Computational Based Designing of a Multi-Epitopes Vaccine against Burkholderia mallei
Abstract
1. Introduction
2. Research Methodology
2.1. Complete Proteome Extraction, BPGA Analysis, and Subtractive Proteomics Filters
2.2. Epitopes Selection Phase
2.3. Multi-Epitopes Vaccine Construction Phase
2.4. Molecular Docking Study
2.5. Molecular Dynamic Simulation
2.6. Binding Free Energies Estimation
3. Results
3.1. Complete Proteome Extraction Phase and Bacterial Pan-Genome Analysis Phase
3.2. BPGA Phase and Subtractive Proteomics Filters
3.3. Epitopes Prediction and Prioritization Phase
3.4. T-Cells Epitopes Prediction
3.5. Epitopes Screening Phase
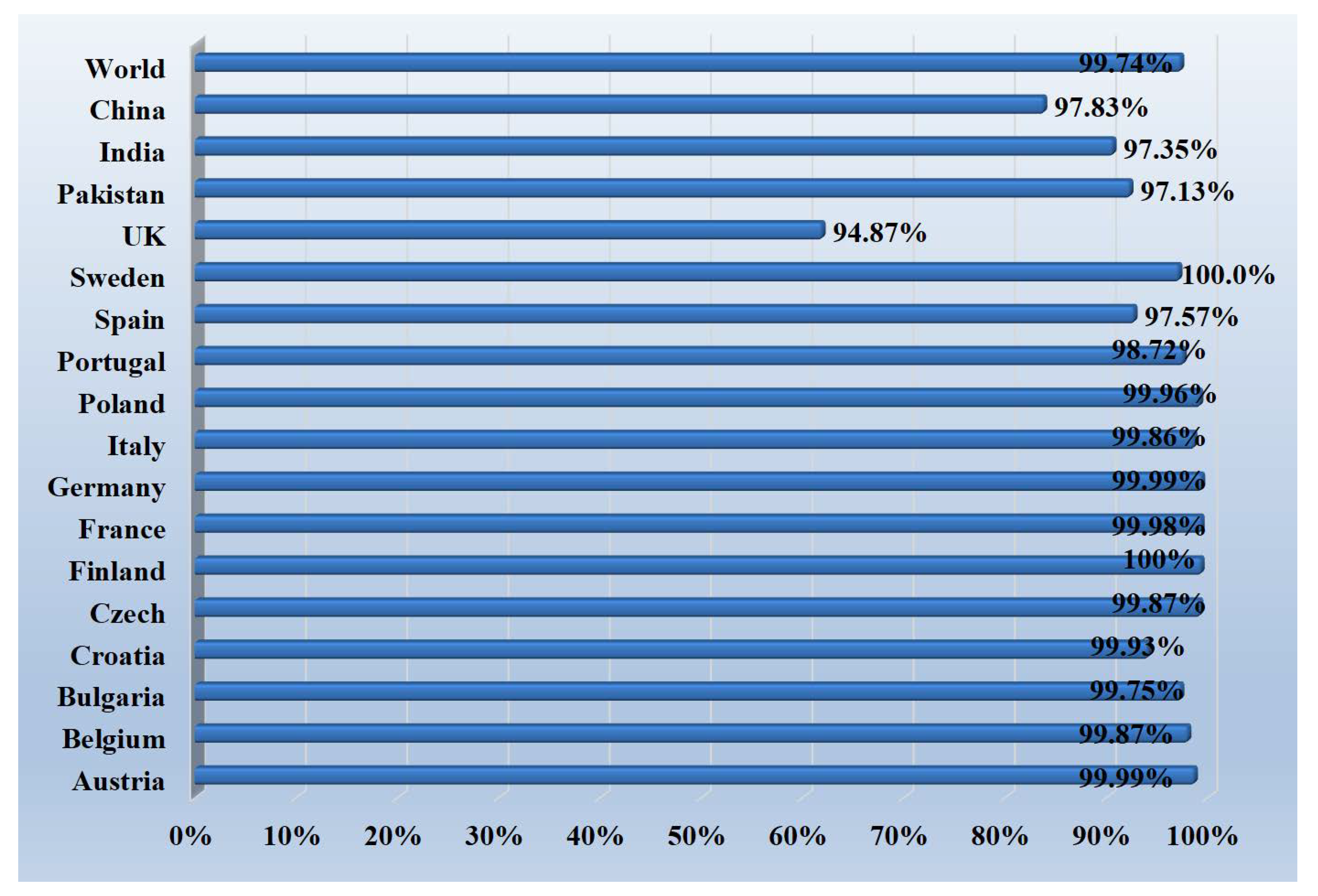
3.6. Population Coverage Analysis
3.7. Multi-Epitopes Vaccine Construction and Processing
3.8. Structure Prediction and Loops Refinement
3.9. Disulfide Engineering and In-Silico Codon Optimization
3.10. Secondary Structure Prediction, Z-Score Calculation and Ramachandran Plot Analysis
3.11. Agreescan3D and CABS-Flex 2.0 Analysis
3.12. Binding Interaction Analysis
3.13. Molecular Dynamic Simulation Analysis
3.14. Binding Free Energy Calculation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hofer, U. The cost of antimicrobial resistance. Nat. Rev. Microbiol. 2019, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, H.P. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: Implications for treatment of melioidosis. Future Microbiol. 2012, 7, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Currie, B.J. Burkholderia pseudomallei and Burkholderia mallei: Melioidosis and glanders. In Mandell, Douglas and Bennett’s Principles and Practice of Infectious Diseases, 7th ed.; Churchill Livingstone Elsevier: Philadelphia, PA, USA, 2010; pp. 2869–2885. [Google Scholar]
- Balaji, V.; Perumalla, S.; Perumal, R.; Inbanathan, F.Y.; Rajamani Sekar, S.K.; Paul, M.M.; Sahni, R.D.; Prakash, J.A.J.; Iyadurai, R. Multi locus sequence typing of Burkholderia pseudomallei isolates from India unveils molecular diversity and confers regional association in Southeast Asia. PLoS Negl. Trop. Dis. 2018, 12, e0006558. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.T.; Sobur, M.A.; Islam, M.S.; Ievy, S.; Hossain, M.J.; El Zowalaty, M.E.; Rahman, A.M.M.T.; Ashour, H.M. Zoonotic diseases: Etiology, impact, and control. Microorganisms 2020, 8, 1405. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, K.A.; Schweizer, H.P. Antibiotic resistance in Burkholderia species. Drug Resist. Updat. 2016, 28, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Saikh, K.U.; Mott, T.M. Innate immune response to Burkholderia mallei. Curr. Opin. Infect. Dis. 2017, 30, 297. [Google Scholar] [CrossRef] [PubMed]
- Vozandychova, V.; Stojkova, P.; Hercik, K.; Rehulka, P.; Stulik, J. The Ubiquitination System within Bacterial Host–Pathogen Interactions. Microorganisms 2021, 9, 638. [Google Scholar] [CrossRef]
