The Transcriptomic Response of the Murine Thyroid Gland to Iodide Overload and the Role of the Nrf2 Antioxidant System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Next-Generation Messenger RNA (mRNA) Sequencing
2.3. Next-Generation miRNA Sequencing
2.4. Pathway Analysis
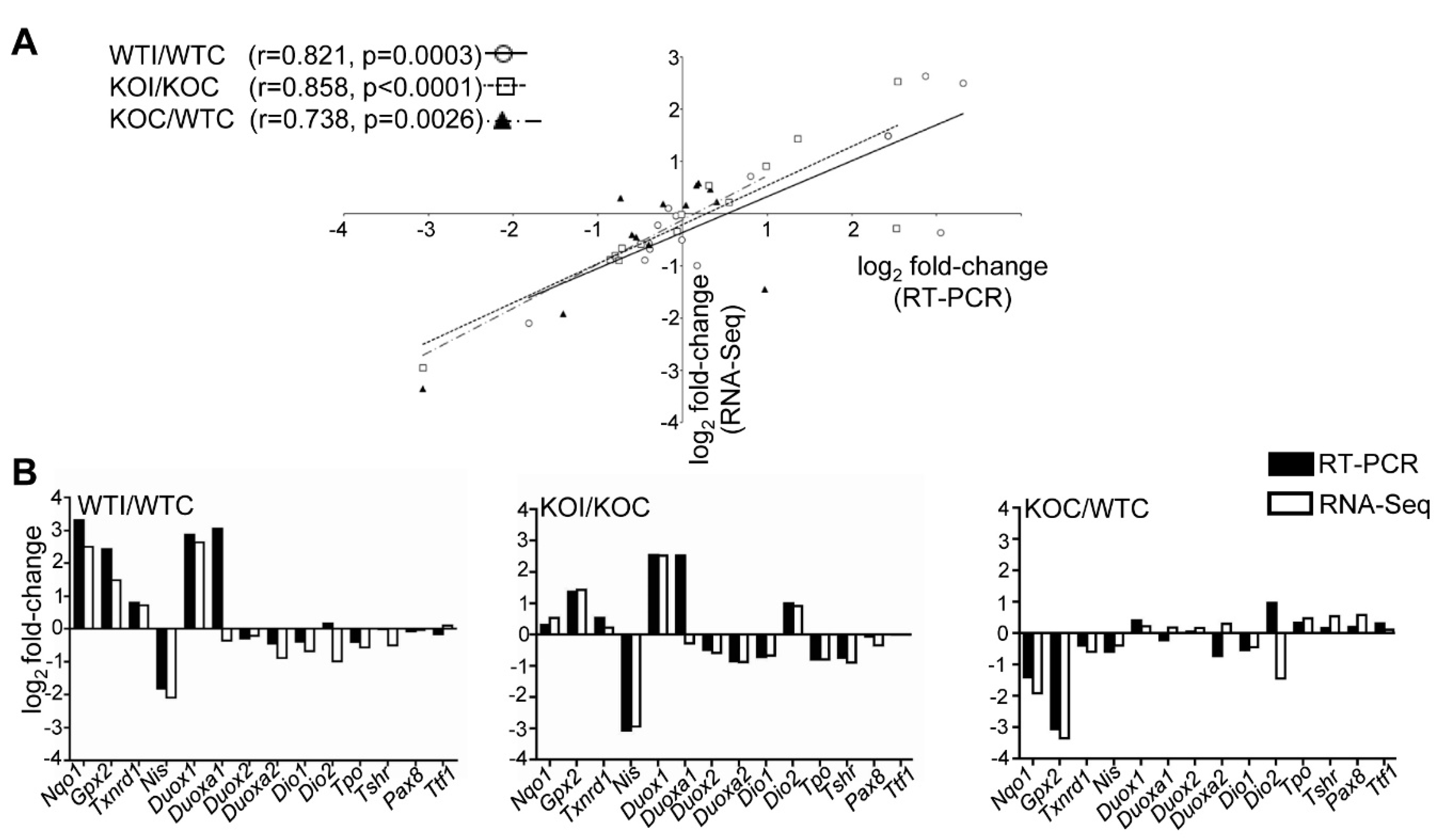
2.5. Real-Time PCR
2.6. Statistics
3. Results
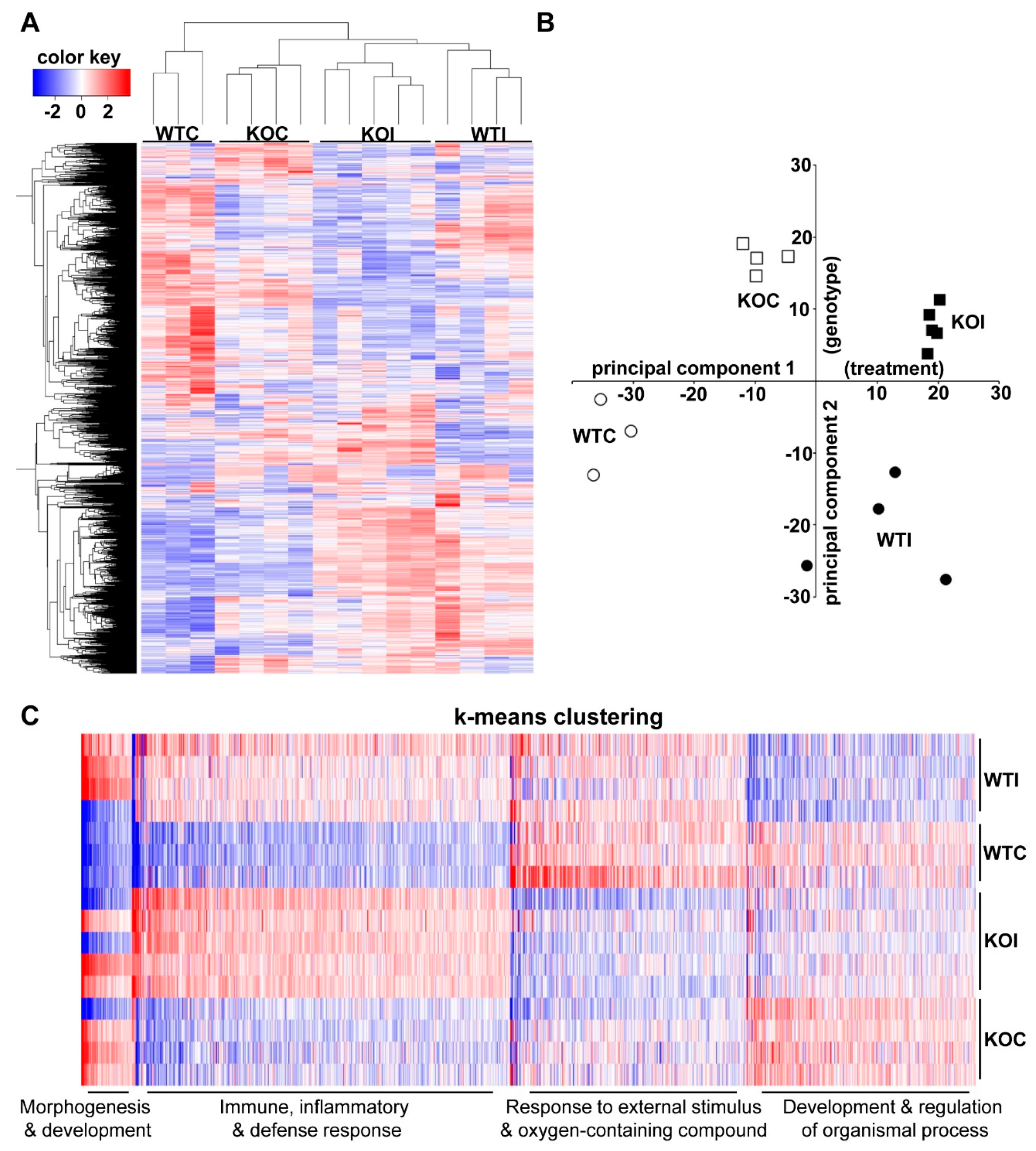
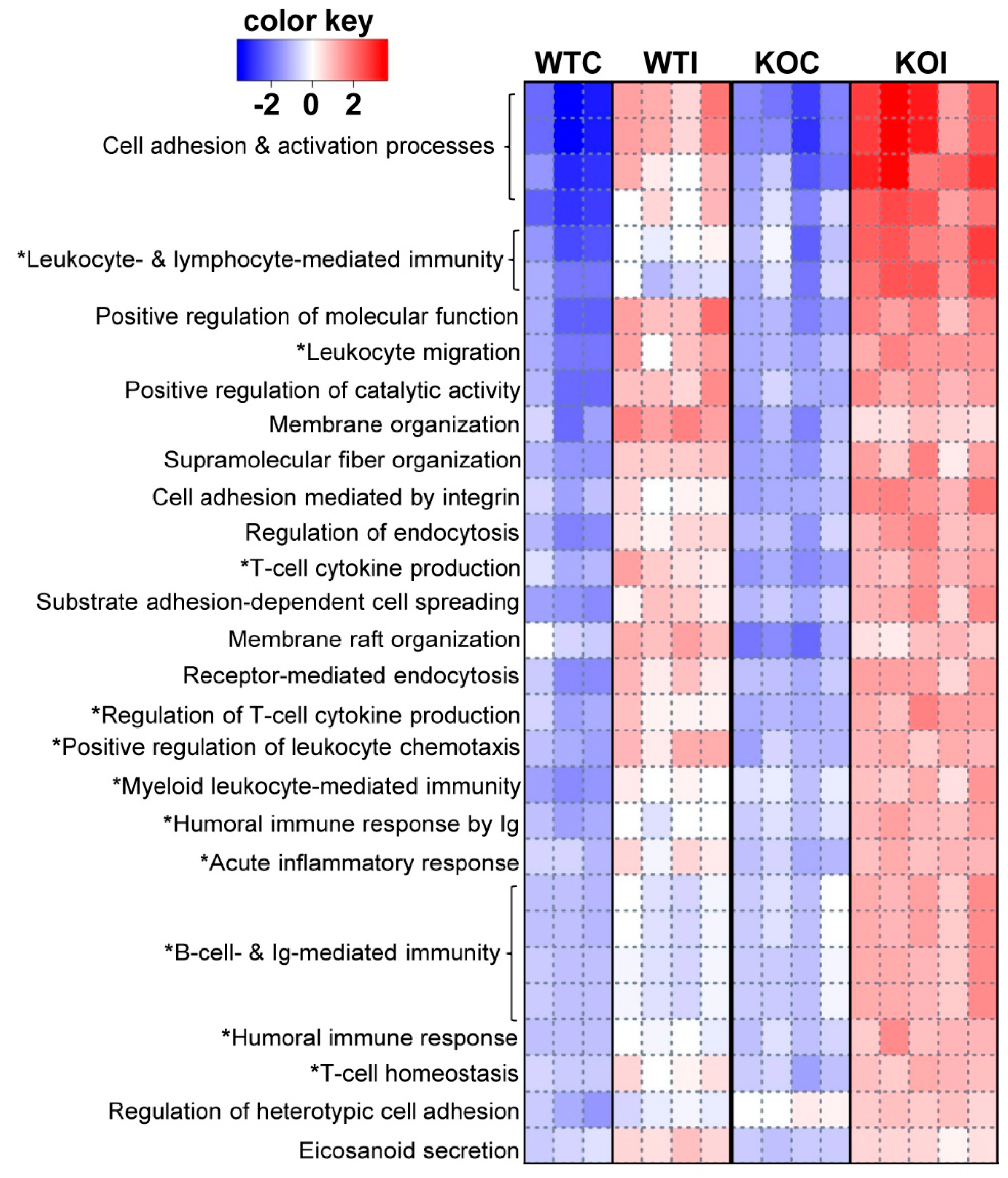
3.1. Iodide Exposure of WT and Nrf2 KO Mice Leads to Distinct Gene Expression Clusters Enriched for Immune, Inflammatory, and Antioxidant Response Functions
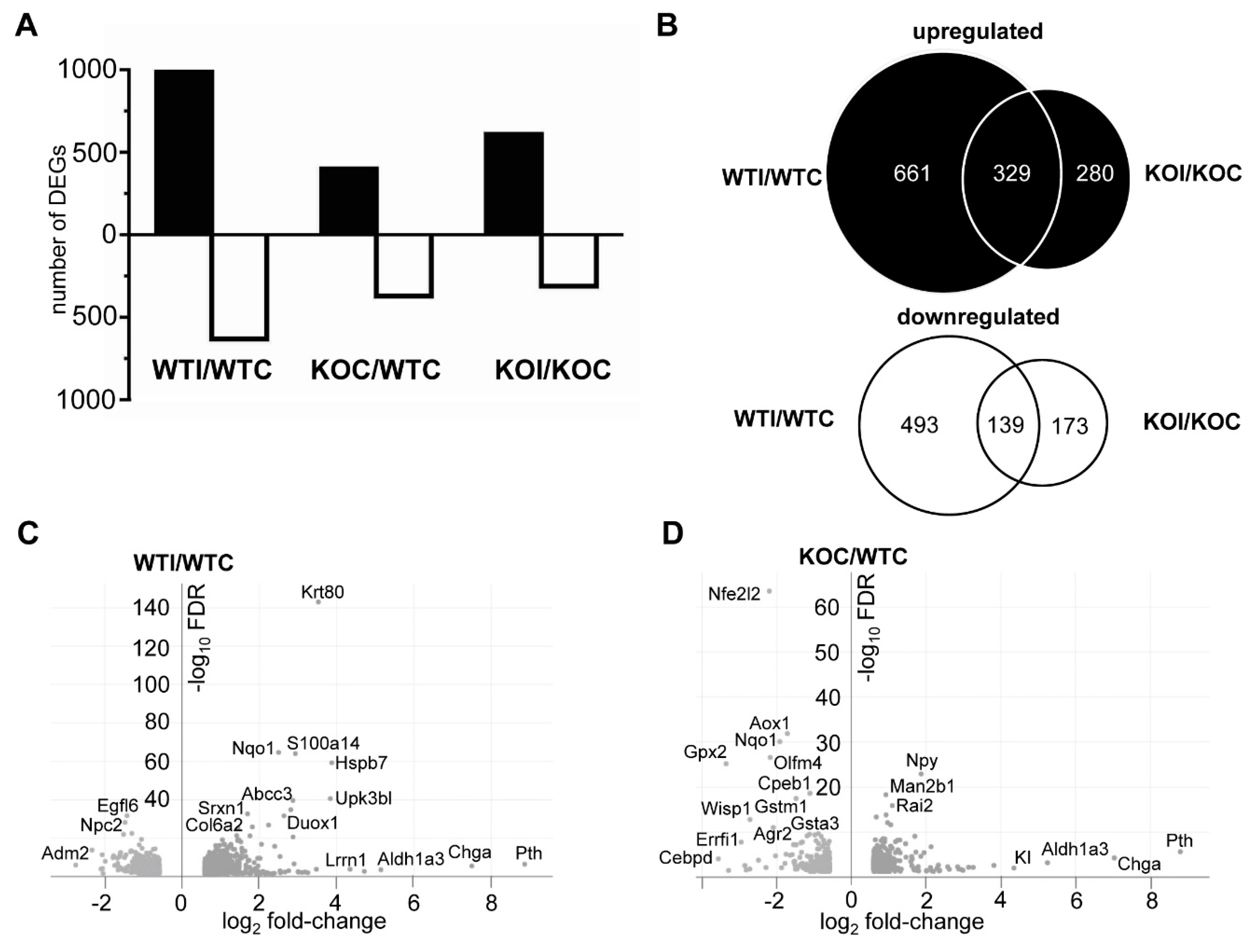
3.2. Iodide Excess Leads to Differential Gene Expression in the Thyroid of WT and Nrf2 KO Mice, in Particular in Cytoprotective/Antioxidant and Thyroid Economy-Related Genes
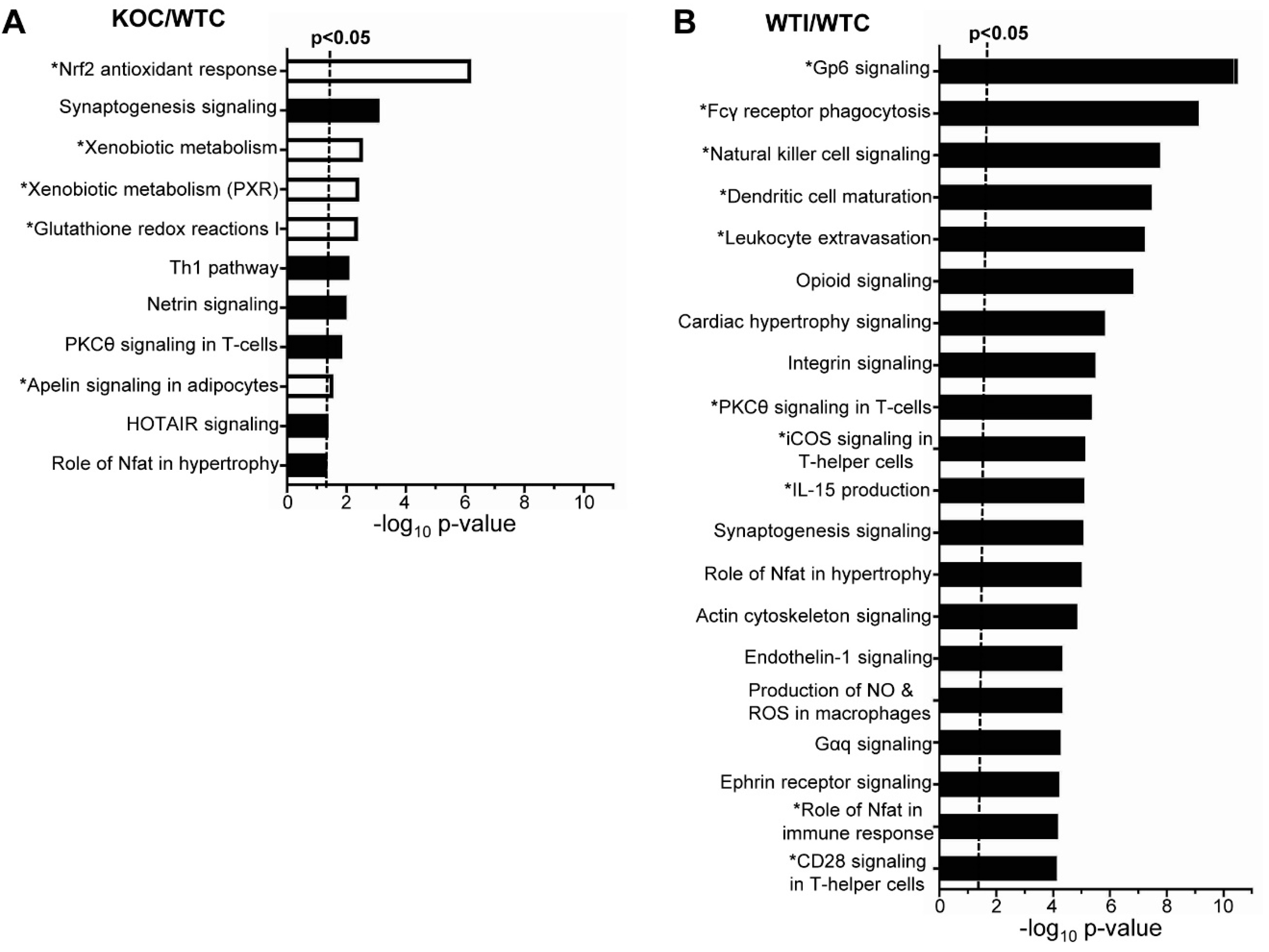
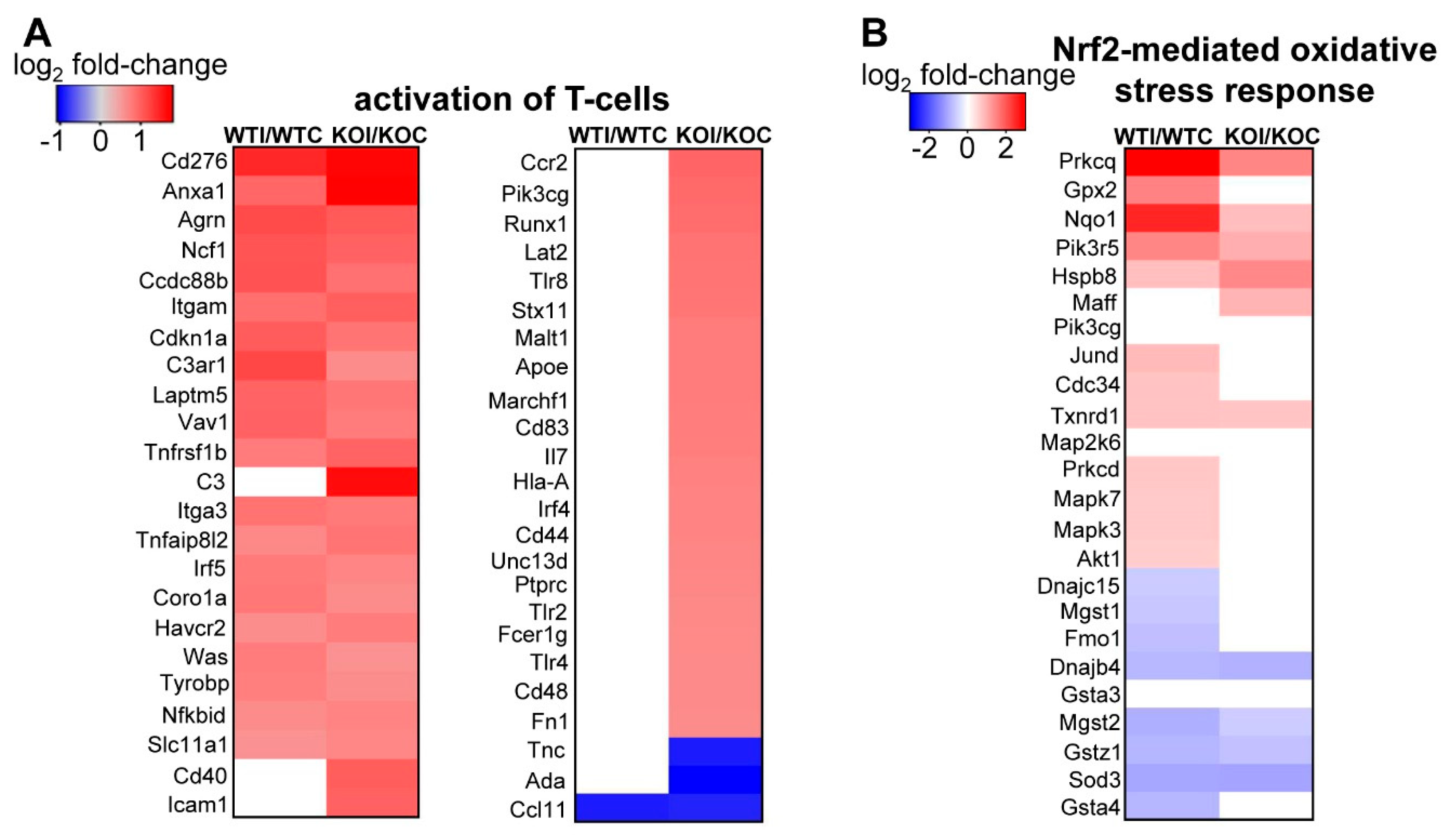
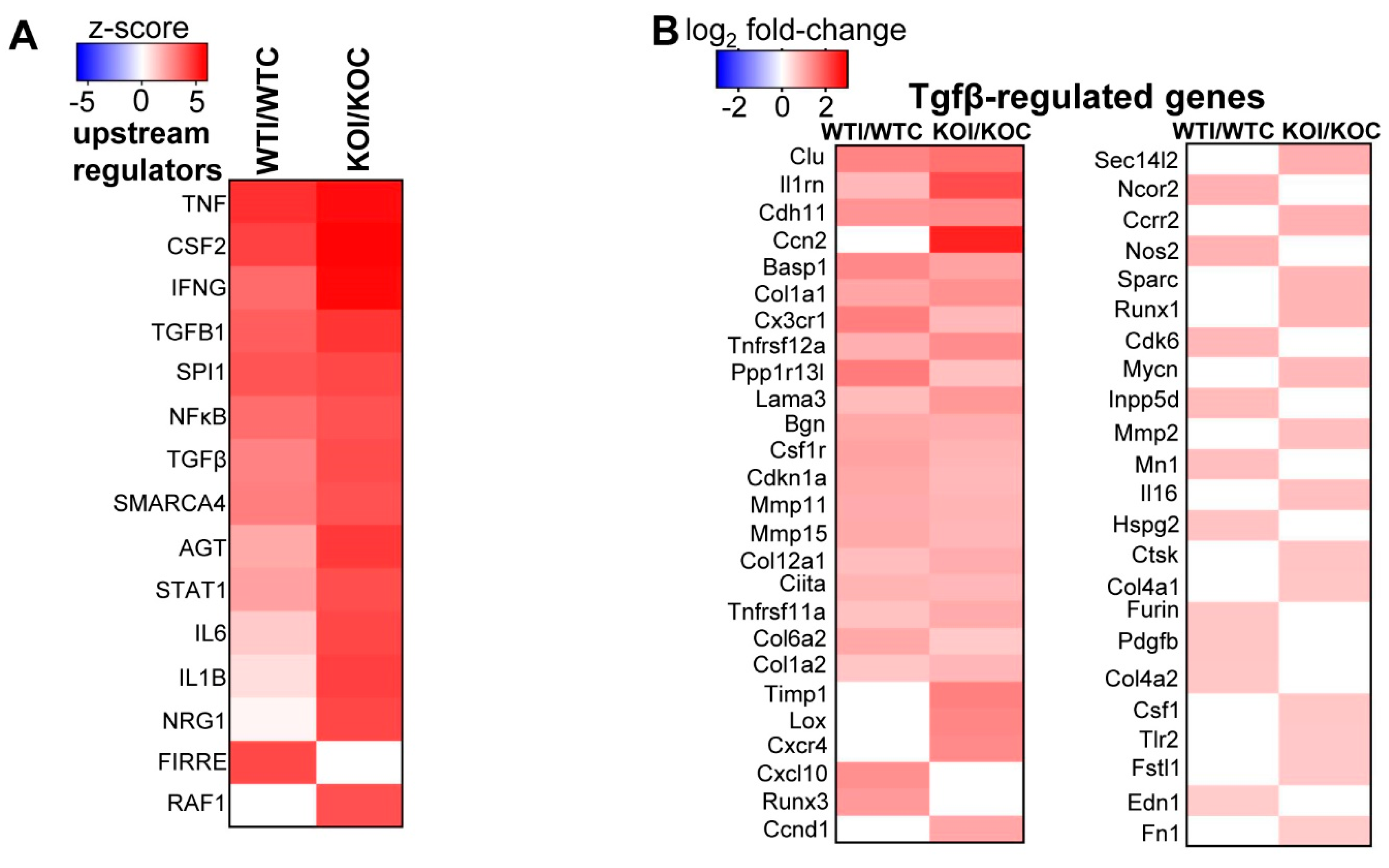
3.3. Nrf2 Mediates the Antioxidant Response and Ameliorates the Inflammatory–Immune–Fibrosis Response to Excess Iodide
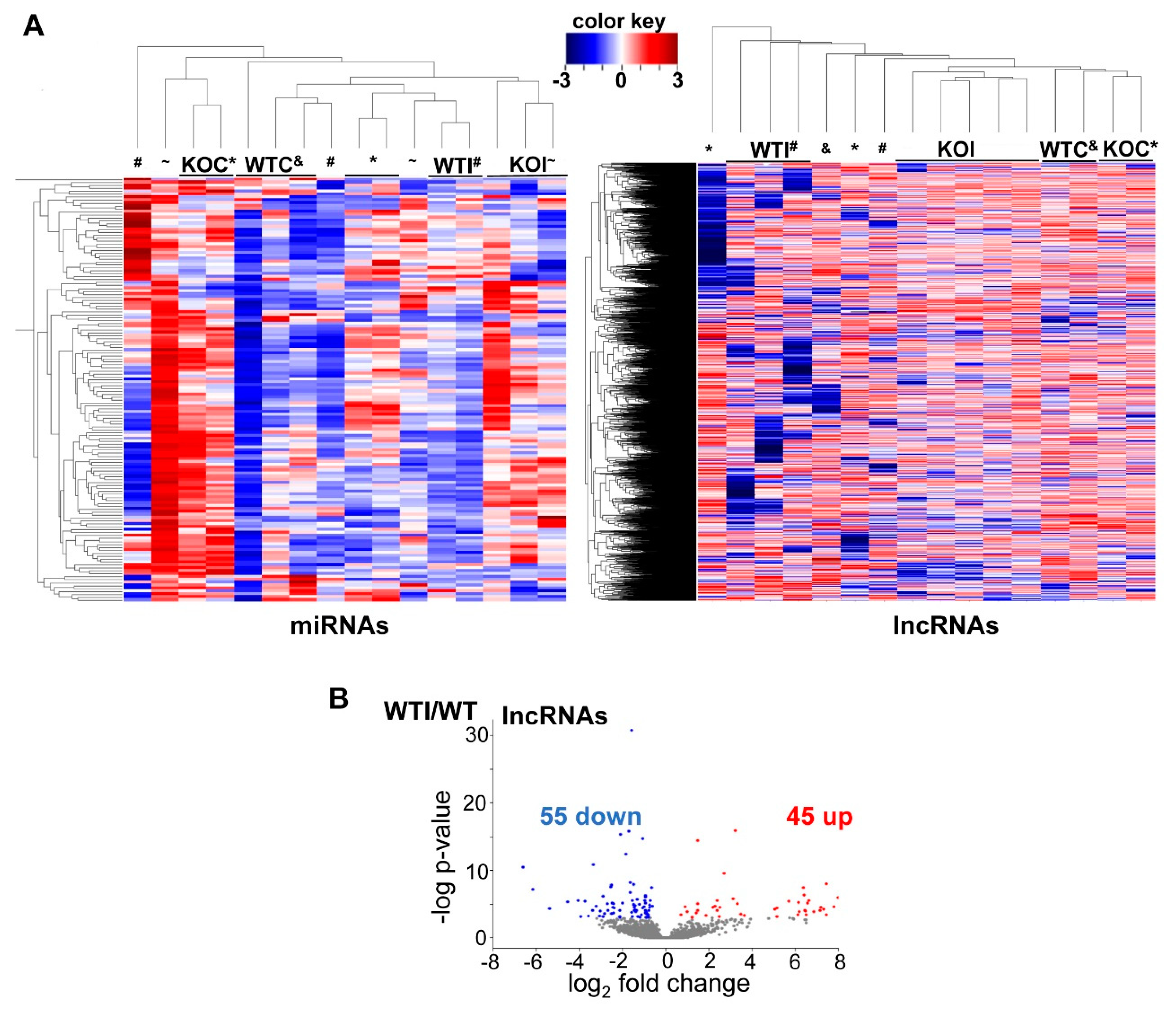
3.4. Iodide Excess Affects the Expression of lncRNAs and miRNAs in Thyroid
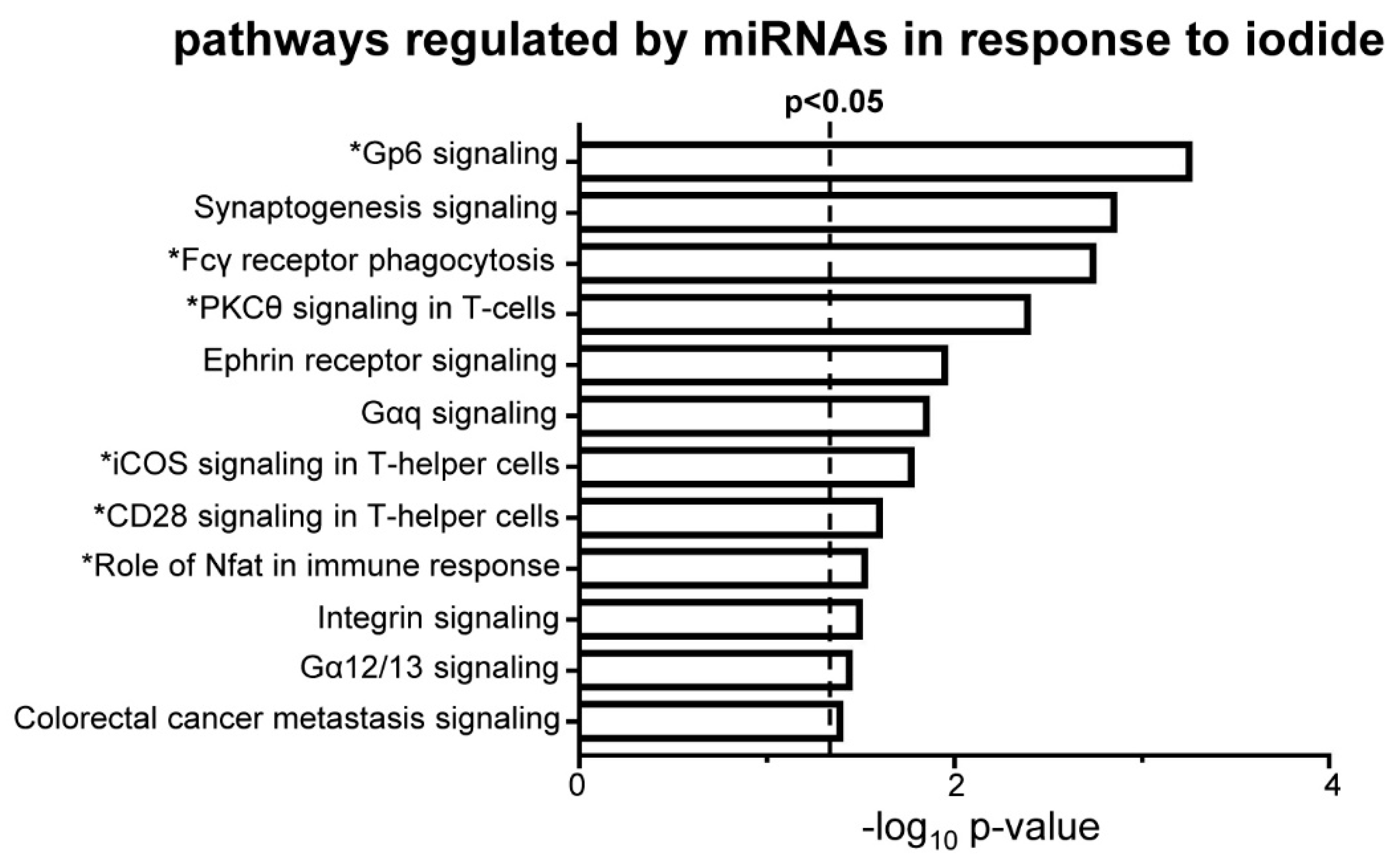
3.5. Iodide Treatment Affects the Expression of miRNAs That Target mRNAs Which Are Part of the Observed Upregulated Inflammatory–Immune Response Pathways
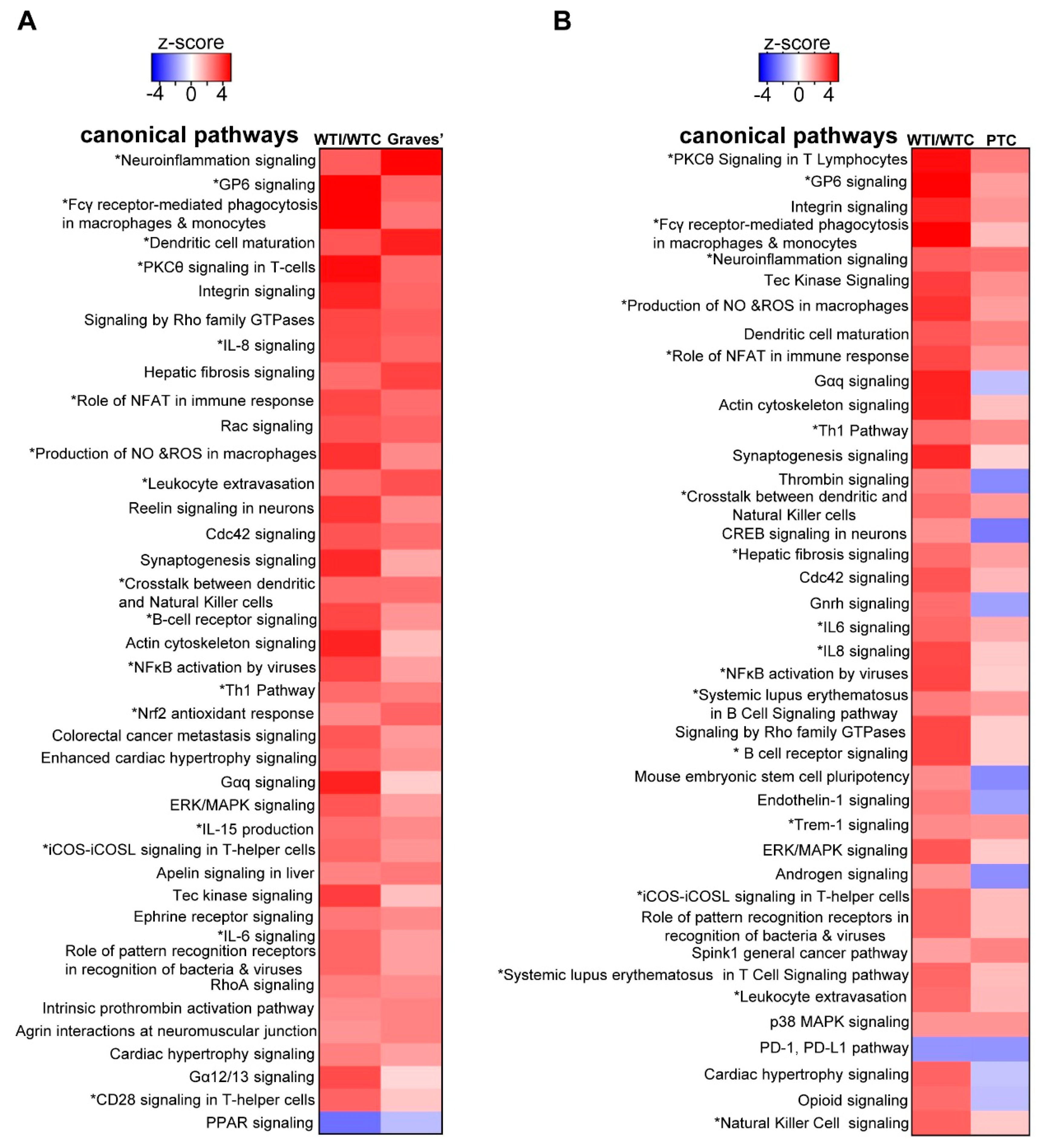
3.6. The Transcriptomic Signature of the Murine Thyroid in Response to Excess Iodide Overlaps Significantly with the Signatures of Graves’ Disease in a Murine Model and of Human Papillary Thyroid Carcinoma (PTC)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Leoni, S.G.; Kimura, E.T.; Santisteban, P.; De la Vieja, A. Regulation of thyroid oxidative state by thioredoxin reductase has a crucial role in thyroid responses to iodide excess. Mol. Endocrinol. 2011, 25, 1924–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziros, P.G.; Habeos, I.G.; Chartoumpekis, D.V.; Ntalampyra, E.; Somm, E.; Renaud, C.O.; Bongiovanni, M.; Trougakos, I.P.; Yamamoto, M.; Kensler, T.W.; et al. NFE2-Related Transcription Factor 2 Coordinates Antioxidant Defense with Thyroglobulin Production and Iodination in the Thyroid Gland. Thyroid 2018, 28, 780–798. [Google Scholar] [CrossRef] [PubMed]
