Identification of Shared Neoantigens in BRCA1-Related Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Identification
2.2. Data Analyses
2.3. Antigenicity Prediction of Recurrent Somatic Mutations
2.4. Allele and Haplotype Frequency Calculations
2.5. Statistical Analysis
3. Results
3.1. Characteristics of BRCA1-Positive and BRCA1-Negative Samples
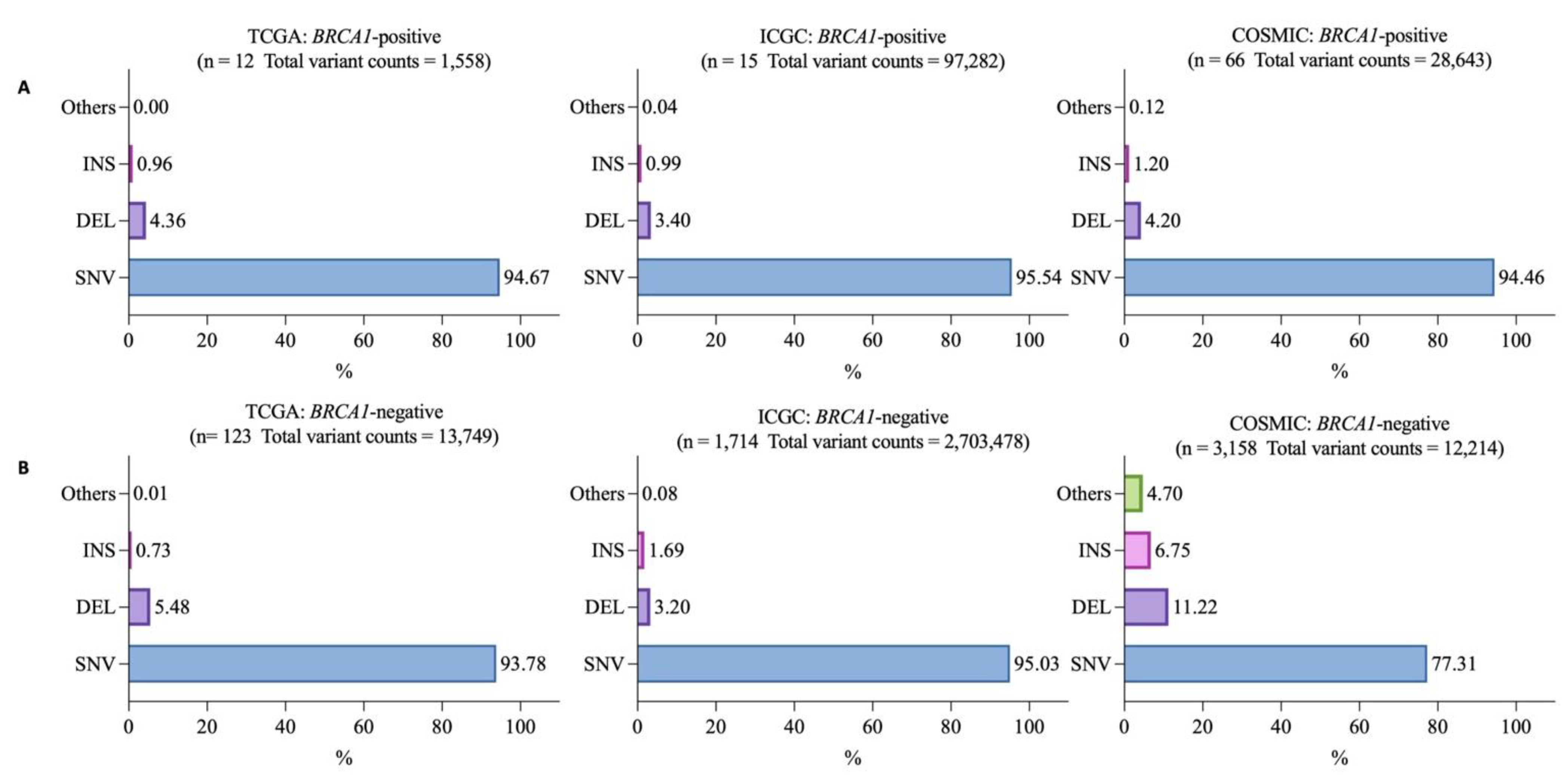
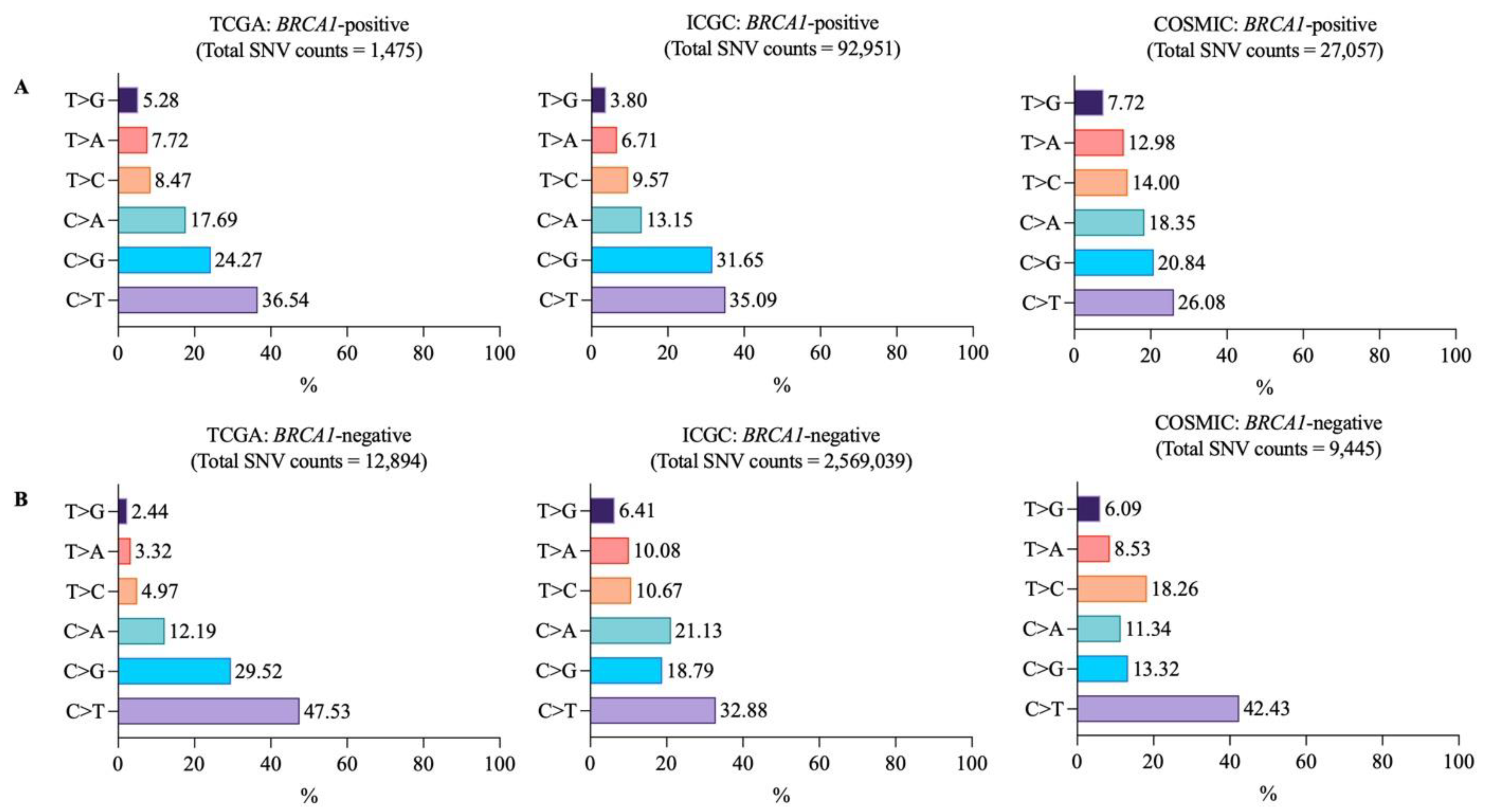
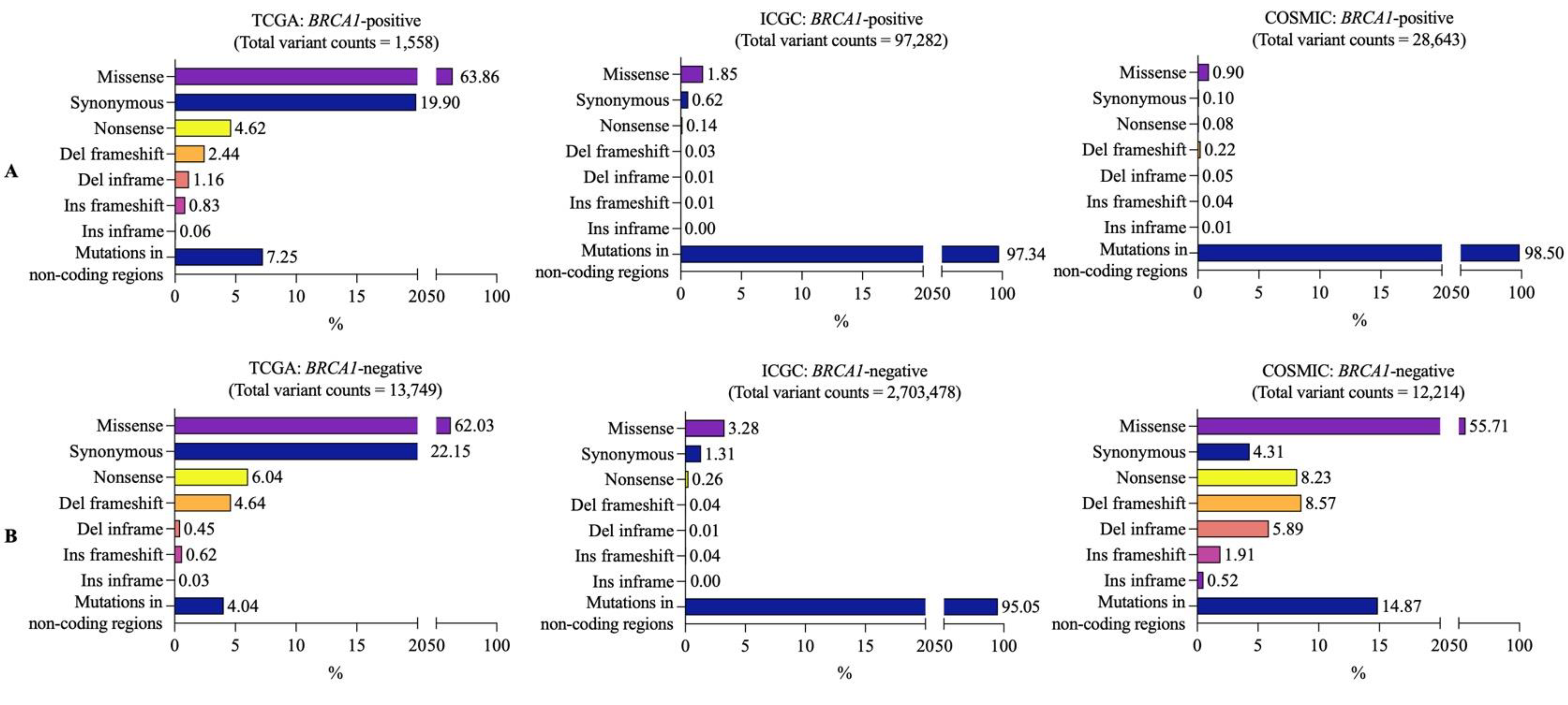
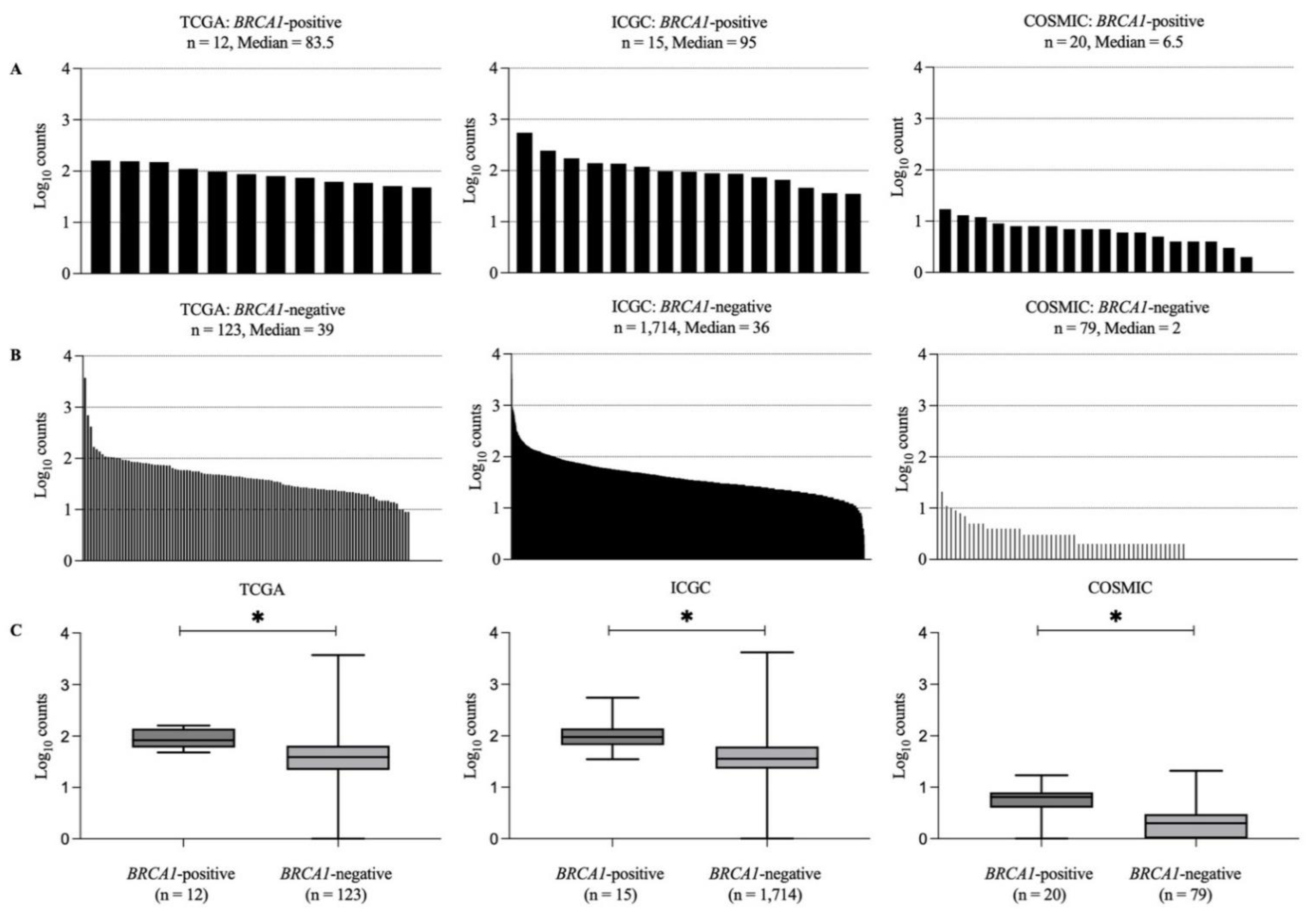
3.2. Mutational Landscapes of BRCA1-Positive and -Negative Samples
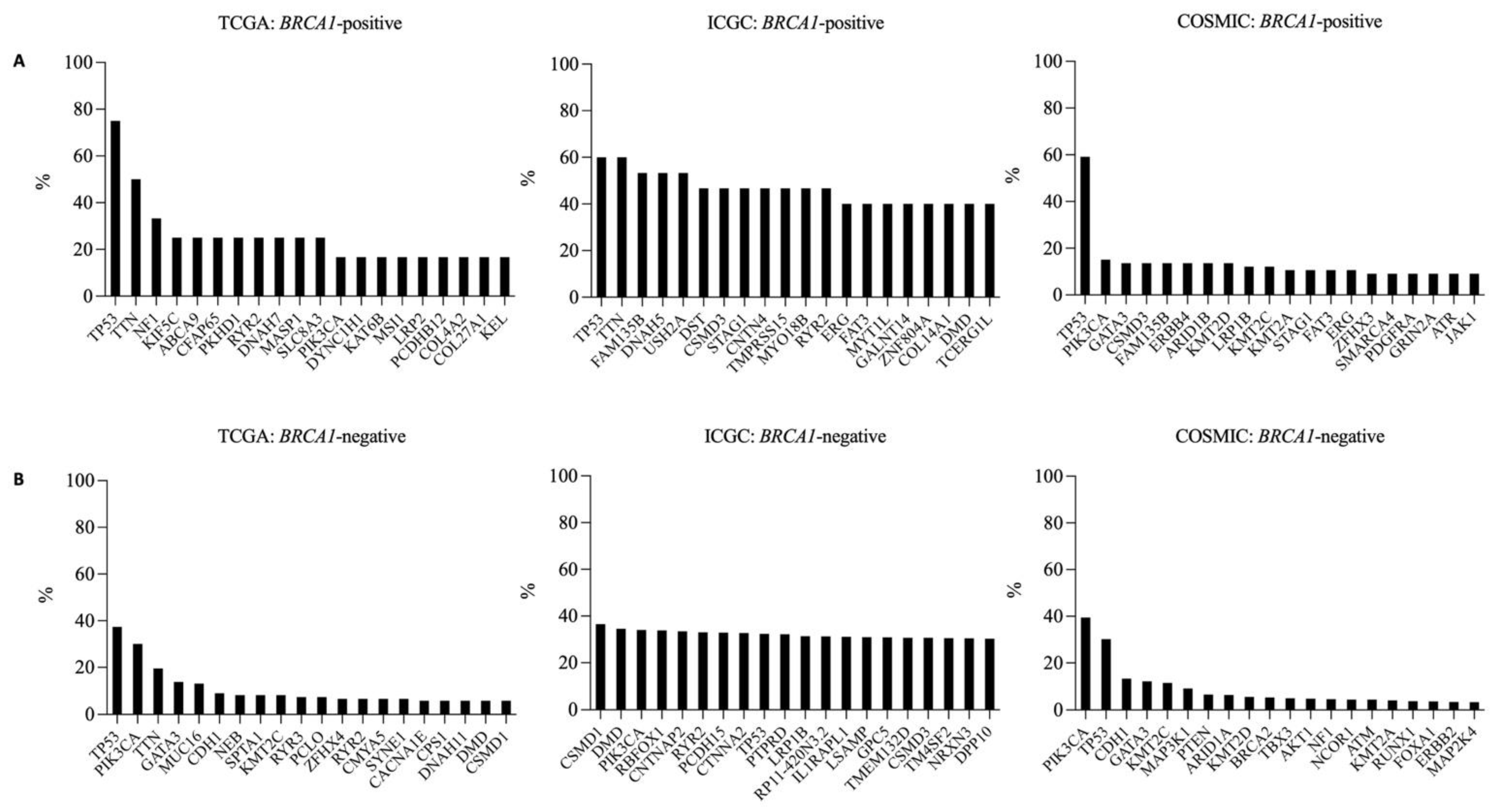
3.3. Frequently Mutated Genes in BRCA1-Positive and -Negative Breast Cancer Samples
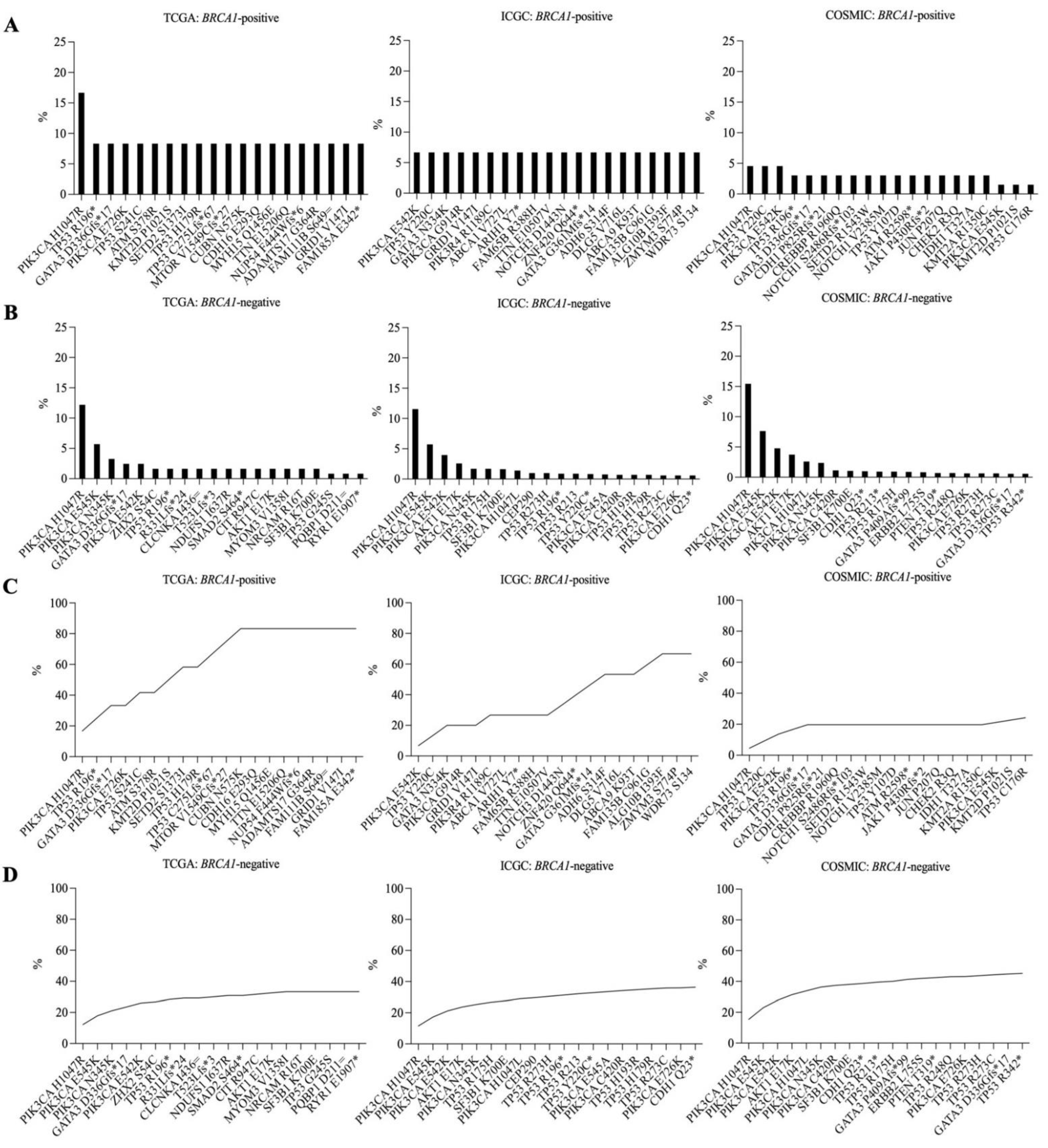
3.4. Recurrent Somatic Mutations in BRCA1-Positive and BRCA1-Negative Breast Cancer Samples
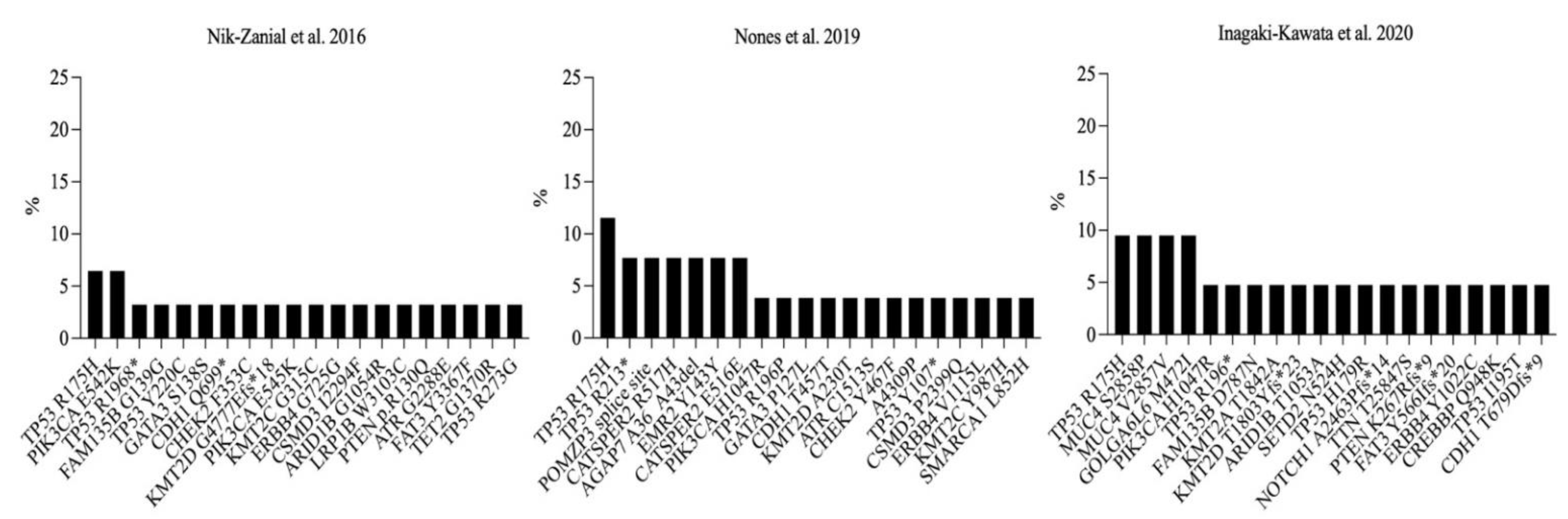
3.5. Recurrent Somatic Mutations in Germline BRCA1-Mutated Breast Cancer Samples
3.6. Predicted Antigenicity of Top Recurrent Mutations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Efremova, M.; Finotello, F.; Rieder, D.; Trajanoski, Z. Neoantigens Generated by Individual Mutations and Their Role in Cancer Immunity and Immunotherapy. Front. Immunol. 2017, 8, 1679. [Google Scholar] [CrossRef] [PubMed]
- Feola, S.; Chiaro, J.; Martins, B.; Cerullo, V. Uncovering the Tumor Antigen Landscape: What to Know about the Discovery Process. Cancers 2020, 12, 1660. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, R.E.; Jansen, K. Turning the corner on therapeutic cancer vaccines. NPJ Vaccines 2019, 4, 7. [Google Scholar] [CrossRef]
- Blass, E.; Ott, P.A. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 2021, 18, 215–229. [Google Scholar] [CrossRef]
- Li, L.; Goedegebuure, S.P.; Gillanders, W.E. Preclinical and clinical development of neoantigen vaccines. Ann. Oncol. 2017, 28, xii11–xii17. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Mo, Y.; Wang, Y.; Wu, P.; Zhang, Y.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; Li, X.; et al. Neoantigen vaccine: An emerging tumor immunotherapy. Mol. Cancer 2019, 18, 128. [Google Scholar] [CrossRef] [PubMed]
