1654–1664. [Google Scholar] [CrossRef]
- Lapuente, D.; Fuchs, J.; Willar, J.; Vieira Antao, A.; Eberlein, V.; Uhlig, N.; Issmail, L.; Schmidt, A.; Oltmanns, F.; Peter, A.S.; et al. Protective mucosal immunity against SARS-CoV-2 after heterologous systemic prime-mucosal boost immunization. Nat. Commun. 2021, 12, 6871. [Google Scholar] [CrossRef] [PubMed]
- Permpoonpattana, P.; Hong, H.A.; Phetcharaburanin, J.; Huang, J.M.; Cook, J.; Fairweather, N.F.; Cutting, S.M. Immunization with Bacillus spores expressing toxin A peptide repeats protects against infection with Clostridium difficile strains producing toxins A and B. Infect. Immun. 2011, 79, 2295–2302. [Google Scholar] [CrossRef] [PubMed]
- Reljic, R.; Sibley, L.; Huang, J.M.; Pepponi, I.; Hoppe, A.; Hong, H.A.; Cutting, S.M. Mucosal vaccination against tuberculosis using inert bioparticles. Infect. Immun. 2013, 81, 4071–4080. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Belitsky, B.R.; Brinker, J.P.; Kerstein, K.O.; Brown, D.W.; Clements, J.D.; Keusch, G.T.; Tzipori, S.; Sonenshein, A.L.; Herrmann, J.E. Development of a Bacillus subtilis-based rotavirus vaccine. Clin. Vaccine Immunol. 2010, 17, 1647–1655. [Google Scholar] [CrossRef] [PubMed]
- Barnes, A.G.; Cerovic, V.; Hobson, P.S.; Klavinskis, L.S. Bacillus subtilis spores: A novel microparticle adjuvant which can instruct a balanced Th1 and Th2 immune response to specific antigen. Eur. J. Immunol. 2007, 37, 1538–1547. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Hong, H.A.; Huang, J.M.; Colenutt, C.; Khang, D.D.; Nguyen, T.V.; Park, S.M.; Shim, B.S.; Song, H.H.; Cheon, I.S.; et al. Killed Bacillus subtilis spores as a mucosal adjuvant for an H5N1 vaccine. Vaccine 2012, 30, 3266–3277. [Google Scholar] [CrossRef]
- Hosseini, S.; Curilovs, A.; Cutting, S.M. Biological Containment of Genetically Modified Bacillus subtilis. Appl. Environ. Microbiol. 2018, 84, e02334-17. [Google Scholar] [CrossRef]
- Ianiro, G.; Rizzatti, G.; Plomer, M.; Lopetuso, L.; Scaldaferri, F.; Franceschi, F.; Cammarota, G.; Gasbarrini, A. Bacillus clausii for the Treatment of Acute Diarrhea in Children: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients 2018, 10, 1074. [Google Scholar] [CrossRef]
- Huang, J.M.; Hong, H.A.; Van Tong, H.; Hoang, T.H.; Brisson, A.; Cutting, S.M. Mucosal delivery of antigens using adsorption to bacterial spores. Vaccine 2010, 28, 1021–1030. [Google Scholar] [CrossRef]
- Harwood, C.R.; Cutting, S.M. (Eds.) Molecular Biological Methods for Bacillus; John Wiley & Sons Ltd.: Chichester, UK, 1990; p. 581. [Google Scholar]
- Christensen, D.; Mortensen, R.; Rosenkrands, I.; Dietrich, J.; Andersen, P. Vaccine-induced Th17 cells are established as resident memory cells in the lung and promote local IgA responses. Mucosal Immunol. 2017, 10, 260–270. [Google Scholar] [CrossRef]
- Brustolin, M.; Rodon, J.; Rodriguez de la Concepcion, M.L.; Avila-Nieto, C.; Cantero, G.; Perez, M.; Te, N.; Noguera-Julian, M.; Guallar, V.; Valencia, A.; et al. Protection against reinfection with D614- or G614-SARS-CoV-2 isolates in golden Syrian hamster. Emerg. Microbes Infect. 2021, 10, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden Syrian hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Tabynov, K.; Turebekov, N.; Babayeva, M.; Fomin, G.; Yerubayev, T.; Yespolov, T.; Li, L.; Renukaradhya, G.J.; Petrovsky, N.; Tabynov, K. An adjuvanted subunit SARS-CoV-2 spike protein vaccine provides protection against COVID-19 infection and transmission. NPJ Vaccines 2022, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Meseda, C.A.; Stauft, C.B.; Selvaraj, P.; Lien, C.Z.; Pedro, C.; Nunez, I.A.; Woerner, A.M.; Wang, T.T.; Weir, J.P. MVA vector expression of SARS-CoV-2 spike protein and protection of adult Syrian hamsters against SARS-CoV-2 challenge. NPJ Vaccines 2021, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.; Martinez, C.I.; Tedjakusuma, S.N.; Peinovich, N.; Dora, E.G.; Birch, S.M.; Kajon, A.E.; Werts, A.D.; Tucker, S.N. Oral Vaccination Protects Against Severe Acute Respiratory Syndrome Coronavirus 2 in a Syrian Hamster Challenge Model. J. Infect. Dis. 2022, 225, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Wang, P.; Wong, Y.C.; Xu, H.; Lau, S.Y.; Liu, L.; Mok, B.W.; Peng, Q.; Liu, N.; Woo, K.F.; et al. Nasal prevention of SARS-CoV-2 infection by intranasal influenza-based boost vaccination in mouse models. EBioMedicine 2022, 75, 103762. [Google Scholar] [CrossRef]
