Epigenetic Regulation of NRF2/KEAP1 by Phytochemicals
Abstract
:1. Introduction
2. Multilayer Regulation of NRF2 Signaling
2.1. Transcriptional Regulation
2.2. Post-Transcriptional Regulation
2.3. Regulation of NRF2 Protein Stability
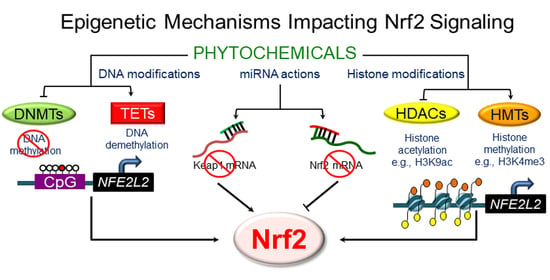
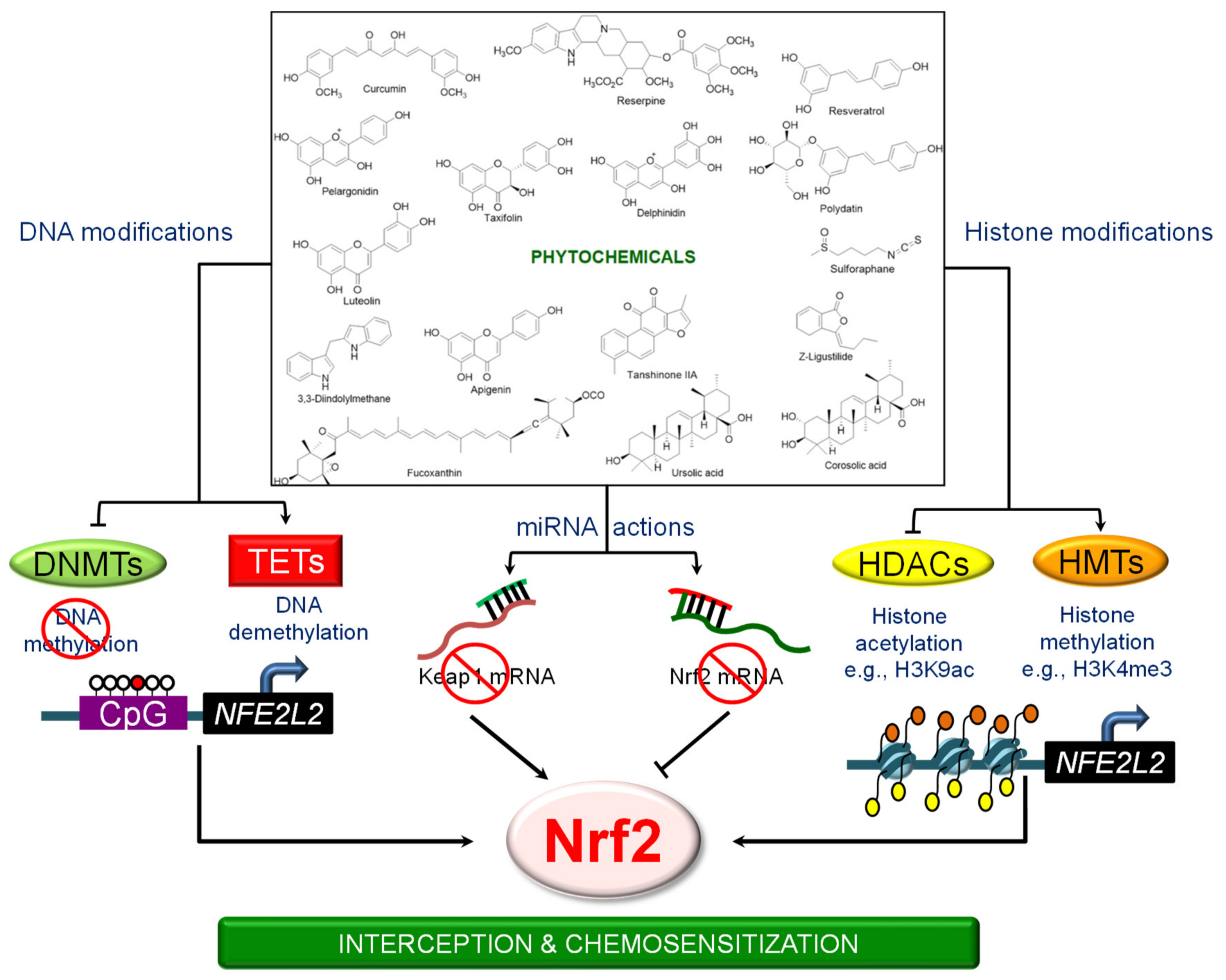
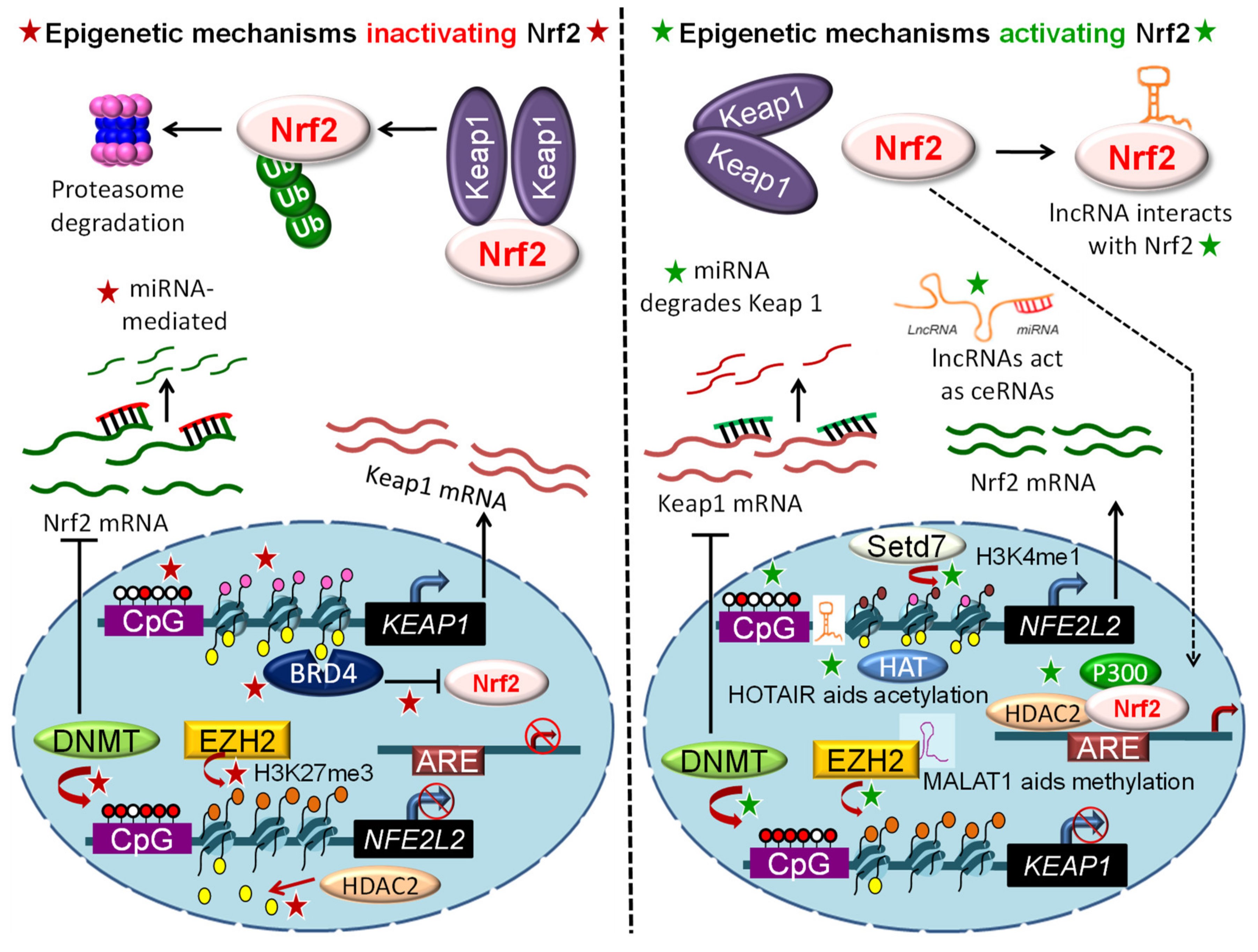
3. Epigenetic Mechanisms Regulating NRF2 Signaling
3.1. DNA Methylation
3.2. Histone Modifications
3.3. Epigenetic “Readers”
3.4. Regulation by Non-Coding RNAs
4. Phytochemicals and the Epigenetic Regulation of NRF2 Signaling
4.1. 3,3′-Diindolylmethane (DIM)
4.2. Apigenin
4.3. Corosolic Acid
4.4. Curcumin
4.5. Delphinidin
4.6. Fucoxanthin
4.7. Luteolin
4.8. Pelargonidin
4.9. Polydatin
4.10. Reserpine
4.11. Resveratrol
4.12. Sulforaphane
4.13. Tanshinone IIA
4.14. Taxifolin
4.15. Ursolic Acid
4.16. γ. Tocopherol–Rich Mixture of Tocopherols (γ-TmT)
4.17. Z-Ligustilide
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Nrf2 | murine nuclear factor erythroid 2-related factor 2 |
| Keap1 | murine Kelch-like ECH-associated protein 1 |
| NRF2 and KEAP1 | the corresponding human proteins |
| NFE2L2 | human gene encoding NRF2 protein |
| Nfe2l2 | murine gene encoding Nrf2 protein |
| KEAP1 | human gene encoding KEAP1 protein |
| Keap1 | murine gene encoding Keap1 protein |
| BET | bromodomain and extraterminal domain |
| miRNA | microRNA |
| lncRNA | long non-coding RNA |
| ARE | antioxidant response element |
| DNMT | DNA methyltransferase |
| HDAC | histone deacetylase |
| TET | ten-eleven translocation |
| AhR | aryl hydrocarbon receptor |
| Arnt | Aryl hydrocarbon receptor nuclear translocator |
| NF-κB | nuclear factor kappa B |
| Cul3 | Cullin 3 |
| Rbx1 | RING-box protein 1 |
| SCF | S-phase kinase-associated protein 1/Cullin/F-box |
| β-TrCP | β-transducin repeats-containing proteins |
| HRD1 | HMG-CoA reductase degradation protein 1 |
| GSK-3β | glycogen synthase kinase-3β |
| ERK | extracellular signal-regulated kinase |
| MAPK | p38 MAP kinase |
| PI3K | phosphoinositide 3-kinase |
| PKC | protein kinase C |
| Hrd-1 | 3-hydroxy-3-methylglutaryl reductase degradation 1 |
| EZH2 | Enhancer of zeste homolog 2 |
| Sp1 | Specificity protein 1 |
| SETD7/Set7/9 | SET Domain Containing 7 |
| HAT | histone acetyltransferase |
| HDAC | histone deacetylase |
| CBP | CREB binding protein |
| ChIP | chromatin immunoprecipitation |
| 3′UTR | 3′untranslated region |
| I3C | indole-3-carbinol |
| DIM | 3,3′-diindolylmethane |
| NAFLD | non-alcoholic fatty liver disease |
| TRAMP | transgenic adenocarcinoma mouse prostate |
| SAHA | suberoylanilide hydroxamic acid |
References
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2–Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [Green Version]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef]
- Yates, M.S.; Kensler, T.W. Chemopreventive promise of targeting the Nrf2 pathway. Drug News Perspect. 2007, 20, 109–117. [Google Scholar] [CrossRef]
- Kensler, T.W.; Chen, J.G.; Egner, P.A.; Fahey, J.W.; Jacobson, L.P.; Stephenson, K.K.; Ye, L.; Coady, J.L.; Wang, J.B.; Wu, Y. Effects of glucosinolate-rich broccoli sprouts on urinary levels of aflatoxin-DNA adducts and phenanthrenetetraols in a randomized clinical trial in HeZuo township, Qidong, People’s Republic of China. Cancer Epidemiol. Biomarkers Prev. 2005, 14, 2605–2613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, M.; Li, Y.; Ye, X.; Zhang, L.; Wang, Z.; Xu, X.; Shen, Y.; Zheng, C. Loss-of-function mutations in KEAP1 drive lung cancer progression via KEAP1/NRF2 pathway activation. Cell Commun. Signal. 2020, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, B.; Tong, K.I.; Ohta, T.; Nakamura, Y.; Scharlock, M.; Ohtsuji, M.; Kang, M.I.; Kobayashi, A.; Yokoyama, S.; Yamamoto, M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006, 21, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Sun, Z.; Villeneuve, N.F.; Zhang, S.; Zhao, F.; Li, Y.; Chen, W.; Yi, X.; Zheng, W.; Wondrak, G.T. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 2008, 29, 1235–1243. [Google Scholar] [CrossRef] [Green Version]
- Harder, B.; Jiang, T.; Wu, T.; Tao, S.; De La Vega, M.R.; Tian, W.; Chapman, E.; Zhang, D.D. Molecular mechanisms of Nrf2 regulation and how these influence chemical modulation for disease intervention. Biochem. Soc. Trans. 2015, 43, 680–686. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.S.; Nam, L.B.; Yoo, O.K.; Keum, Y.S. Molecular mechanisms and systemic targeting of NRF2 dysregulation in cancer. Biochem. Pharmacol. 2020, 114002. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N. Nrf2: Friend or foe for chemoprevention? Carcinogenesis 2010, 31, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Lu, H.; Bai, Y. Nrf2 in cancers: A double-edged sword. Cancer Med. 2019, 8, 2252–2267. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Li, J.; Dashwood, R.H. Emerging crosstalk between long non-coding RNAs and Nrf2 signaling. Cancer Lett. 2020, 490, 154–164, [published online ahead of print, 24 July 2020]. [Google Scholar] [PubMed]
- Dodson, M.; De La Vega, M.R.; Cholanians, A.B.; Schmidlin, C.J.; Chapman, E.; Zhang, D.D. Modulating NRF2 in disease: Timing is everything. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 555–575. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [Green Version]