- Vitale, M.; Di Matola, T.; D’Ascoli, F.; Salzano, S.; Bogazzi, F.; Fenzi, G.; Martino, E.; Rossi, G. Iodide excess induces apoptosis in thyroid cells through a p53-independent mechanism involving oxidative stress. Endocrinology 2000, 141, 598–605. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sihvola, V.; Levonen, A.L. Keap1 as the redox sensor of the antioxidant response. Arch Biochem. Biophys. 2017, 617, 94–100. [Google Scholar] [CrossRef]
- Wakabayashi, N.; Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Kang, M.I.; Kobayashi, A.; Yamamoto, M.; Kensler, T.W.; Talalay, P. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 2004, 101, 2040–2045. [Google Scholar] [CrossRef] [Green Version]
- Renaud, C.O.; Ziros, P.G.; Chartoumpekis, D.V.; Bongiovanni, M.; Sykiotis, G.P. Keap1/Nrf2 Signaling: A New Player in Thyroid Pathophysiology and Thyroid Cancer. Front. Endocrinol. 2019, 10, 510. [Google Scholar] [CrossRef]
- Matana, A.; Ziros, P.G.; Chartoumpekis, D.V.; Renaud, C.O.; Polasek, O.; Hayward, C.; Zemunik, T.; Sykiotis, G.P. Rare and common genetic variations in the Keap1/Nrf2 antioxidant response pathway impact thyroglobulin gene expression and circulating levels, respectively. Biochem. Pharmacol. 2020, 173, 113605. [Google Scholar] [CrossRef]
- Wang, T.; Liang, X.; Abeysekera, I.R.; Iqbal, U.; Duan, Q.; Naha, G.; Lin, L.; Yao, X. Activation of the Nrf2-Keap 1 Pathway in Short-Term Iodide Excess in Thyroid in Rats. Oxid. Med. Cell Longev. 2017, 2017, 4383652. [Google Scholar] [CrossRef]
- Ziros, P.G.; Manolakou, S.D.; Habeos, I.G.; Lilis, I.; Chartoumpekis, D.V.; Koika, V.; Soares, P.; Kyriazopoulou, V.E.; Scopa, C.D.; Papachristou, D.J.; et al. Nrf2 is commonly activated in papillary thyroid carcinoma, and it controls antioxidant transcriptional responses and viability of cancer cells. J. Clin. Endocrinol. Metab. 2013, 98, E1422–E1427. [Google Scholar] [CrossRef] [Green Version]