- Bambini, S.; Rappuoli, R. The use of genomics in microbial vaccine development. Drug Discov. Today 2009, 14, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Qiu, M.; Zou, S.; Li, Y.; Luo, K.; Chen, R.; Sun, Y.; Wang, K.; Zhuang, X.; Zhang, S. Multi-epitope vaccine design using an immunoinformatics approach for 2019 novel coronavirus in China (SARS-CoV-2). BioRxiv 2020. [Google Scholar] [CrossRef]
- Bidmos, F.A.; Siris, S.; Gladstone, C.A.; Langford, P.R. Bacterial vaccine antigen discovery in the reverse vaccinology 2.0 era: Progress and challenges. Front. Immunol. 2018, 9, 2315. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Azam, S.S. A novel approach of virulome based reverse vaccinology for exploring and validating peptide-based vaccine candidates against the most troublesome nosocomial pathogen: Acinetobacter baumannii. J. Mol. Graph. Model. 2018, 83, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yero, D.; Conchillo-Solé, O.; Daura, X. Antigen discovery in bacterial panproteomes. In Vaccine Delivery Technology; Springer: Berlin/Heidelberg, Germany, 2021; pp. 43–62. [Google Scholar]
- Dalsass, M.; Brozzi, A.; Medini, D.; Rappuoli, R. Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front. Immunol. 2019, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, M.; Alshammari, A.; Alasmari, A.F.; Alharbi, S.M.; Tahir ul Qamar, M.; Ullah, A.; Ahmad, S.; Irfan, M.; Khalil, A.A.K. Designing of a Recombinant Multi-Epitopes Based Vaccine against Enterococcus mundtii Using Bioinformatics and Immunoinformatics Approaches. Int. J. Environ. Res. Public Health 2022, 19, 3729. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.M.; dos Santos Barbosa, M.; Tiwari, S.; Seyffert, N.; de Carvalho Azevedo, V.A.; Nascimento, R.J.M.; de Paula Castro, T.L.; Marchioro, S.B. Immunoinformatic approach for the evaluation of sortase C and E proteins as vaccine targets against caseous lymphadenitis. Inform. Med. Unlocked 2021, 26, 00718. [Google Scholar] [CrossRef]
- Geer, L.Y.; Marchler-Bauer, A.; Geer, R.C.; Han, L.; He, J.; He, S.; Liu, C.; Shi, W.; Bryant, S.H. The NCBI biosystems database. Nucleic Acids Res. 2010, 38, D492–D496. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Tusnady, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef] [PubMed]
- Zaharieva, N.; Dimitrov, I.; Flower, D.R.; Doytchinova, I. Immunogenicity prediction by VaxiJen: A ten year overview. J. Proteom. Bioinform. 2017, 10, 298–310. [Google Scholar]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v. 2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef] [PubMed]
- ProtParam, E. ExPASy-ProtParam Tool. 2017. Available online: https://web.expasy.org/protparam/ (accessed on 15 March 2022).