- Miao, W.; Hu, L.; Scrivens, P.J.; Batist, G. Transcriptional regulation of NF-E2 p45-related Factor (NRF2) expression by the Aryl Hydrocarbon Receptor-Xenobiotic Response Element signaling pathway direct cross-talk between phase I and II drug-metabolizing enzymes. J. Biol. Chem. 2005, 280, 20340–20348. [Google Scholar] [CrossRef] [Green Version]
- Rushworth, S.A.; Zaitseva, L.; Murray, M.Y.; Shah, N.M.; Bowles, K.M.; MacEwan, D.J. The high Nrf2 expression in human acute myeloid leukemia is driven by NF-κB and underlies its chemo-resistance. Blood 2012, 120, 5188–5198. [Google Scholar] [CrossRef] [Green Version]
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Kenneth, H.Y.; Yeo, C.J.; Calhoun, E.S. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, N.; Skoko, J.J.; Chartoumpekis, D.V.; Kimura, S.; Slocum, S.L.; Noda, K.; Palliyaguru, D.L.; Fujimuro, M.; Boley, P.A.; Tanaka, Y. Notch-Nrf2 axis: Regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol. Cell. Biol. 2014, 34, 653–663. [Google Scholar] [CrossRef] [Green Version]
- Kwak, M.K.; Itoh, K.; Yamamoto, M.; Kensler, T.W. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: Role of antioxidant response element-like sequences in the Nrf2 promoter. Mol. Cell. Biol. 2002, 22, 2883–2892. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, L.D.; Lee, J.; Gnad, F.; Klijn, C.; Schaub, A.; Reeder, J.; Daemen, A.; Bakalarski, C.E.; Holcomb, T.; Shames, D.S. Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep. 2016, 16, 2605–2617. [Google Scholar] [CrossRef] [Green Version]
- Poganik, J.R.; Long, M.J.; Disare, M.T.; Liu, X.; Chang, S.H.; Hla, T.; Aye, Y. Post-transcriptional regulation of Nrf2-mRNA by the mRNA-binding proteins HuR and AUF1. FASEB J. 2019, 33, 14636–14652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011, 31, 1121–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Zhao, F.; Gao, B.; Tan, C.; Yagishita, N.; Nakajima, T.; Wong, P.K.; Chapman, E.; Fang, D.; Zhang, D.D. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014, 28, 708–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002, 277, 42769–42774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Misra, V.; Thimmulappa, R.K.; Lee, H.; Ames, S.; Hoque, M.O.; Herman, J.G.; Baylin, S.B.; Sidransky, D.; Gabrielson, E. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS. Med. 2006, 3, e420. [Google Scholar] [CrossRef] [Green Version]
- Solis, L.M.; Behrens, C.; Dong, W.; Suraokar, M.; Ozburn, N.C.; Moran, C.A.; Corvalan, A.H.; Biswal, S.; Swisher, S.G.; Bekele, B.N. Nrf2 and Keap1 abnormalities in non–small cell lung carcinoma and association with clinicopathologic features. Clin. Cancer Res. 2010, 16, 3743–3753. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Wu, B.; Yan, J.; Li, X.; Sun, H.; Zhou, D. A possible gene silencing mechanism: Hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells. Biochem. Biophys. Res. Commun. 2012, 428, 80–85. [Google Scholar] [CrossRef]
- Guo, Y.; Yu, S.; Zhang, C.; Kong, A.N.T. Epigenetic regulation of Keap1-Nrf2 signaling. Free. Radic. Biol. Med. 2015, 88, 337–349. [Google Scholar] [CrossRef]
- Cheng, D.; Wu, R.; Guo, Y.; Kong, A.N.T. Regulation of Keap1–Nrf2 signaling: The role of epigenetics. Curr. Opin. Toxicol. 2016, 1, 134–138. [Google Scholar] [CrossRef] [Green Version]
- Fabrizio, F.P.; Sparaneo, A.; Trombetta, D.; Muscarella, L.A. Epigenetic versus genetic deregulation of the KEAP1/NRF2 axis in solid tumors: Focus on methylation and noncoding RNAs. Oxid. Med. Cell. Longev. 2018, 2018, 2492063. [Google Scholar] [CrossRef]
- Yu, S.; Khor, T.O.; Cheung, K.L.; Li, W.; Wu, T.Y.; Huang, Y.; Foster, B.A.; Kan, Y.W.; Kong, A.N. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS ONE 2010, 5, e8579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khor, T.O.; Fuentes, F.; Shu, L.; Paredes-Gonzalez, X.; Yang, A.Y.; Liu, Y.; Smiraglia, D.J.; Yegnasubramanian, S.; Nelson, W.G.; Kong, A.N.T. Epigenetic DNA methylation of antioxidative stress regulator NRF2 in human prostate cancer. Cancer Prev. Res. 2014, 7, 1186–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muscarella, L.A.; Barbano, R.; D’Angelo, V.; Copetti, M.; Coco, M.; Balsamo, T.; la Torre, A.; Notarangelo, A.; Troiano, M.; Parisi, S. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics 2011, 6, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbano, R.; Muscarella, L.A.; Pasculli, B.; Valori, V.M.; Fontana, A.; Coco, M.; la Torre, A.; Balsamo, T.; Poeta, M.L.; Marangi, G.F. Aberrant Keap1 methylation in breast cancer and association with clinicopathological features. Epigenetics 2013, 8, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Muscarella, L.A.; Parrella, P.; D’Alessandro, V.; la Torre, A.; Barbano, R.; Fontana, A.; Tancredi, A.; Guarnieri, V.; Balsamo, T.; Coco, M. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics 2011, 6, 710–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Yuan, F.; Che, G.; Xiao, X.; Nie, X.; Wang, Y.; Jia, J.; Kong, A.-N.; Zhang, L. Epigenetic modifications but not genetic polymorphisms regulate KEAP1 expression in colorectal cancer. J. Cell. Biochem. 2019, 120, 12311–12320. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, F.P.; Costantini, M.; Copetti, M.; la Torre, A.; Sparaneo, A.; Fontana, A.; Poeta, L.; Gallucci, M.; Sentinelli, S.; Graziano, P. Keap1/Nrf2 pathway in kidney cancer: Frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma. Oncotarget 2017, 8, 11187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Xu, J.; Li, C.; Shi, S.; Ji, S.; Xu, W.; Liu, J.; Jin, K.; Liang, D.; Liang, C. MBD1 is an epigenetic regulator of KEAP1 in pancreatic cancer. Curr. Mol. Med. 2016, 16, 404–411. [Google Scholar] [CrossRef]