- Danilovic, D.L.S.; de Mello, E.S.; Frazzato, E.S.T.; Wakamatsu, A.; de Lima Jorge, A.A.; Hoff, A.O.; Marui, S. Oncogenic mutations in KEAP1 disturbing inhibitory Nrf2-Keap1 interaction: Activation of antioxidative pathway in papillary thyroid carcinoma. Head Neck 2018, 40, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.D.; Vucic, E.A.; Pikor, L.A.; Thu, K.L.; Hubaux, R.; Lam, W.L. Frequent concerted genetic mechanisms disrupt multiple components of the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in thyroid cancer. Mol. Cancer 2013, 12, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuadrado, A.; Rojo, A.I.; Wells, G.; Hayes, J.D.; Cousin, S.P.; Rumsey, W.L.; Attucks, O.C.; Franklin, S.; Levonen, A.L.; Kensler, T.W.; et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug. Discov. 2019, 18, 295–317. [Google Scholar] [CrossRef] [Green Version]
- Paunkov, A.; Chartoumpekis, D.V.; Ziros, P.G.; Chondrogianni, N.; Kensler, T.W.; Sykiotis, G.P. Impact of Antioxidant Natural Compounds on the Thyroid Gland and Implication of the Keap1/Nrf2 Signaling Pathway. Curr. Pharm. Des. 2019, 25, 1828–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrano-Nascimento, C.; Salgueiro, R.B.; Vitzel, K.F.; Pantaleao, T.; Correa da Costa, V.M.; Nunes, M.T. Iodine excess exposure during pregnancy and lactation impairs maternal thyroid function in rats. Endocr. Connect. 2017, 6, 510–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Mao, C.; Dong, L.; Kang, P.; Ding, C.; Zheng, T.; Wang, X.; Xiao, Y. Excessive Iodine Promotes Pyroptosis of Thyroid Follicular Epithelial Cells in Hashimoto’s Thyroiditis through the ROS-NF-kappaB-NLRP3 Pathway. Front. Endocrinol. 2019, 10, 778. [Google Scholar] [CrossRef]
- Yamazaki, K.; Yamada, E.; Kanaji, Y.; Yanagisawa, T.; Kato, Y.; Takano, K.; Obara, T.; Sato, K. Genes regulated by thyrotropin and iodide in cultured human thyroid follicles: Analysis by cDNA microarray. Thyroid 2003, 13, 149–158. [Google Scholar] [CrossRef]