- Roudko, V.; Greenbaum, B.; Bhardwaj, N. Computational Prediction and Validation of Tumor-Associated Neoantigens. Front. Immunol. 2020, 11, 27. [Google Scholar] [CrossRef]
- Shemesh, C.S.; Hsu, J.C.; Hosseini, I.; Shen, B.Q.; Rotte, A.; Twomey, P.; Girish, S.; Wu, B. Personalized Cancer Vaccines: Clinical Landscape, Challenges, and Opportunities. Mol. Ther. 2021, 29, 555–570. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Hu, C.; Polley, E.C.; Yadav, S.; Lilyquist, J.; Shimelis, H.; Na, J.; Hart, S.N.; Goldgar, D.E.; Shah, S.; Pesaran, T.; et al. The Contribution of Germline Predisposition Gene Mutations to Clinical Subtypes of Invasive Breast Cancer From a Clinical Genetic Testing Cohort. J. Natl. Cancer Inst. 2020, 112, 1231–1241. [Google Scholar] [CrossRef]
- Kurian, A.W. BRCA1 and BRCA2 mutations across race and ethnicity: Distribution and clinical implications. Curr. Opin. Obstet. Gynecol. 2010, 22, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Waddell, N.; Arnold, J.; Cocciardi, S.; da Silva, L.; Marsh, A.; Riley, J.; Johnstone, C.N.; Orloff, M.; Assie, G.; Eng, C.; et al. Subtypes of familial breast tumours revealed by expression and copy number profiling. Breast Cancer Res. Treat. 2010, 123, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Barrowdale, D.; Andrulis, I.L.; Domchek, S.M.; Eccles, D.; Nevanlinna, H.; Ramus, S.J.; Spurdle, A.; Robson, M.; Sherman, M.; et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: Results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol. Biomark. Prev. 2012, 21, 134–147. [Google Scholar] [CrossRef]
- Atchley, D.P.; Albarracin, C.T.; Lopez, A.; Valero, V.; Amos, C.I.; Gonzalez-Angulo, A.M.; Hortobagyi, G.N.; Arun, B.K. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J. Clin. Oncol. 2008, 26, 4282–4288. [Google Scholar] [CrossRef]
- Tung, N.M.; Robson, M.E.; Ventz, S.; Santa-Maria, C.A.; Nanda, R.; Marcom, P.K.; Shah, P.D.; Ballinger, T.J.; Yang, E.S.; Vinayak, S.; et al. TBCRC 048: Phase II Study of Olaparib for Metastatic Breast Cancer and Mutations in Homologous Recombination-Related Genes. J. Clin. Oncol. 2020, 38, 4274–4282. [Google Scholar] [CrossRef]