- Li, Z.; Xu, L.; Tang, N.; Xu, Y.; Ye, X.; Shen, S.; Niu, X.; Lu, S.; Chen, Z. The polycomb group protein EZH2 inhibits lung cancer cell growth by repressing the transcription factor Nrf2. FEBS Lett. 2014, 588, 3000–3007. [Google Scholar] [CrossRef] [Green Version]
- Mishra, M.; Zhong, Q.; Kowluru, R.A. Epigenetic modifications of Keap1 regulate its interaction with the protective factor Nrf2 in the development of diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 2014, 55, 7256–7265. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Chin, Y.E.; Zhang, D.D. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol. Cell. Biol. 2009, 29, 2658–2672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.H.; Qu, J.; Shen, X. NF-κB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim. Biophys. Acta. 2008, 1783, 713–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercado, N.; Thimmulappa, R.; Thomas, C.M.; Fenwick, P.S.; Chana, K.K.; Donnelly, L.E.; Biswal, S.; Ito, K.; Barnes, P.J. Decreased histone deacetylase 2 impairs Nrf2 activation by oxidative stress. Biochem. Biophys. Res. Commun. 2011, 406, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, N.; Bohmann, D. BET-ting on Nrf2: How Nrf2 Signaling can Influence the Therapeutic Activities of BET Protein Inhibitors. Bioessays 2018, 40, 1800007. [Google Scholar] [CrossRef]
- Hussong, M.; Börno, S.T.; Kerick, M.; Wunderlich, A.; Franz, A.; Sültmann, H.; Timmermann, B.; Lehrach, H.; Hirsch-Kauffmann, M.; Schweiger, M.R. The bromodomain protein BRD4 regulates the KEAP1/NRF2-dependent oxidative stress response. Cell. Death. Dis. 2014, 5, e1195. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, N.; Wu, H.J.; Bennett, R.L.; Troche, C.; Licht, J.D.; Weber, J.D.; Maggi, L.B., Jr.; Tomasson, M.H. Sabotaging of the oxidative stress response by an oncogenic noncoding RNA. FASEB J. 2017, 31, 482–490. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T.X.; Wang, J.K.; Shen, L.J.; Long, C.L.; Liu, B.; Wei, Y.; Han, L.D.; Wei, Y.X.; Wu, S.D.; Wei, G.H. Increased m6A RNA modification is related to the inhibition of the Nrf2-mediated antioxidant response in di-(2-ethylhexyl) phthalate-induced prepubertal testicular injury. Environ. Pollut. 2020, 259, 113911. [Google Scholar] [CrossRef] [PubMed]
- Kurinna, S.; Werner, S. NRF2 and microRNAs: New but awaited relations. Biochem. Soc. Trans. 2015, 43, 595–601. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Ahmadi, Z.; Samarghandian, S.; Mohammadinejad, R.; Yaribeygi, H.; Sathyapalan, T.; Sahebkar, A. MicroRNA-mediated regulation of Nrf2 signaling pathway: Implications in disease therapy and protection against oxidative stress. Life Sci. 2020, 244, 117329. [Google Scholar] [CrossRef]
- Eades, G.; Yang, M.; Yao, Y.; Zhang, Y.; Zhou, Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J. Biol. Chem. 2011, 286, 40725–40733. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wu, W.; Jiao, G.; Li, C.; Liu, H. MiR-455-3p activates Nrf2/ARE signaling via HDAC2 and protects osteoblasts from oxidative stress. Int. J. Biol. Macromol. 2018, 107, 2094–2101. [Google Scholar] [CrossRef] [PubMed]
- Van Jaarsveld, M.T.M.; Helleman, J.; Boersma, A.W.M.; Van Kuijk, P.F.; Van Ijcken, W.F.; Despierre, E.; Vergote, I.; Mathijssen, R.H.J.; Berns, E.; Verweij, J. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene 2013, 32, 4284–4293. [Google Scholar] [CrossRef] [PubMed]
- Kabaria, S.; Choi, D.C.; Chaudhuri, A.D.; Jain, M.R.; Li, H.; Junn, E. MicroRNA-7 activates Nrf2 pathway by targeting Keap1 expression. Free. Radic. Biol. Med. 2015, 89, 548–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akdemir, B.; Nakajima, Y.; Inazawa, J.; Inoue, J. miR-432 induces NRF2 stabilization by directly targeting KEAP1. Mol. Cancer Res. 2017, 15, 1570–1578. [Google Scholar] [CrossRef] [Green Version]
- Kwak, M.K.; Kensler, T.W. Targeting NRF2 signaling for cancer chemoprevention. Toxicol. Appl. Pharmacol. 2010, 244, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, A.; Li, Y.; Sarkar, F.H. The bounty of nature for changing the cancer landscape. Mol. Nutr. Food. Res. 2016, 60, 1251–1263. [Google Scholar] [CrossRef]
- Higdon, J.V.; Delage, B.; Williams, D.E.; Dashwood, R.H. Cruciferous vegetables and human cancer risk: Epidemiologic evidence and mechanistic basis. Pharmacol. Res. 2007, 55, 224–236. [Google Scholar] [CrossRef] [Green Version]
- Grose, K.R.; Bjeldanes, L.F. Oligomerization of indole-3-carbinol in aqueous acid. Chem. Res. Toxicol. 1992, 5, 188–193. [Google Scholar] [CrossRef]
- Dashwood, R.H.; Fong, A.T.; Arbogast, D.N.; Bjeldanes, L.F.; Hendricks, J.D.; Bailey, G.S. Anticarcinogenic activity of indole-3-carbinol acid products: Ultrasensitive bioassay by trout embryo microinjection. Cancer Res. 1994, 54, 3617–3619. [Google Scholar]
- Wattenberg, L.W. Chemoprevention of cancer. Cancer Res. 1985, 45, 1–8. [Google Scholar] [CrossRef]
- Dashwood, R.H.; Arbogast, D.N.; Fong, A.T.; Pereira, C.; Hendricks, J.D.; Bailey, G.S. Quantitative inter-relationships between aflatoxin B1 carcinogen dose, indole-3-carbinol anti-carcinogen dose, target organ DNA adduction and final tumor response. Carcinogenesis 1989, 10, 175–181. [Google Scholar] [CrossRef]