- Yamada, E.; Yamazaki, K.; Takano, K.; Obara, T.; Sato, K. Iodide inhibits vascular endothelial growth factor-A expression in cultured human thyroid follicles: A microarray search for effects of thyrotropin and iodide on angiogenesis factors. Thyroid 2006, 16, 545–554. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, K.; Tanigawa, K.; Suzuki, K.; Yamada, E.; Yamada, T.; Takano, K.; Obara, T.; Sato, K. Iodide-induced chemokines and genes related to immunological function in cultured human thyroid follicles in the presence of thyrotropin. Thyroid 2010, 20, 67–76. [Google Scholar] [CrossRef]
- Muller, K.; Krohn, K.; Eszlinger, M.; Ludgate, M.; Fuhrer, D. Effect of iodine on early stage thyroid autonomy. Genomics 2011, 97, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Nascimento, C.; Morillo-Bernal, J.; Rosa-Ribeiro, R.; Nunes, M.T.; Santisteban, P. Impaired Gene Expression Due to Iodine Excess in the Development and Differentiation of Endoderm and Thyroid Is Associated with Epigenetic Changes. Thyroid 2020, 30, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Ziros, P.G.; Chartoumpekis, D.V.; Sykiotis, G.P. A Simple Protocol for High Efficiency Protein Isolation After RNA Isolation from Mouse Thyroid and Other Very Small Tissue Samples. Methods Mol. Biol. 2016, 1449, 383–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Nioi, P.; McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: Reassessment of the ARE consensus sequence. Biochem. J. 2003, 374, 337–348. [Google Scholar] [CrossRef]
- Soriano, F.X.; Leveille, F.; Papadia, S.; Higgins, L.G.; Varley, J.; Baxter, P.; Hayes, J.D.; Hardingham, G.E. Induction of sulfiredoxin expression and reduction of peroxiredoxin hyperoxidation by the neuroprotective Nrf2 activator 3H-1,2-dithiole-3-thione. J. Neurochem. 2008, 107, 533–543. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Liu, J.; Li, C.; Peng, X.; Li, H.; Li, Z. Identification and validation of potential target genes in papillary thyroid cancer. Eur. J. Pharmacol. 2019, 843, 217–225. [Google Scholar] [CrossRef] [PubMed]