- Blast, N. Basic local alignment search tool. Natl. Libr. Med. Natl. Cent. Biotechnol. Inf. 2015, 43, D6–D17. [Google Scholar]
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.; Ahmad, S.; Azam, S.S. Vaccinomics to design a novel single chimeric subunit vaccine for broad-spectrum immunological applications targeting nosocomial Enterobacteriaceae pathogens. Eur. J. Pharm. Sci. 2020, 146, 105258. [Google Scholar] [CrossRef]
- Gul, S.; Ahmad, S.; Ullah, A.; Ismail, S.; Khurram, M.; Tahir ul Qamar, M.; Hakami, A.R.; Alkhathami, A.G.; Alrumaihi, F.; Allemailem, K.S. Designing a Recombinant Vaccine against Providencia rettgeri Using Immunoinformatics Approach. Vaccines 2022, 10, 189. [Google Scholar] [CrossRef]
- Ullah, A.; Ahmad, S.; Ismail, S.; Afsheen, Z.; Khurram, M.; Tahir ul Qamar, M.; AlSuhaymi, N.; Alsugoor, M.H.; Allemailem, K.S. Towards A Novel Multi-Epitopes Chimeric Vaccine for Simulating Strong Immune Responses and Protection against Morganella morganii. Int. J. Environ. Res. Public Health 2021, 18, 10961. [Google Scholar] [CrossRef]
- ul Qamar, M.T.; Shahid, F.; Aslam, S.; Ashfaq, U.A.; Aslam, S.; Fatima, I.; Fareed, M.M.; Zohaib, A.; Chen, L.-L. Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2. Infect. Dis. Poverty 2020, 9, 1–14. [Google Scholar] [CrossRef]
- Cheng, J.; Randall, A.Z.; Sweredoski, M.J.; Baldi, P. SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 2005, 33, W72–W76. [Google Scholar] [CrossRef]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef]
- Dorosti, H.; Eslami, M.; Negahdaripour, M.; Ghoshoon, M.B.; Gholami, A.; Heidari, R.; Dehshahri, A.; Erfani, N.; Nezafat, N.; Ghasemi, Y. Vaccinomics approach for developing multi-epitope peptide pneumococcal vaccine. J. Biomol. Struct. Dyn. 2019, 37, 3524–3535. [Google Scholar] [CrossRef]
- Sachdeva, G.; Kumar, K.; Jain, P.; Ramachandran, S.; Ramachandran, S. SPAAN: A software for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics 2004, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Albekairi, T.H.; Alshammari, A.; Alharbi, M.; Alshammary, A.F.; Tahir ul Qamar, M.; Ullah, A.; Irfan, M.; Ahmad, S. Designing of a Novel Multi-Antigenic Epitope-Based Vaccine against E. hormaechei: An Intergraded Reverse Vaccinology and Immunoinformatics Approach. Vaccines 2022, 10, 665. [Google Scholar] [CrossRef]
- Zahroh, H.; Ma’rup, A.; Tambunan, U.S.F.; Parikesit, A.A. Immunoinformatics approach in designing epitopebased vaccine against meningitis-inducing bacteria (Streptococcus pneumoniae, Neisseria meningitidis, and Haemophilus influenzae type b). Drug Target Insights 2016, 10, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Pearlman, D.A.; Case, D.A.; Caldwell, J.W.; Ross, W.S.; Cheatham III, T.E.; DeBolt, S.; Ferguson, D.; Seibel, G.; Kollman, P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995, 91, 1–41. [Google Scholar] [CrossRef]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham III, T.E.; Darden, T.A.; Duke, R.E.; Gohlke, H. The FF14SB force field. Amber 2014, 14, 29–31. [Google Scholar]