- Dashwood, R.H.; Fong, A.T.; Williams, D.E.; Hendricks, J.D.; Bailey, G.S. Promotion of aflatoxin B1 carcinogenesis by the natural tumor modulator indole-3-carbinol: Influence of dose, duration, and intermittent exposure on indole-3-carbinol promotional potency. Cancer Res. 1991, 51, 2362–2365. [Google Scholar] [PubMed]
- Guo, D.; Schut, H.A.; Davis, C.D.; Snyderwine, E.G.; Bailey, G.S.; Dashwood, R.H. Protection by chlorophyllin and indole-3-carbinol against 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)-induced DNA adducts and colonic aberrant crypts in the F344 rat. Carcinogenesis 1995, 16, 2931–2937. [Google Scholar] [CrossRef]
- Xu, M.; Bailey, A.C.; Hernaez, J.F.; Taoka, C.R.; Schut, H.A.; Dashwood, R.H. Protection by green tea, black tea, and indole-3-carbinol against 2-amino-3-methylimidazo[4,5-f]quinoline-induced DNA adducts and colonic aberrant crypts in the F344 rat. Carcinogenesis 1996, 17, 1429–1434. [Google Scholar] [CrossRef] [Green Version]
- Dashwood, R.H. Indole-3-carbinol: Anticarcinogen or tumor promoter in brassica vegetables? Chem. Biol. Interact. 1998, 110, 1–5. [Google Scholar] [CrossRef]
- Xu, M.; Orner, G.A.; Bailey, G.S.; Stoner, G.D.; Horio, D.T.; Dashwood, R.H. Post-initiation effects of chlorophyllin and indole-3-carbinol in rats given 1,2-dimethylhydrazine or 2-amino-3-methylimidazo[4,5-f]quinoline. Carcinogenesis 2001, 22, 309–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, C.A.; Xu, M.; Orner, G.A.; Fong, A.T.; Bailey, G.S.; Stoner, G.D.; Horio, D.T.; Dashwood, R.H. beta-Catenin mutation in rat colon tumors initiated by 1,2-dimethylhydrazine and 2-amino-3-methylimidazo[4,5-f]quinoline, and the effect of post-initiation treatment with chlorophyllin and indole-3-carbinol. Carcinogenesis 2001, 22, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Beaver, L.M.; Yu, T.W.; Sokolowski, E.I.; Williams, D.E.; Dashwood, R.H.; Ho, E. 3,3′-Diindolylmethane, but not indole-3-carbinol, inhibits histone deacetylase activity in prostate cancer cells. Toxicol. Appl. Pharmacol. 2012, 263, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Shorey, L.E.; Madeen, E.P.; Atwell, L.L.; Ho, E.; Löhr, C.V.; Pereira, C.B.; Dashwood, R.H.; Williams, D.E. Differential modulation of dibenzo[def,p]chrysene transplacental carcinogenesis: Maternal diets rich in indole-3-carbinol versus sulforaphane. Toxicol. Appl. Pharmacol. 2013, 270, 60–69. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.P.; Hsu, A.; Buchanan, A.; Palomera-Sanchez, Z.; Beaver, L.M.; Houseman, E.A.; Williams, D.E.; Dashwood, R.H.; Ho, E. Effects of sulforaphane and 3,3′-diindolylmethane on genome-wide promoter methylation in normal prostate epithelial cells and prostate cancer cells. PLoS ONE 2014, 9, e86787. [Google Scholar] [CrossRef] [Green Version]
- Kolluri, S.K.; Jin, U.H.; Safe, S. Role of the aryl hydrocarbon receptor in carcinogenesis and potential as an anti-cancer drug target. Arch. Toxicol. 2017, 91, 2497–2513. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, N.; Fritz, V.; Upadhyaya, P.; Kassie, F.; Hecht, S.S. Research on cruciferous vegetables, indole-3-carbinol, and cancer prevention: A tribute to Lee W. Wattenberg. Mol. Nutr. Food. Res. 2016, 60, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Yerushalmi, R.; Bargil, S.; Bar, Y.; Ozlavo, R.; Tuval, S.; Rapson, Y.; Pomerantz, A.; Zoref, D.; Sharon, E.; Caspi, O.; et al. 3,3-Diindolylmethane (DIM): A nutritional intervention and its impact on breast density in healthy BRCA carriers. A prospective clinical trial. Carcinogenesis 2020. [published online ahead of print 27 May 2020]. [Google Scholar] [CrossRef]
- Bradlow, H.L.; Michnovicz, J.J.; Halper, M.; Miller, D.G.; Wong, G.Y.; Osborne, M.P. Long-term responses of women to indole-3-carbinol or a high fiber diet. Cancer. Epidemiol. Biomarkers. Prev. 1994, 3, 591–595. [Google Scholar]
- Bradlow, H.L.; Sepkovic, D.W.; Telang, N.T.; Osborne, M.P. Indole-3-carbinol. A novel approach to breast cancer prevention. Ann. N.Y. Acad. Sci. 1995, 768, 180–200. [Google Scholar] [CrossRef] [PubMed]
- Michnovicz, J.J. Increased estrogen 2-hydroxylation in obese women using oral indole-3-carbinol. Int. J. Obes. Relat. Metab. Disord. 1998, 22, 227–229. [Google Scholar] [CrossRef] [Green Version]
- Bell, M.C.; Crowley-Nowick, P.; Bradlow, H.L.; Sepkovic, D.W.; Schmidt-Grimminger, D.; Howell, P.; Mayeaux, E.J.; Tucker, A.; Turbat-Herrera, E.A.; Mathis, J.M. Placebo-controlled trial of indole-3-carbinol in the treatment of CIN. Gynecol. Oncol. 2000, 78, 123–129. [Google Scholar] [CrossRef] [Green Version]
- Sepkovic, D.W.; Bradlow, H.L.; Bell, M. Quantitative determination of 3,3′-diindolylmethane in urine of individuals receiving indole-3-carbinol. Nutr. Cancer. 2001, 41, 57–63. [Google Scholar] [CrossRef]
- Wu, T.Y.; Khor, T.O.; Su, Z.Y.; Saw, C.L.L.; Shu, L.; Cheung, K.L.; Huang, Y.; Yu, S.; Kong, A.N.T. Epigenetic modifications of Nrf2 by 3, 3′-diindolylmethane in vitro in TRAMP C1 cell line and in vivo TRAMP prostate tumors. AAPS J. 2013, 15, 864–874. [Google Scholar] [CrossRef] [Green Version]
- Pandey, M.; Kaur, P.; Shukla, S.; Abbas, A.; Fu, P.; Gupta, S. Plant flavone apigenin inhibits HDAC and remodels chromatin to induce growth arrest and apoptosis in human prostate cancer cells: In vitro and in vivo study. Mol. Carcinog. 2012, 51, 952–962. [Google Scholar] [CrossRef] [Green Version]