- McWalter, G.K.; Higgins, L.G.; McLellan, L.I.; Henderson, C.J.; Song, L.; Thornalley, P.J.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Transcription factor Nrf2 is essential for induction of NAD(P)H:quinone oxidoreductase 1, glutathione S-transferases, and glutamate cysteine ligase by broccoli seeds and isothiocyanates. J. Nutr. 2004, 134, 3499S–3506S. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.C.; Cui, J.Y.; Klaassen, C.D. Effect of graded Nrf2 activation on phase-I and -II drug metabolizing enzymes and transporters in mouse liver. PLoS ONE 2012, 7, e39006. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.K.; Finkelman, F.D.; Li, C.W.; Concepcion, E.; Smith, E.; Jacobson, E.; Latif, R.; Keddache, M.; Zhang, W.; Tomer, Y. Genetically driven target tissue overexpression of CD40: A novel mechanism in autoimmune disease. J. Immunol. 2012, 189, 3043–3053. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Jazdzewski, K.; Li, W.; Liyanarachchi, S.; Nagy, R.; Volinia, S.; Calin, G.A.; Liu, C.G.; Franssila, K.; Suster, S.; et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 19075–19080. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, K.; Kensler, T.W. Nrf2 in liver toxicology. Arch. Pharm. Res. 2020, 43, 337–349. [Google Scholar] [CrossRef]
- Ziros, P.G.; Renaud, C.O.; Chartoumpekis, D.V.; Bongiovanni, M.; Habeos, I.; Liao, X.H.; Refetoff, S.; Kopp, P.A.; Brix, K.; Sykiotis, G.P. Mice hypomorphic for Keap1, a negative regulator of the Nrf2 antioxidant response, show age-dependent diffuse goiter with elevated thyrotropin levels. Thyroid 2020. [Google Scholar] [CrossRef]
- Yoh, K.; Itoh, K.; Enomoto, A.; Hirayama, A.; Yamaguchi, N.; Kobayashi, M.; Morito, N.; Koyama, A.; Yamamoto, M.; Takahashi, S. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int. 2001, 60, 1343–1353. [Google Scholar] [CrossRef] [Green Version]