- Sargsyan, K.; Grauffel, C.; Lim, C. How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Jaydari, A.; Nazifi, N.; Forouharmehr, A. Computational design of a novel multi-epitope vaccine against Coxiella burnetii. Hum. Immunol. 2020, 81, 596–605. [Google Scholar] [CrossRef] [PubMed]
- Galyov, E.E.; Brett, P.J.; DeShazer, D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu. Rev. Microbiol. 2010, 64, 495–517. [Google Scholar] [CrossRef]
- Sette, A.; Rappuoli, R. Reverse vaccinology: Developing vaccines in the era of genomics. Immunity 2010, 33, 530–541. [Google Scholar] [CrossRef]
- ul Qamar, M.T.; Ahmad, S.; Fatima, I.; Ahmad, F.; Shahid, F.; Naz, A.; Abbasi, S.W.; Khan, A.; Mirza, M.U.; Ashfaq, U.A. Designing multi-epitope vaccine against Staphylococcus aureus by employing subtractive proteomics, reverse vaccinology and immuno-informatics approaches. Comput. Biol. Med. 2021, 132, 104389. [Google Scholar] [CrossRef]




| Target Proteins | Predicted B-Cells Epitopes |
|---|---|
| core/3507/1/Org1_Gene145(type VI secretion system tube protein Hcp | ASQPGAMASGSGGNAGKASF |
| KQYWQQNDNGGKGAEVSVGWNIKE | |
| core/426/1/Org1_Gene4503 (type IV pilus secretin PilQ protein) | EAVASLPPLPVGAPFGWSASASVGAAGRAPLPEAAAPQWRFDSARDPVAGAPSPDVDGGAPAAEFAGEAMPERMP AAPTAEPARSTSADAGTSSAVASAGLQAQ EAALEGPPVPLAPAQRMSDESDEHRSSPPAAGAVSTASVAGTGTETGDPSGDNRPISINLQQAS |
| VAELAERERQRFDAHARAAQLEPLASRG | |
| LAGSAGQRILSKRGSVLA | |
| RGFSRNLGARLALRAPDAGERATGIVAGRNGTLAELAARPISGFDAATAGLTLFAARASRL | |
| SDDRDDVTRVPLL |
| Major Histocompatibility Complex II (MHC-II) | Percentile Score | Major Histocompatibility Complex I(MHC-I) | Percentile Score |
|---|---|---|---|
| ASQPGAMASGSGGN | 6 | ASQPGAMAS | 3.5 |
| AMASGSGGNAGKASF | 8 | ASGSGGNAGK | 0.7 |
| GGKGAEVSVGWNIK | 26 | KGAEVSVGWN | 2.8 |
| QYWQQNDNGGKGAEV | 34 | DNGGKGAEV | 4.2 |
| KQYWQQNDNGGKGAE | 32 | KQYWQQNDN | 9.3 |
| PVGAPFGWSASASVGA | 18 | APFGWSASA | 0.25 |
| GAAGRAPLPEAAAPQWR | 13 | LPEAAAPQW | 0.01 |
| FDSARDPVAGAPSPDVDGG | 10 | DSARDPVAGA | 0.29 |
| DGGAPAAEFAGEAMPERMPAA | 0.58 | AMPERMPAA | 0.06 |
| DAGTSSAVASAGLQAQEAALE | 8.16 | GLQAQEAAL | 0.48 |
| GPPVPLAPAQRMSDESDE | 23 | VPLAPAQRM | 0.01 |
| ESDEHRSSPPAAGAVSTAS | 8.56 | RSSPPAAGA | 0.33 |
| SGDNRPISINLQQAS | 5.4 | DNRPISINL | 0.49 |
| AERERQRFDAHARA | 15 | RQRFDAHAR | 0.21 |
| HARAAQLEPLASRG | 22 | AQLEPLASR | 0.6 |
| AGQRILSKRGSVLA | 2.7 | ILSKRGSVL | 0.7 |
| RGFSRNLGARLALR | 0.01 | RGFSRNLGAR | 0.4 |
| APDAGERATGIVAGRNG | 79 | ERATGIVAGR | 0.6 |
| TLAELAARPISGFD | 20 | LAARPISGF | 0.49 |
| AGLTLFAARASRL | 0.68 | TLFAARASR | 0.01 |