- Tseng, T.H.; Chien, M.H.; Lin, W.L.; Wen, Y.C.; Chow, J.M.; Chen, C.K.; Kuo, T.C.; Lee, W.J. Inhibition of MDA-MB-231 breast cancer cell proliferation and tumor growth by apigenin through induction of G2/M arrest and histone H3 acetylation-mediated p21WAF1/CIP1 expression. Environ. Toxicol. 2017, 32, 434–444. [Google Scholar] [CrossRef]
- Yang, J.; Wu, R.; Li, W.; Gao, L.; Yang, Y.; Li, P.; Kong, A.-N. The triterpenoid corosolic acid blocks transformation and epigenetically reactivates Nrf2 in TRAMP-C1 prostate cells. Mol. Carcinog. 2018, 57, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Khor, T.O.; Huang, Y.; Wu, T.Y.; Shu, L.; Lee, J.; Kong, A.N.T. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem. Pharmacol. 2011, 82, 1073–1078. [Google Scholar] [CrossRef]
- Kuo, H.; Wu, R.; Li, S.; Yang, A.Y.; Kong, A.N. Anthocyanin delphinidin prevents neoplastic transformation of mouse skin JB6 P+ cells: Epigenetic re-activation of Nrf2-ARE pathway. AAPS J. 2019, 21, 83. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, I.; Cao, M.; Su, Z.; Wu, R.; Guo, Y.; Fang, M.; Kong, A.N. Fucoxanthin elicits epigenetic modifications, Nrf2 activation and blocking transformation in mouse skin JB6 P+ cells. AAPS J. 2018, 20, 32. [Google Scholar] [CrossRef]
- Zuo, Q.; Wu, R.; Xiao, X.; Yang, C.; Yang, Y.; Wang, C.; Lin, L.; Kong, A.N. The dietary flavone luteolin epigenetically activates the Nrf2 pathway and blocks cell transformation in human colorectal cancer HCT116 cells. J. Cell. Biochem. 2018, 119, 9573–9582. [Google Scholar] [CrossRef]
- Kang, K.A.; Piao, M.J.; Hyun, Y.J.; Zhen, A.X.; Cho, S.J.; Ahn, M.J.; Yi, J.M.; Hyun, J.W. Luteolin promotes apoptotic cell death via upregulation of Nrf2 expression by DNA demethylase and the interaction of Nrf2 with p53 in human colon cancer cells. Exp. Mol. Med. 2019, 51, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Li, W.; Wang, C.; Wu, R.; Yin, R.; Kuo, H.C.; Wang, L.; Kong, A.N. Pelargonidin reduces the TPA induced transformation of mouse epidermal cells–potential involvement of Nrf2 promoter demethylation. Chem. Biol. Interact. 2019, 309, 108701. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.J.; Yu, H.W.; Yang, Y.Z.; Wu, W.Y.; Chen, T.Y.; Jia, K.K.; Kang, L.L.; Jiao, R.Q.; Kong, L.D. Polydatin prevents fructose-induced liver inflammation and lipid deposition through increasing miR-200a to regulate Keap1/Nrf2 pathway. Redox Biol. 2018, 18, 124–137. [Google Scholar] [CrossRef]
- Hong, B.; Su, Z.; Zhang, C.; Yang, Y.; Guo, Y.; Li, W.; Kong, A.N.T. Reserpine inhibit the JB6 P+ cell transformation through epigenetic reactivation of Nrf2-mediated anti-oxidative stress pathway. AAPS J. 2016, 18, 659–669. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, H.; Teimouri, M.; Shabani, M.; Koushki, M.; Khorzoughi, R.B.; Namvarjah, F.; Izadi, P.; Meshkani, R. Resveratrol alleviates non-alcoholic fatty liver disease through epigenetic modification of the Nrf2 signaling pathway. Int. J. Biochem. Cell. Biol. 2020, 119, 105667. [Google Scholar] [CrossRef]
- Singh, B.; Shoulson, R.; Chatterjee, A.; Ronghe, A.; Bhat, N.K.; Dim, D.C.; Bhat, H.K. Resveratrol inhibits estrogen-induced breast carcinogenesis through induction of NRF2-mediated protective pathways. Carcinogenesis 2014, 35, 1872–1880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Jiang, X.; Meng, L.; Dong, X.; Shen, Y.; Xin, Y. Anticancer activity of sulforaphane: The epigenetic mechanisms and the Nrf2 signaling pathway. Oxid. Med. Cell. Longev. 2018, 2018, 5438179. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.S.; Li, J.; Beaver, L.M.; Dashwood, W.M.; Sun, D.; Rajendran, P.; Williams, D.E.; Ho, E.; Dashwood, R.H. A functional pseudogene, NMRAL2P, is regulated by Nrf2 and serves as a coactivator of NQO1 in sulforaphane-treated colon cancer cells. Mol. Nutr. Food. Res. 2017, 61, 1600769. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, C.; Guo, Y.; Su, Z.Y.; Yang, Y.; Shu, L.; Kong, A.N.T. Blocking of JB6 cell transformation by tanshinone IIA: Epigenetic reactivation of Nrf2 antioxidative stress pathway. AAPS J. 2014, 16, 1214–1225. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Liu, L.; Zhang, X.; Jiang, X.; Wang, L. Tanshinone IIA prevents rifampicin-induced liver injury by regulating BSEP/NTCP expression via epigenetic activation of NRF2. Liver Int. 2020, 40, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Kuang, H.; Tang, Z.; Zhang, C.; Wang, Z.; Li, W.; Yang, C.; Wang, Q.; Yang, B.; Kong, A.N. Taxifolin Activates the Nrf2 Anti-Oxidative Stress Pathway in Mouse Skin Epidermal JB6 P+ Cells through Epigenetic Modifications. Int. J. Mol. Sci. 2017, 18, 1546. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Ramirez, C.N.; Su, Z.Y.; Kong, A.N.T. Epigenetic modifications of triterpenoid ursolic acid in activating Nrf2 and blocking cellular transformation of mouse epidermal cells. J. Nutr. Biochem. 2016, 33, 54–62. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Shu, L.; Zhang, C.; Li, W.; Wu, R.; Guo, Y.; Yang, Y.; Kong, A.N. Histone methyltransferase Setd7 regulates Nrf2 signaling pathway by phenethyl isothiocyanate and ursolic acid in human prostate cancer cells. Mol. Nutr. Food Res. 2018, 62, e1700840. [Google Scholar] [CrossRef]
- Huang, Y.; Khor, T.O.; Shu, L.; Saw, C.L.L.; Wu, T.Y.; Suh, N.; Yang, C.S.; Kong, A.N.T. A γ-tocopherol-rich mixture of tocopherols maintains Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation. J. Nutr. 2012, 142, 818–823. [Google Scholar] [CrossRef]