- Swisher, E.M.; Lin, K.K.; Oza, A.M.; Scott, C.L.; Giordano, H.; Sun, J.; Konecny, G.E.; Coleman, R.L.; Tinker, A.V.; O’Malley, D.M.; et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): An international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017, 18, 75–87. [Google Scholar] [CrossRef]
- Zhao, W.; Wu, J.; Chen, S.; Zhou, Z. Shared neoantigens: Ideal targets for off-the-shelf cancer immunotherapy. Pharmacogenomics 2020, 21, 637–645. [Google Scholar] [CrossRef]
- Klebanoff, C.A.; Wolchok, J.D. Shared cancer neoantigens: Making private matters public. J. Exp. Med. 2018, 215, 5–7. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, T.; Bunse, L.; Pusch, S.; Sahm, F.; Wiestler, B.; Quandt, J.; Menn, O.; Osswald, M.; Oezen, I.; Ott, M.; et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature 2014, 512, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, S.; Qu, R.; Li, B. Recurrent Neoantigens in Colorectal Cancer as Potential Immunotherapy Targets. BioMed Res. Int. 2020, 2020, 2861240. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, Q.; Wu, R.; Li, B.; Chen, Q.; Zhang, X.; Shi, C. A Comprehensive Survey of Genomic Alterations in Gastric Cancer Reveals Recurrent Neoantigens as Potential Therapeutic Targets. BioMed Res. Int. 2019, 2019, 2183510. [Google Scholar] [CrossRef] [PubMed]
- Pozdeyev, N.; Erickson, T.A.; Zhang, L.; Ellison, K.; Rivard, C.J.; Sams, S.; Hirsch, F.R.; Haugen, B.R.; French, J.D. Comprehensive Immune Profiling of Medullary Thyroid Cancer. Thyroid 2020, 30, 1263–1279. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Wald, A.I.; Roy, S.; Durso, M.B.; Nikiforov, Y.E. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J. Clin. Endocrinol. Metab. 2013, 98, E1852–E1860. [Google Scholar] [CrossRef]
- Roudko, V.; Bozkus, C.C.; Orfanelli, T.; McClain, C.B.; Carr, C.; O’Donnell, T.; Chakraborty, L.; Samstein, R.; Huang, K.L.; Blank, S.V.; et al. Shared Immunogenic Poly-Epitope Frameshift Mutations in Microsatellite Unstable Tumors. Cell 2020, 183, 1634–1649.e17. [Google Scholar] [CrossRef]