- Cuburu, N.; Kim, R.; Guittard, G.C.; Thompson, C.D.; Day, P.M.; Hamm, D.E.; Pang, Y.S.; Graham, B.S.; Lowy, D.R.; Schiller, J.T. A Prime-Pull-Amplify Vaccination Strategy To Maximize Induction of Circulating and Genital-Resident Intraepithelial CD8(+) Memory T Cells. J. Immunol. 2019, 202, 1250–1264. [Google Scholar] [CrossRef]
- Nicholson, W.L. Roles of Bacillus endospores in the environment. Cell. Mol. Life Sci. 2002, 59, 410–416. [Google Scholar] [CrossRef]
- Shaffer, B.T.; Lighthart, B. Survey of Culturable Airborne Bacteria at Four Diverse Locations in Oregon: Urban, Rural, Forest, and Coastal. Microb. Ecol. 1997, 34, 167–177. [Google Scholar] [CrossRef]
- Hong, H.A.; To, E.; Fakhry, S.; Baccigalupi, L.; Ricca, E.; Cutting, S.M. Defining the natural habitat of Bacillus spore-formers. Res. Microbiol. 2009, 160, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Steidler, L.; Neirynck, S.; Huyghebaert, N.; Snoeck, V.; Vermeire, A.; Goddeeris, B.; Cox, E.; Remon, J.P.; Remaut, E. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 2003, 21, 785–789. [Google Scholar] [CrossRef] [PubMed]
- de Souza, R.D.; Batista, M.T.; Luiz, W.B.; Cavalcante, R.C.; Amorim, J.H.; Bizerra, R.S.; Martins, E.G.; Ferreira, L.C. Bacillus subtilis spores as vaccine adjuvants: Further insights into the mechanisms of action. PLoS ONE 2014, 9, e87454. [Google Scholar] [CrossRef] [PubMed]
- Rhee, K.J.; Sethupathi, P.; Driks, A.; Lanning, D.K.; Knight, K.L. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J. Immunol. 2004, 172, 1118–1124. [Google Scholar] [CrossRef]
- Huang, J.M.; La Ragione, R.M.; Cooley, W.A.; Todryk, S.; Cutting, S.M. Cytoplasmic delivery of antigens, by Bacillus subtilis enhances Th1 responses. Vaccine 2008, 26, 6043–6052. [Google Scholar] [CrossRef]
- Huang, J.M.; La Ragione, R.M.; Nunez, A.; Cutting, S.M. Immunostimulatory activity of Bacillus spores. FEMS Immunol. Med. Microbiol. 2008, 53, 195–203. [Google Scholar] [CrossRef]
- James, J.; Meyer, S.M.; Hong, H.A.; Dang, C.; Linh, H.T.Y.; Ferreira, W.; Katsande, P.M.; Vo, L.; Hynes, D.; Love, W.; et al. Intranasal Treatment of Ferrets with Inert Bacterial Spores Reduces Disease Caused by a Challenging H7N9 Avian Influenza Virus. Vaccines 2022, 10, 1559. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katsande, P.M.; Fernández-Bastit, L.; Ferreira, W.T.; Vergara-Alert, J.; Hess, M.; Lloyd-Jones, K.; Hong, H.A.; Segales, J.; Cutting, S.M. Heterologous Systemic Prime–Intranasal Boosting Using a Spore SARS-CoV-2 Vaccine Confers Mucosal Immunity and Cross-Reactive Antibodies in Mice as well as Protection in Hamsters. Vaccines 2022, 10, 1900. https://doi.org/10.3390/vaccines10111900
Katsande PM, Fernández-Bastit L, Ferreira WT, Vergara-Alert J, Hess M, Lloyd-Jones K, Hong HA, Segales J, Cutting SM. Heterologous Systemic Prime–Intranasal Boosting Using a Spore SARS-CoV-2 Vaccine Confers Mucosal Immunity and Cross-Reactive Antibodies in Mice as well as Protection in Hamsters. Vaccines. 2022; 10(11):1900. https://doi.org/10.3390/vaccines10111900
Chicago/Turabian StyleKatsande, Paidamoyo M., Leira Fernández-Bastit, William T. Ferreira, Júlia Vergara-Alert, Mateusz Hess, Katie Lloyd-Jones, Huynh A. Hong, Joaquim Segales, and Simon M. Cutting. 2022. "Heterologous Systemic Prime–Intranasal Boosting Using a Spore SARS-CoV-2 Vaccine Confers Mucosal Immunity and Cross-Reactive Antibodies in Mice as well as Protection in Hamsters" Vaccines 10, no. 11: 1900. https://doi.org/10.3390/vaccines10111900
APA StyleKatsande, P. M., Fernández-Bastit, L., Ferreira, W. T., Vergara-Alert, J., Hess, M., Lloyd-Jones, K., Hong, H. A., Segales, J., & Cutting, S. M. (2022). Heterologous Systemic Prime–Intranasal Boosting Using a Spore SARS-CoV-2 Vaccine Confers Mucosal Immunity and Cross-Reactive Antibodies in Mice as well as Protection in Hamsters. Vaccines, 10(11), 1900. https://doi.org/10.3390/vaccines10111900