- Su, Z.Y.; Khor, T.O.; Shu, L.; Lee, J.H.; Saw, C.L.L.; Wu, T.Y.; Huang, Y.; Suh, N.; Yang, C.S.; Conney, A.H.; et al. Epigenetic reactivation of Nrf2 in murine prostate cancer TRAMP C1 cells by natural phytochemicals Z-ligustilide and Radix angelica sinensis via promoter CpG demethylation. Chem. Res. Toxicol. 2013, 26, 477–485. [Google Scholar] [CrossRef]
- Madunić, J.; Madunić, I.V.; Gajski, G.; Popić, J.; Garaj-Vrhovac, V. Apigenin: A dietary flavonoid with diverse anticancer properties. Cancer Lett. 2018, 413, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, R.; Datt, M.; Liu, X.; Gupta, S. Dietary Flavones as Dual Inhibitors of DNA Methyltransferases and Histone Methyltransferases. PLoS ONE 2016, 11, e0162956. [Google Scholar]
- Chen, L.; Xie, W.; Xie, W.; Zhuang, W.; Jiang, C.; Liu, N. Apigenin attenuates isoflurane-induced cognitive dysfunction via epigenetic regulation and neuroinflammation in aged rats. Arch. Gerontol. Geriatr. 2017, 73, 29–36. [Google Scholar] [CrossRef]
- Paredes-Gonzalez, X.; Fuentes, F.; Su, Z.Y.; Kong, A.N.T. Apigenin reactivates Nrf2 anti-oxidative stress signaling in mouse skin epidermal JB6 P+ cells through epigenetics modifications. AAPS J. 2014, 16, 727–735. [Google Scholar] [CrossRef] [Green Version]
- Gao, A.M.; Zhang, X.Y.; Ke, Z.P. Apigenin sensitizes BEL-7402/ADM cells to doxorubicin through inhibiting miR-101/Nrf2 pathway. Oncotarget 2017, 8, 82085–82091. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Wang, G.; Peng, W.; Xu, Y.; Zhang, Y.; Ge, Y.; Jing, Y.; Gong, Z. Corosolic acid isolated from Eriobotrya japonica leaves reduces glucose level in human hepatocellular carcinoma cells, zebrafish and rats. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Chen, Y.; Ge, Z.; Huang, S.; Zhou, L.; Zhai, C.; Chen, Y.; Hu, Q.; Cao, W.; Weng, Y.; Li, Y. Delphinidin attenuates pathological cardiac hypertrophy via the AMPK/NOX/MAPK signaling pathway. Aging 2020, 12, 5362. [Google Scholar] [CrossRef]
- Afaq, F.; Syed, D.N.; Malik, A.; Hadi, N.; Sarfaraz, S.; Kweon, M.-H.; Khan, N.; Zaid, M.A.; Mukhtar, H. Delphinidin, an anthocyanidin in pigmented fruits and vegetables, protects human HaCaT keratinocytes and mouse skin against UVB-mediated oxidative stress and apoptosis. J. Invest. Dermatol. 2007, 127, 222–232. [Google Scholar] [CrossRef] [Green Version]
- Bae, M.; Kim, M.B.; Park, Y.K.; Lee, J.Y. Health benefits of fucoxanthin in the prevention of chronic diseases. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids. 2020, 1865, 158618. [Google Scholar] [CrossRef]
- Imran, M.; Rauf, A.; Abu-Izneid, T.; Nadeem, M.; Shariati, M.A.; Khan, I.A.; Imran, A.; Orhan, I.E.; Rizwan, M.; Atif, M. Luteolin, a flavonoid, as an anticancer agent: A review. Biomed. Pharmacother. 2019, 112, 108612. [Google Scholar] [CrossRef]
- Karthi, N.; Karthiga, A.; Kalaiyarasu, T.; Stalin, A.; Manju, V.; Singh, S.K.; Cyril, R.; Lee, S.M. Exploration of cell cycle regulation and modulation of the DNA methylation mechanism of pelargonidin: Insights from the molecular modeling approach. Comput. Biol. Chem. 2017, 70, 175–185. [Google Scholar] [CrossRef]
- Huang, K.; Chen, C.; Hao, J.; Huang, J.; Wang, S.; Liu, P.; Huang, H. Polydatin promotes Nrf2-ARE anti-oxidative pathway through activating Sirt1 to resist AGEs-induced upregulation of fibronectin and transforming growth factor-β1 in rat glomerular mesangial cells. Mol. Cell. Endocrinol. 2015, 399, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A.; Sinclair, D.A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug. Discov. 2006, 5, 493–506. [Google Scholar] [CrossRef]
- Bonkowski, M.S.; Sinclair, D.A. Slowing ageing by design: The rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell. Biol. 2016, 17, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Yagishita, Y.; Fahey, J.W.; Dinkova-Kostova, A.T.; Kensler, T.W. Broccoli or sulforaphane: Is it the source or dose that matters? Molecules 2019, 24, 3593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Talalay, P.; Cho, C.G.; Posner, G.H. A major inducer of anticarcinogenic protective enzymes from broccoli: Isolation and elucidation of structure. Proc. Natl. Acad. Sci. USA 1992, 89, 2399–2403. [Google Scholar] [CrossRef] [Green Version]
- Houghton, C.A. Sulforaphane: Its “Coming of Age” as a clinically relevant nutraceutical in the prevention and treatment of chronic disease. Oxid. Med. Cell. Longev. 2019, 2019, 2716870. [Google Scholar] [CrossRef] [Green Version]
- Ho, E.; Beaver, L.M.; Williams, D.E.; Dashwood, R.H. Dietary factors and epigenetic regulation for prostate cancer prevention. Adv. Nutr. 2011, 2, 497–510. [Google Scholar] [CrossRef] [Green Version]
- Nian, H.; Delage, B.; Ho, E.; Dashwood, R.H. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: Studies with sulforaphane and garlic organosulfur compounds. Environ. Mol. Mutagen. 2009, 50, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Dashwood, R.H.; Ho, E. Dietary agents as histone deacetylase inhibitors: Sulforaphane and structurally related isothiocyanates. Nutr. Rev. 2008, 66, S36–S38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, J.D.; Dashwood, R.H.; Ho, E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008, 269, 291–304. [Google Scholar] [CrossRef] [Green Version]