- Sharma, A.; Koldovsky, U.; Xu, S.; Mick, R.; Roses, R.; Fitzpatrick, E.; Weinstein, S.; Nisenbaum, H.; Levine, B.L.; Fox, K.; et al. HER-2 pulsed dendritic cell vaccine can eliminate HER-2 expression and impact ductal carcinoma in situ. Cancer 2012, 118, 4354–4362. [Google Scholar] [CrossRef]
- Czerniecki, B.J.; Koski, G.K.; Koldovsky, U.; Xu, S.; Cohen, P.A.; Mick, R.; Nisenbaum, H.; Pasha, T.; Xu, M.; Fox, K.R.; et al. Targeting HER-2/neu in early breast cancer development using dendritic cells with staged interleukin-12 burst secretion. Cancer Res. 2007, 67, 1842–1852. [Google Scholar] [CrossRef]
- Blixt, O.; Bueti, D.; Burford, B.; Allen, D.; Julien, S.; Hollingsworth, M.; Gammerman, A.; Fentiman, I.; Taylor-Papadimitriou, J.; Burchell, J.M. Autoantibodies to aberrantly glycosylated MUC1 in early stage breast cancer are associated with a better prognosis. Breast Cancer Res. 2011, 13, R25. [Google Scholar] [CrossRef] [Green Version]
- Goydos, J.S.; Elder, E.; Whiteside, T.L.; Finn, O.J.; Lotze, M.T. A phase I trial of a synthetic mucin peptide vaccine. Induction of specific immune reactivity in patients with adenocarcinoma. J. Surg. Res. 1996, 63, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Liu, S.; Zhao, L.; Sun, H.X. A Comprehensive Survey of Genomic Mutations in Breast Cancer Reveals Recurrent Neoantigens as Potential Therapeutic Targets. Front. Oncol. 2022, 12, 786438. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef]
- Nones, K.; Johnson, J.; Newell, F.; Patch, A.M.; Thorne, H.; Kazakoff, S.H.; de Luca, X.M.; Parsons, M.T.; Ferguson, K.; Reid, L.E.; et al. Whole-genome sequencing reveals clinically relevant insights into the aetiology of familial breast cancers. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Inagaki-Kawata, Y.; Yoshida, K.; Kawaguchi-Sakita, N.; Kawashima, M.; Nishimura, T.; Senda, N.; Shiozawa, Y.; Takeuchi, Y.; Inoue, Y.; Sato-Otsubo, A.; et al. Genetic and clinical landscape of breast cancers with germline BRCA1/2 variants. Commun. Biol. 2020, 3, 578. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Jurtz, V.; Paul, S.; Andreatta, M.; Marcatili, P.; Peters, B.; Nielsen, M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017, 199, 3360–3368. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef]