- Thimmulappa, R.K.; Lee, H.; Rangasamy, T.; Reddy, S.P.; Yamamoto, M.; Kensler, T.W.; Biswal, S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Investig. 2006, 116, 984–995. [Google Scholar] [CrossRef] [Green Version]
- Yuasa, R.; Eggo, M.C.; Meinkoth, J.; Dillmann, W.H.; Burrow, G.N. Iodide induces transforming growth factor beta 1 (TGF-beta 1) mRNA in sheep thyroid cells. Thyroid 1992, 2, 141–145. [Google Scholar] [CrossRef]
- Meakin, P.J.; Chowdhry, S.; Sharma, R.S.; Ashford, F.B.; Walsh, S.V.; McCrimmon, R.J.; Dinkova-Kostova, A.T.; Dillon, J.F.; Hayes, J.D.; Ashford, M.L. Susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis upon consumption of a high-fat diet is associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes but not with insulin resistance. Mol. Cell Biol. 2014, 34, 3305–3320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.S.; Harrison, D.J.; Kisielewski, D.; Cassidy, D.M.; McNeilly, A.D.; Gallagher, J.R.; Walsh, S.V.; Honda, T.; McCrimmon, R.J.; Dinkova-Kostova, A.T.; et al. Experimental Nonalcoholic Steatohepatitis and Liver Fibrosis Are Ameliorated by Pharmacologic Activation of Nrf2 (NF-E2 p45-Related Factor 2). Cell Mol. Gastroenterol. Hepatol. 2018, 5, 367–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, X.P.; Park, M.; Hershman, J.M. Transforming growth factor-beta blocks protein kinase-A-mediated iodide transport and protein kinase-C-mediated DNA synthesis in FRTL-5 rat thyroid cells. Endocrinology 1992, 131, 45–50. [Google Scholar] [CrossRef]
- Nicolussi, A.; D’Inzeo, S.; Santulli, M.; Colletta, G.; Coppa, A. TGF-beta control of rat thyroid follicular cells differentiation. Mol. Cell Endocrinol. 2003, 207, 1–11. [Google Scholar] [CrossRef]
- Grubeck-Loebenstein, B.; Buchan, G.; Sadeghi, R.; Kissonerghis, M.; Londei, M.; Turner, M.; Pirich, K.; Roka, R.; Niederle, B.; Kassal, H.; et al. Transforming growth factor beta regulates thyroid growth. Role in the pathogenesis of nontoxic goiter. J. Clin. Investig. 1989, 83, 764–770. [Google Scholar] [CrossRef]