- Myzak, M.C.; Dashwood, R.H. Chemoprotection by sulforaphane: Keep one eye beyond Keap1. Cancer Lett. 2006, 233, 208–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dashwood, R.H.; Myzak, M.C.; Ho, E. Dietary HDAC inhibitors: Time to rethink weak ligands in cancer chemoprevention? Carcinogenesis 2006, 27, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Myzak, M.C.; Karplus, P.A.; Chung, F.L.; Dashwood, R.H. A novel mechanism of chemoprotection by sulforaphane: Inhibition of histone deacetylase. Cancer Res. 2004, 64, 5767–5774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Cole, R.N.; Itoh, K.; Wakabayashi, N.; Katoh, Y.; Yamamoto, M.; Talalay, P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 2002, 99, 11908–11913. [Google Scholar] [CrossRef] [Green Version]
- Myzak, M.C.; Dashwood, R.H. Histone deacetylases as targets for dietary cancer preventive agents: Lessons learned with butyrate, diallyl disulfide, and sulforaphane. Curr. Drug Targets. 2006, 7, 443–452. [Google Scholar] [CrossRef]
- Dashwood, R.H.; Ho, E. Dietary histone deacetylase inhibitors: From cells to mice to man. Semin. Cancer Biol. 2007, 17, 363–369. [Google Scholar] [CrossRef] [Green Version]
- Gerhauser, C. Epigenetic impact of dietary isothiocyanates in cancer chemoprevention. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 405–410. [Google Scholar] [CrossRef]
- Tollefsbol, T.O. Dietary epigenetics in cancer and aging. Cancer Treat. Res. 2014, 159, 257–267. [Google Scholar]
- Myzak, M.C.; Ho, E.; Dashwood, R.H. Dietary agents as histone deacetylase inhibitors. Mol. Carcinog. 2006, 45, 443–446. [Google Scholar] [CrossRef] [Green Version]
- Hsu, A.; Wong, C.P.; Yu, Z.; Williams, D.E.; Dashwood, R.H.; Ho, E. Promoter de-methylation of cyclin D2 by sulforaphane in prostate cancer cells. Clin. Epigenetics 2011, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Beaver, L.M.; Kuintzle, R.; Buchanan, A.; Wiley, M.W.; Glasser, S.T.; Wong, C.P.; Johnson, G.S.; Chang, J.H.; Löhr, C.V.; Williams, D.E.; et al. Long noncoding RNAs and sulforaphane: A target for chemoprevention and suppression of prostate cancer. J. Nutr. Biochem. 2017, 42, 72–83. [Google Scholar] [CrossRef] [Green Version]
- Rafiei, H.; Ashrafizadeh, M.; Ahmadi, Z. MicroRNAs as novel targets of sulforaphane in cancer therapy: The beginning of a new tale? Phytother. Res. 2020, 34, 721–728. [Google Scholar] [CrossRef]
- Gao, L.; Cheng, D.; Yang, J.; Wu, R.; Li, W.; Kong, A.N. Sulforaphane epigenetically demethylates the CpG sites of the miR-9-3 promoter and reactivates miR-9-3 expression in human lung cancer A549 cells. J. Nutr. Biochem. 2018, 56, 109–115. [Google Scholar] [CrossRef]
- Martin, S.L.; Kala, R.; Tollefsbol, T.O. Mechanisms for the inhibition of colon cancer cells by sulforaphane through epigenetic modulation of microRNA-21 and human Telomerase Reverse Transcriptase (hTERT) down-regulation. Curr. Cancer Drug Targets 2018, 18, 97–106. [Google Scholar] [CrossRef]
- Manigandan, K.; Manimaran, D.; Jayaraj, R.L.; Elangovan, N.; Dhivya, V.; Kaphle, A. Taxifolin curbs NF-κB-mediated Wnt/β-catenin signaling via up-regulating Nrf2 pathway in experimental colon carcinogenesis. Biochimie 2015, 119, 103–112. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, C.; Li, W.; Wu, R.; Guo, Y.; Cheng, D.; Yang, Y.; Androulakis, I.P.; Kong, A.N. Pharmacokinetics and pharmacodynamics of the triterpenoid ursolic acid in regulating the antioxidant, anti-inflammatory, and epigenetic gene responses in rat leukocytes. Mol. Pharm. 2017, 14, 3709–3717. [Google Scholar] [CrossRef] [Green Version]
- Brigelius-Flohé, R.; Traber, M.G. Vitamin E: Function and metabolism. FASEB J. 1999, 13, 1145–1155. [Google Scholar] [CrossRef]
- Burton, G.W.; Traber, M.G. Vitamin E: Antioxidant activity, biokinetics, and bioavailability. Annu. Rev. Nutr. 1990, 10, 357–382. [Google Scholar] [CrossRef]
- Podda, M.; Weber, C.; Traber, M.G.; Packer, L. Simultaneous determination of tissue tocopherols, tocotrienols, ubiquinols, and ubiquinones. J. Lipid. Res. 1996, 37, 893–901. [Google Scholar]
- Ju, J.; Picinich, S.C.; Yang, Z.; Zhao, Y.; Suh, N.; Kong, A.N.; Yang, C.S. Cancer-preventive activities of tocopherols and tocotrienols. Carcinogenesis 2010, 31, 533–542. [Google Scholar] [CrossRef] [Green Version]
- Kayden, H.J.; Traber, M.G. Absorption, lipoprotein transport, and regulation of plasma concentrations of vitamin E in humans. J. Lipid. Res. 1993, 34, 343–358. [Google Scholar]
- Traber, M.G.; Sies, H. Vitamin E in humans: Demand and delivery. Annu. Rev. Nutr. 1996, 16, 321–347. [Google Scholar] [CrossRef]
- Traber, M.G.; Ramakrishnan, R.; Kayden, H.J. Human plasma vitamin E kinetics demonstrate rapid recycling of plasma RRR-alpha-tocopherol. Proc. Natl. Acad. Sci. USA 1994, 91, 10005–10008. [Google Scholar] [CrossRef] [Green Version]
- Burton, G.W.; Traber, M.G.; Acuff, R.V.; Walters, D.N.; Kayden, H.; Hughes, L.; Ingold, K.U. Human plasma and tissue alpha-tocopherol concentrations in response to supplementation with deuterated natural and synthetic vitamin E. Am. J. Clin. Nutr. 1998, 67, 669–684. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.S.; Luo, P.; Zeng, Z.; Wang, H.; Malafa, M.; Suh, N. Vitamin E and cancer prevention: Studies with different forms of tocopherols and tocotrienols. Mol. Carcinog. 2020, 59, 365–389. [Google Scholar] [CrossRef]