- Greenbaum, J.; Sidney, J.; Chung, J.; Brander, C.; Peters, B.; Sette, A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 2011, 63, 325–335. [Google Scholar] [CrossRef]
- Gonzalez-Galarza, F.F.; McCabe, A.; Santos, E.; Jones, J.; Takeshita, L.; Ortega-Rivera, N.D.; Cid-Pavon, G.M.D.; Ramsbottom, K.; Ghattaoraya, G.; Alfirevic, A.; et al. Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020, 48, D783–D788. [Google Scholar] [CrossRef]
- Nolan, E.; Savas, P.; Policheni, A.N.; Darcy, P.K.; Vaillant, F.; Mintoff, C.P.; Dushyanthen, S.; Mansour, M.; Pang, J.B.; Fox, S.B.; et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci. Transl. Med. 2017, 9, eaal4922. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Saez, O.; Chic, N.; Pascual, T.; Adamo, B.; Vidal, M.; Gonzalez-Farre, B.; Sanfeliu, E.; Schettini, F.; Conte, B.; Braso-Maristany, F.; et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020, 22, 45. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Holstege, H.; van der Gulden, H.; Treur-Mulder, M.; Zevenhoven, J.; Velds, A.; Kerkhoven, R.M.; van Vliet, M.H.; Wessels, L.F.; Peterse, J.L.; et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 12111–12116. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.T.; Chien, Y.C.; Lin, Y.H.; Wu, H.H.; Lee, D.F.; Yu, Y.L. The Function of the Mutant p53-R175H in Cancer. Cancers 2021, 13, 4088. [Google Scholar] [CrossRef]
- Hsiue, E.H.; Wright, K.M.; Douglass, J.; Hwang, M.S.; Mog, B.J.; Pearlman, A.H.; Paul, S.; DiNapoli, S.R.; Konig, M.F.; Wang, Q.; et al. Targeting a neoantigen derived from a common TP53 mutation. Science 2021, 371, eabc8697. [Google Scholar] [CrossRef]
- Iiizumi, S.; Ohtake, J.; Murakami, N.; Kouro, T.; Kawahara, M.; Isoda, F.; Hamana, H.; Kishi, H.; Nakamura, N.; Sasada, T. Identification of Novel HLA Class II-Restricted Neoantigens Derived from Driver Mutations. Cancers 2019, 11, 266. [Google Scholar] [CrossRef]