| SDDRDDVTRVPLL | 19 | DDVTRVPLL | 1.3 |
| Selected Epitopes | Predicted IC50 Value (nM) Score | Antigenicity | Allergenicity | Water Solubility | Toxicity |
|---|---|---|---|---|---|
| EAMPERMPAA | 6.28 | 0.7304 | Non-allergen | Good water soluble | Non-toxin |
| RSSPPAAGA | 6.41 | 0.8995 | |||
| DNRPISINL | 17.38 | 1.1305 | |||
| RQRFDAHAR | 9.27 | 0.8286 | |||
| AERERQRFDA | 23.55 | 0.8414 | |||
| HARAAQLEPL | 4.72 | 1.1458 |
| Model | RMSD | Mol Probity | Clash Score | Poor Rotamers | Rama Favored | GALAXY Energy |
|---|---|---|---|---|---|---|
| Initial | 0.000 | 3.689 | 124.8 | 6.6 | 91.4 | 28,723.56 |
| MODEL 1 | 3.679 | 1.487 | 2.9 | 0.0 | 93.8 | −3390.36 |
| MODEL 2 | 3.058 | 1.594 | 3.7 | 0.0 | 93.3 | −3373.64 |
| MODEL 3 | 2.887 | 1.548 | 2.6 | 0.0 | 91.4 | −3361.08 |
| MODEL 4 | 0.992 | 1.654 | 3.4 | 0.0 | 90.9 | −3353.07 |
| MODEL 5 | 3.741 | 1.691 | 4.0 | 0.6 | 91.4 | −3352.74 |
| MODEL 6 | 1.194 | 1.521 | 3.7 | 0.0 | 94.7 | −3348.47 |
| MODEL 7 | 2.506 | 1.642 | 4.3 | 0.0 | 93.3 | −3344.80 |
| MODEL 8 | 0.939 | 1.406 | 2.9 | 0.0 | 95.2 | −3343.12 |
| MODEL 9 | 0.929 | 1.466 | 3.1 | 0.0 | 94.7 | −3341.38 |
| MODEL 10 | 3.104 | 1.494 | 3.4 | 0.0 | 94.7 | −3339.60 |
| Pair of Amino Acid Residues | Chi3 Value | Energy |
|---|---|---|
| PHE9-ALA31 | −65.14 | 5.86 |
| SER16-THR27 | 98.73 | 1.12 |
| ILE38-LEU41 | 87.1 | 2.09 |
| VAL71-GLY75 | 125.46 | 5.24 |
| TRP109-LYS112 | 114.42 | 3.8 |
| ALA123-ALA153 | 72.87 | 3.43 |
| GLU125-ALA131 | 115.02 | 3.97 |
| ALA128-ALA131 | 111.84 | 2.53 |
| PRO143-PRO149 | −66.45 | 4.67 |
| PRO155-ASN160 | 94.44 | 3.65 |
| ILE165-ALA196 | 123.98 | 8.9 |
| GLY168-PRO171 | −114.69 | 4.22 |
| ALA178-ALA187 | 117.98 | 7.08 |
| ALA178-ARG191 | −93.98 | 0.38 |
| GLY182-GLY186 | 103.77 | 0.3 |
| ASP195-PRO198 | 99.88 | 1.93 |
| Cluster | Members | Representative | Weighted Score |
|---|---|---|---|
| 0 | 51 | Center | −773.1 |
| Lowest Energy | −944.1 | ||
| 1 | 47 | Center | −760.8 |
| Lowest Energy | −824.1 | ||
| 2 | 44 | Center | −783.9 |
| Lowest Energy | −798.3 | ||
| 3 | 35 | Center | −753.1 |
| Lowest Energy | −890.3 | ||
| 4 | 34 | Center | −760.4 |
| Lowest Energy | −904.0 | ||
| 5 | 32 | Center | −752.1 |
| Lowest Energy | −952.1 | ||
| 6 | 32 | Center | −833.9 |
| Lowest Energy | −1027.7 | ||
| 7 | 30 | Center | −841.1 |
| Lowest Energy | −841.1 | ||
| 8 | 28 | Center | −725.5 |
| Lowest Energy | −860.2 | ||
| 9 | 26 | Center | −862.9 |
| Lowest Energy | −942.2 | ||
| 10 | 25 | Center | −722.8 |
| Lowest Energy | −933.1 |
| Cluster | Members | Representative | Weighted Score |
|---|---|---|---|
| 0 | 99 | Center | −895.4 |
| Lowest Energy | −975.5 | ||
| 1 | 79 | Center | −920.5 |
| Lowest Energy | −1108.2 | ||
| 2 | 71 | Center | −938.3 |
| Lowest Energy | −1076.4 | ||
| 3 | 62 | Center | −937.8 |