- Liao, T.; Qu, N.; Shi, R.L.; Guo, K.; Ma, B.; Cao, Y.M.; Xiang, J.; Lu, Z.W.; Zhu, Y.X.; Li, D.S.; et al. BRAF-activated LncRNA functions as a tumor suppressor in papillary thyroid cancer. Oncotarget 2017, 8, 238–247. [Google Scholar] [CrossRef] [Green Version]
- Pan, W.; Zhou, L.; Ge, M.; Zhang, B.; Yang, X.; Xiong, X.; Fu, G.; Zhang, J.; Nie, X.; Li, H.; et al. Whole exome sequencing identifies lncRNA GAS8-AS1 and LPAR4 as novel papillary thyroid carcinoma driver alternations. Hum. Mol. Genet. 2016, 25, 1875–1884. [Google Scholar] [CrossRef] [Green Version]
- Landa, I.; Ibrahimpasic, T.; Boucai, L.; Sinha, R.; Knauf, J.A.; Shah, R.H.; Dogan, S.; Ricarte-Filho, J.C.; Krishnamoorthy, G.P.; Xu, B.; et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J. Clin. Investig. 2016, 126, 1052–1066. [Google Scholar] [CrossRef] [Green Version]
- Tartey, S.; Takeuchi, O. Chromatin Remodeling and Transcriptional Control in Innate Immunity: Emergence of Akirin2 as a Novel Player. Biomolecules 2015, 5, 1618–1633. [Google Scholar] [CrossRef] [Green Version]
- Mazeh, H.; Deutch, T.; Karas, A.; Bogardus, K.A.; Mizrahi, I.; Gur-Wahnon, D.; Ben-Dov, I.Z. Next-Generation Sequencing Identifies a Highly Accurate miRNA Panel That Distinguishes Well-Differentiated Thyroid Cancer from Benign Thyroid Nodules. Cancer Epidemiol. Biomark. Prev. 2018, 27, 858–863. [Google Scholar] [CrossRef] [Green Version]
- Labourier, E.; Shifrin, A.; Busseniers, A.E.; Lupo, M.A.; Manganelli, M.L.; Andruss, B.; Wylie, D.; Beaudenon-Huibregtse, S. Molecular Testing for miRNA, mRNA, and DNA on Fine-Needle Aspiration Improves the Preoperative Diagnosis of Thyroid Nodules With Indeterminate Cytology. J. Clin. Endocrinol. Metab. 2015, 100, 2743–2750. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Li, X.; Ma, J.; Shi, C.; Zhu, H.; Xi, Q.; Zhang, J.; Zhao, X.; Gu, M. MiR-204-5p suppresses cell proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma. Biochem. Biophys. Res. Commun. 2015, 457, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Chen, L.; Wang, Y. miR-218 overexpression suppresses tumorigenesis of papillary thyroid cancer via inactivation of PTEN/PI3K/AKT pathway by targeting Runx2. Onco. Targets Ther. 2018, 11, 6305–6316. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.Z.; Bin, H.; Zang, Z.N. MiR-128 suppresses the growth of thyroid carcinoma by negatively regulating SPHK1. Biomed. Pharm. 2019, 109, 1960–1966. [Google Scholar] [CrossRef] [PubMed]
- Leoni, S.G.; Galante, P.A.; Ricarte-Filho, J.C.; Kimura, E.T. Differential gene expression analysis of iodide-treated rat thyroid follicular cell line PCCl3. Genomics 2008, 91, 356–366. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chartoumpekis, D.V.; Ziros, P.G.; Georgakopoulos-Soares, I.; Smith, A.A.T.; Marques, A.C.; Ibberson, M.; A. Kopp, P.; Habeos, I.; Trougakos, I.P.; Khoo, N.K.H.; et al. The Transcriptomic Response of the Murine Thyroid Gland to Iodide Overload and the Role of the Nrf2 Antioxidant System. Antioxidants 2020, 9, 884. https://doi.org/10.3390/antiox9090884
Chartoumpekis DV, Ziros PG, Georgakopoulos-Soares I, Smith AAT, Marques AC, Ibberson M, A. Kopp P, Habeos I, Trougakos IP, Khoo NKH, et al. The Transcriptomic Response of the Murine Thyroid Gland to Iodide Overload and the Role of the Nrf2 Antioxidant System. Antioxidants. 2020; 9(9):884. https://doi.org/10.3390/antiox9090884
Chicago/Turabian StyleChartoumpekis, Dionysios V., Panos G. Ziros, Ilias Georgakopoulos-Soares, Adam A. T. Smith, Ana Claudia Marques, Mark Ibberson, Peter A. Kopp, Ioannis Habeos, Ioannis P. Trougakos, Nicholas K. H. Khoo, and et al. 2020. "The Transcriptomic Response of the Murine Thyroid Gland to Iodide Overload and the Role of the Nrf2 Antioxidant System" Antioxidants 9, no. 9: 884. https://doi.org/10.3390/antiox9090884
APA StyleChartoumpekis, D. V., Ziros, P. G., Georgakopoulos-Soares, I., Smith, A. A. T., Marques, A. C., Ibberson, M., A. Kopp, P., Habeos, I., Trougakos, I. P., Khoo, N. K. H., & Sykiotis, G. P. (2020). The Transcriptomic Response of the Murine Thyroid Gland to Iodide Overload and the Role of the Nrf2 Antioxidant System. Antioxidants, 9(9), 884. https://doi.org/10.3390/antiox9090884