- Chandran, S.S.; Ma, J.; Klatt, M.G.; Dundar, F.; Bandlamudi, C.; Razavi, P.; Wen, H.Y.; Weigelt, B.; Zumbo, P.; Fu, S.N.; et al. Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA. Nat. Med. 2022, 28, 946–957. [Google Scholar] [CrossRef]
| TCGA | ICGC | COSMIC | |
|---|---|---|---|
| Sample size (n) | 12 | 15 | 66 |
| Age | |||
| Mean | 59.58 (±12.86) | 58.73 (±16.26) | 54.14 (±14.91) |
| Unknown | 0 | 0 | 38 |
| Sex | |||
| Female | 12 (100%) | 15 (100%) | 65 (98.48%) |
| Male | 0 | 0 | 1 (1.52%) |
| The American Joint Committee on Cancer (AJCC) stage | |||
| I | 3 (25.00%) | 1 (6.67%) | 0 |
| II | 8 (66.67%) | 9 (60.00%) | 3 (4.55%) |
| III | 0 | 5 (33.33%) | 7 (10.61%) |
| IV | 0 | 0 | 4 (6.06%) |
| Unknown | 1 (8.33%) | 0 | 52 (78.78%) |
| Histology type (count/%) | |||
| Infiltrating ductal carcinoma, NOS | 11 (91.67%) | 14 (93.33%) | 20 (30.30%) |
| Lobular carcinoma, NOS | 0 | 0 | 3 (4.55%) |
| Metaplastic carcinoma, NOS | 1 (8.33%) | 1 (6.67%) | 0 |
| Acini cell carcinoma | 0 | 0 | 1 (1.51%) |
| Phyllodes tumor | 0 | 0 | 1 (1.51%) |
| Unknown | 0 | 0 | 42 (63.63%) |
| Molecular subtype (count/%) | |||
| ER –, HER2 – | 0 | 2 (13.33%) | 5 (7.58%) |
| ER –, HER2 + | 0 | 2 (13.33%) | 1 (1.51%) |
| ER +, HER2 – | 0 | 1 (6.67%) | 3 (4.55%) |
| ER +, HER2 + | 0 | 0 | 0 |
| Hormone receptor + | 0 | 0 | 29 (43.94%) |
| Hormone receptor − | 0 | 0 | 0 |
| Unknown | 12 (100%) | 10 (66.67%) | 28 (42.42%) |
| Sequencing data type | |||
| WGS | 0 | 7 (46.67%) | 11 (16.67%) |
| WES | 12 (100%) | 8 (53.33%) | 9 (13.64%) |
| Target sequencing | 0 | 0 | 46 (69.69%) |
| TCGA | ICGC | COSMIC | |
|---|---|---|---|
| Sample size | 123 | 1714 | 3158 |
| Age | |||
| Mean | 58.34 (±13.68) | 56.67 (±13.85) | 59.39 (±12.49) |
| Unknown | 3 | 116 | 2953 |
| Sex | |||
| Female | 119 (96.74%) | 1699 (99.12%) | 2989 (94.65%) |
| Male | 4 (3.25%) | 15 (0.87%) | 169 (5.35%) |
| The American Joint Committee on Cancer (AJCC) stage | |||
| I | 16 (13.00%) | 51 (2.97%) | 314 (9.94%) |
| II | 65 (52.84%) | 96 (5.60%) | 257 (8.14%) |
| III | 36 (29.26%) | 31 (1.80%) | 158 (5.00%) |
| IV | 4 (3.25%) | 4 (0.23%) | 185 (5.86%) |
| Unknown | 2 (1.62%) | 1532 (89.38%) | 2244 (71.06%) |
| Histology type (count/%) | |||