| Lowest Energy | −1232.3 | ||
| 4 | 34 | Center | −990.6 |
| Lowest Energy | −990.6 | ||
| 5 | 30 | Center | −929.9 |
| Lowest Energy | −1043.2 | ||
| 6 | 25 | Center | −984.5 |
| Lowest Energy | −989.1 | ||
| 7 | 22 | Center | −837.2 |
| Lowest Energy | −940.4 | ||
| 8 | 18 | Center | −838.7 |
| Lowest Energy | −953.3 | ||
| 9 | 17 | Center | −995.2 |
| Lowest Energy | −995.2 | ||
| 10 | 17 | Center | −837.4 |
| Lowest Energy | −985.0 |
| Cluster | Members | Representative | Weighted Score |
|---|---|---|---|
| 0 | 89 | Center | −859.1 |
| Lowest Energy | −1067.3 | ||
| 1 | 50 | Center | −888.9 |
| Lowest Energy | −1002.0 | ||
| 2 | 49 | Center | −845.6 |
| Lowest Energy | −946.1 | ||
| 3 | 45 | Center | −859.9 |
| Lowest Energy | −1003.6 | ||
| 4 | 35 | Center | −915.8 |
| Lowest Energy | −1021.9 | ||
| 5 | 28 | Center | −871.6 |
| Lowest Energy | −941.0 | ||
| 6 | 26 | Center | −920.1 |
| Lowest Energy | −966.6 | ||
| 7 | 26 | Center | −853.4 |
| Lowest Energy | −1032.2 | ||
| 8 | 24 | Center | −838.6 |
| Lowest Energy | −974.0 | ||
| 9 | 24 | Center | −906.1 |
| Lowest Energy | −999.5 | ||
| 10 | 23 | Center | −868.2 |
| Lowest Energy | −971.4 |
| Energy Parameter | TLR-4-Vaccine Complex | MHC-I-Vaccine Complex | MHC-II-Vaccine Complex |
|---|---|---|---|
| MM-GBSA | |||
| VDWAALS | −33.5184 | −26.2334 | −22.3071 |
| EEL | −153.63 | −12.0301 | −225.981 |
| EGB | 167.4479 | 24.8686 | 235.4421 |
| ESURF | −4.2856 | −3.4476 | −2.6587 |
| Delta G gas | −187.149 | −38.2635 | −248.288 |
| Delta G solv | 163.1624 | 21.421 | 232.7834 |
| Delta Total | −23.9861 | −16.8425 | −15.5046 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irfan, M.; Khan, S.; Hameed, A.R.; Al-Harbi, A.I.; Abideen, S.A.; Ismail, S.; Ullah, A.; Abbasi, S.W.; Ahmad, S. Computational Based Designing of a Multi-Epitopes Vaccine against Burkholderia mallei. Vaccines 2022, 10, 1580. https://doi.org/10.3390/vaccines10101580
Irfan M, Khan S, Hameed AR, Al-Harbi AI, Abideen SA, Ismail S, Ullah A, Abbasi SW, Ahmad S. Computational Based Designing of a Multi-Epitopes Vaccine against Burkholderia mallei. Vaccines. 2022; 10(10):1580. https://doi.org/10.3390/vaccines10101580
Chicago/Turabian StyleIrfan, Muhammad, Saifullah Khan, Alaa R. Hameed, Alhanouf I. Al-Harbi, Syed Ainul Abideen, Saba Ismail, Asad Ullah, Sumra Wajid Abbasi, and Sajjad Ahmad. 2022. "Computational Based Designing of a Multi-Epitopes Vaccine against Burkholderia mallei" Vaccines 10, no. 10: 1580. https://doi.org/10.3390/vaccines10101580
APA StyleIrfan, M., Khan, S., Hameed, A. R., Al-Harbi, A. I., Abideen, S. A., Ismail, S., Ullah, A., Abbasi, S. W., & Ahmad, S. (2022). Computational Based Designing of a Multi-Epitopes Vaccine against Burkholderia mallei. Vaccines, 10(10), 1580. https://doi.org/10.3390/vaccines10101580