| Infiltrating ductal carcinoma, NOS | 102 (82.92%) | 485 (28.29%) | 1304 (41.29%) |
| Lobular carcinoma, NOS | 15 (12.19%) | 36 (2.10%) | 258 (8.17%) |
| Infiltrating ductal and lobular carcinoma | 2 (1.62%) | 0 | 76 (2.41%) |
| Tubular carcinoma | 0 | 6 (0.35%) | 0 |
| Metaplastic carcinoma, NOS | 2 (1.62%) | 4 (0.23%) | 16 (0.51%) |
| Papillary carcinoma | 1 (0.81%) | 18 (1.05%) | 0 |
| Adenocarcinoma, NOS | 0 | 6 (0.35%) | 0 |
| Mucinous carcinoma | 1 (0.81%) | 13 (0.75%) | 0 |
| Adenoid cystic carcinoma | 0 | 1 (0.05%) | 0 |
| Acini cell carcinoma | 0 | 0 | 13 (0.41%) |
| Carcinoma with neuroendocrine features | 0 | 1 (0.05%) | 0 |
| Unknown | 0 | 1144 (66.74%) | 1491 (47.21%) |
| Molecular subtype (count/%) | |||
| ER−, HER2− | 0 | 0 | 386 (12.22%) |
| ER−, HER2+ | 0 | 0 | 52 (1.65%) |
| ER+, HER2− | 0 | 477 (27.82%) | 0 |
| ER+, HER2+ | 0 | 0 | 19 (0.60%) |
| Hormone receptor+ | 0 | 0 | 1132 (35.85%) |
| Hormone receptor− | 0 | 0 | 0 |
| Unknown | 123 (100%) | 1237 (72.17%) | 1569 (49.68%) |
| Sequencing data type | |||
| WGS | 0 | 679 (49.61%) | 70 (2.22%) |
| WES | 123 (100%) | 1035 (60.38%) | 9 (0.28%) |
| Target sequencing | 0 | 0 | 3079 (97.50%) |
| Somatic Mutation | Peptide | Length | IC50 | Percentile Rank | HLA Type | Allele Frequency |
|---|---|---|---|---|---|---|
| (NetMHCpan BA) | (NetMHCpan EL) | (%) | ||||
| PIK3CA H1047R | YFMKQMNDAR | 10 | 51.2 | 0.51 | HLA-A*33:03 | 3.28 |
| EYFMKQMNDAR | 11 | 37.03 | 0.08 | HLA-A*33:01 | 1.9 | |
| EYFMKQMNDAR | 11 | 64.4 | 0.21 | HLA-A*33:03 | 3.28 | |
| FMKQMNDAR | 9 | 68.14 | 0.54 | HLA-A*33:03 | 3.28 | |
| YFMKQMNDAR | 10 | 65.48 | 0.41 | HLA-A*33:01 | 1.9 | |
| PIK3CA E542K | KITEQEKDFLW | 11 | 73.72 | 0.18 | HLA-B*58:01 | 3.53 |
| PIK3CA E545K | SEITKQEKDFLW | 12 | 47.3 | 0.06 | HLA-B*44:03 | 3.42 |
| ITKQEKDFLW | 10 | 87.18 | 0.35 | HLA-B*15:17 | 1.15 | |
| ITKQEKDFLW | 10 | 14.11 | 0.01 | HLA-B*57:01 | 2.33 | |
| ITKQEKDFLW | 10 | 83.49 | 0.06 | HLA-B*57:02 | 0.28 | |
| ITKQEKDFLW | 10 | 29.83 | 0.03 | HLA-B*57:03 | 0.51 | |
| ITKQEKDFLW | 10 | 49.48 | 0.02 | HLA-B*57:04 | 0.19 | |
| ITKQEKDFLW | 10 | 13.03 | 0.04 | HLA-B*58:01 | 3.53 | |
| SEITKQEKDFLW | 12 | 37.97 | 0.03 | HLA-B*44:02 | 3.76 | |
| PIK3CA N345K | ATYVKVNIR | 9 | 18.66 | 0.04 | HLA-A*31:01 | 2.35 |
| ATYVKVNIR | 9 | 95.92 | 0.34 | HLA-A*68:01 | 3.2 | |
| CATYVKVNIR | 10 | 49.13 | 2.3 | HLA-A*68:01 | 3.2 | |
| IKILCATYVK | 10 | 87.01 | 2.9 | HLA-A*03:02 | 2.33 | |
| IKILCATYVK | 10 | 91.23 | 3 | HLA-A*11:01 | 12.96 | |
| IKILCATYVK | 10 | 91.23 | 3 | HLA-A*11:02 | 1.75 | |
| ILCATYVKV | 9 | 49.53 | 0.7 | HLA-A*02:03 | 3.28 | |
| ILCATYVKV | 9 | 53.62 | 0.8 | HLA-A*02:02 | 1.87 | |
| ILCATYVKV | 9 | 80.57 | 0.58 | HLA-A*02:01 | 15.67 | |
| KILCATYVK | 9 | 16.81 | 0.42 | HLA-A*11:01 | 12.96 | |
| KILCATYVK | 9 | 16.81 | 0.42 | HLA-A*11:02 | 1.75 | |
| KILCATYVK | 9 | 18.78 | 0.32 | HLA-A*03:02 | 2.33 | |
| KILCATYVK | 9 | 39.61 | 0.53 | HLA-A*03:01 | 7.29 | |
| KILCATYVK | 9 | 48.12 | 0.56 | HLA-A*30:01 | 2.98 | |
| KILCATYVK | 9 | 98.38 | 2.1 | HLA-A*31:01 | 2.35 | |
| KILCATYVKV | 10 | 83.05 | 3.1 | HLA-A*02:06 | 1.82 | |
| RIKILCATYVK | 11 | 59.05 | 0.64 | HLA-A*30:01 | 2.98 | |
| TP53 R175H | - | - | - | - | - | - |
| TP53 R196* | - | - | - | - | - | - |
| TP53 Y220C | VVPCEPPEV | 9 | 142.34 | 0.28 | HLA-A*02:06 | 1.82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruangapirom, L.; Sutivijit, N.; Teerapakpinyo, C.; Mutirangura, A.; Doungkamchan, C. Identification of Shared Neoantigens in BRCA1-Related Breast Cancer. Vaccines 2022, 10, 1597. https://doi.org/10.3390/vaccines10101597
Ruangapirom L, Sutivijit N, Teerapakpinyo C, Mutirangura A, Doungkamchan C. Identification of Shared Neoantigens in BRCA1-Related Breast Cancer. Vaccines. 2022; 10(10):1597. https://doi.org/10.3390/vaccines10101597
Chicago/Turabian StyleRuangapirom, Lucksica, Nannapat Sutivijit, Chinachote Teerapakpinyo, Apiwat Mutirangura, and Chatchanan Doungkamchan. 2022. "Identification of Shared Neoantigens in BRCA1-Related Breast Cancer" Vaccines 10, no. 10: 1597. https://doi.org/10.3390/vaccines10101597
APA StyleRuangapirom, L., Sutivijit, N., Teerapakpinyo, C., Mutirangura, A., & Doungkamchan, C. (2022). Identification of Shared Neoantigens in BRCA1-Related Breast Cancer. Vaccines, 10(10), 1597. https://doi.org/10.3390/vaccines10101597