- Montagnani, M.M.; Marzagalli, M.; Fontana, F.; Raimondi, M.; Moretti, R.M.; Limonta, P. Anticancer properties of tocotrienols: A review of cellular mechanisms and molecular targets. J. Cell. Physiol. 2019, 234, 1147–1164. [Google Scholar] [CrossRef] [Green Version]
- Abraham, A.; Kattoor, A.J.; Saldeen, T.; Mehta, J.L. Vitamin E and its anticancer effects. Crit. Rev. Food. Sci. Nutr. 2019, 59, 2831–2838. [Google Scholar] [CrossRef]
- Jiang, Q. Natural forms of vitamin E as effective agents for cancer prevention and therapy. Adv. Nutr. 2017, 8, 850–867. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, Y.; Huang, X.; Xie, Y.; Qu, Y.; Long, H.; Gu, N.; Jiang, W. Z-Ligustilide protects vascular endothelial cells from oxidative stress and rescues high fat diet-induced atherosclerosis by activating multiple NRF2 downstream genes. Atherosclerosis 2019, 284, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Uchi, H.; Morino-Koga, S.; Shi, W.; Furue, M. Z-ligustilide ameliorated ultraviolet B-induced oxidative stress and inflammatory cytokine production in human keratinocytes through upregulation of Nrf2/HO-1 and suppression of NF-κB pathway. Exp. Dermatol. 2015, 24, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Zhao, P.; Lu, Y.P.; Chen, M.M.; Sun, H.; Wu, X.M.; Zhu, L. Z-ligustilide activates the Nrf2/HO-1 pathway and protects against cerebral ischemia-reperfusion injury in vivo and in vitro. Brain. Res. 2013, 1520, 168–177. [Google Scholar] [CrossRef]
- Mohammad, H.P.; Barbash, O.; Creasy, C.L. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418. [Google Scholar] [CrossRef]
- Gerhauser, C. Cancer chemoprevention and nutriepigenetics: State of the art and future challenges. Top. Curr. Chem. 2013, 329, 73–132. [Google Scholar]
- Ganesan, A.; Arimondo, P.B.; Rots, M.G.; Jeronimo, C.; Berdasco, M. The timeline of epigenetic drug discovery: From reality to dreams. Clin. Epigenetics 2019, 11, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiMarco-Crook, C.; Xiao, H. Diet-based strategies for cancer chemoprevention: The role of combination regimens using dietary bioactive components. Annu. Rev. Food. Sci. Technol. 2015, 6, 505–526. [Google Scholar] [CrossRef]
- Zhang, Y.; Kutateladze, T.G. Diet and the epigenome. Nat. Commun. 2018, 9, 3375. [Google Scholar] [CrossRef]
- Stefanson, A.L.; Bakovic, M. Dietary regulation of Keap1/Nrf2/ARE pathway: Focus on plant-derived compounds and trace minerals. Nutrients 2014, 6, 3777–3801. [Google Scholar] [CrossRef] [Green Version]
- Fernández, Ó.; Giovannoni, G.; Fox, R.J.; Gold, R.; Phillips, J.T.; Potts, J.; Okwuokenye, M.; Marantz, J.L. Efficacy and safety of delayed-release dimethyl fumarate for relapsing-remitting multiple sclerosis in prior interferon users: An integrated analysis of DEFINE and CONFIRM. Clin. Ther. 2017, 39, 1671–1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.G.; Otterson, G.A. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr. Med. Chem. Anti-cancer. Agents 2003, 3, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Brocks, D.; Schmidt, C.R.; Daskalakis, M.; Jang, H.S.; Shah, N.M.; Li, D.; Li, J.; Zhang, B.; Hou, Y.; Laudato, S.; et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat. Genet. 2017, 49, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.; Dashwood, W.M.; Li, L.; Kang, Y.; Kim, E.; Johnson, G.; Fischer, K.A.; Löhr, C.V.; Williams, D.E.; Ho, E.; et al. Nrf2 status affects tumor growth, HDAC3 gene promoter associations, and the response to sulforaphane in the colon. Clin. Epigenetics 2015, 7, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajendran, P.; Johnson, G.; Li, L.; Chen, Y.S.; Dashwood, M.; Nguyen, N.; Ulusan, A.; Ertem, F.; Zhang, M.; Li, J. Acetylation of CCAR2 establishes a BET/BRD9 acetyl switch in response to combined deacetylase and bromodomain inhibition. Cancer Res. 2019, 79, 918–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Sl No. | Phytochemical | Chemical Name | Epigenetic Mechanism of Nrf2 Regulation | Molecular Targets | Cell Type | Reference |
|---|---|---|---|---|---|---|
| 1. | 3,3′-diindolylmethane | 3,3′-Methylenebis(1H-indole) | Decreased methylation of CpG sites in the promoter region of mouse Nfe2l2 | Suppressed mRNA and protein expression of Dnmt1, Dnmt3a, and Dnmt3b; inhibited protein expression of Hdac2 and Hdac3 | TRAMP-C1 prostate cells | [79] |
| 2. | Apigenin | 5,7-Dihydroxy-2-(4hydroxy-phenyl)-4H-chromen-4-one | Decreased Nfe2l2 hyper-methylation; induced expression of miR-101, targeting Nfe2l2 mRNA | Inhibited Dnmt1, Dnmt3a and Dnmt3b; inhibited Hdacs; induced miR101 | Mouse epidermal JB6 P+ cells BEL-7402/ADM cells | [80,81] |
| 3. | Corosolic acid | 2α,3β-2,3-dihydroxyurs-12-en-28-oic acid | Decreased Nfe2l2 hypermethylation; increased histone H3 lysine 27 acetylation; decreased H3 lysine 27 trimethylation | Decreased levels of Dnmt1, Dnmt3a and Dnmt3b; reduced levels of Hdac1, Hdac2, Hdac3, Hdac4, Hdac7 and Hdac8 | TRAMP-C1 prostate cells | [82] |
| 4. | Curcumin | 1, 7-bis (4-hydroxy-3-methoxy-phenyl)-1, 6 heptadiene-3, 5-dione | Decreased Nfe2l2 hypermethylation | Inhibited enzymatic activity of Dnmt enzymes | TRAMP-C1 prostate cells | [83] |
| 5. | Delphinidin | 3,5,7-Trihydroxy-2-(3,4,5-trihydroxyphenyl) chromenium | Demethylation of 15 CpG sites in the mouse Nfe2l2 promoter region | Decreased protein expression of Dnmt1, Dnmt3a, and class I/II Hdacs | Mouse epidermal JB6 P+ cells | [84] |
| 6. | Fucoxanthin | 3,5′-Dihydroxy-8-oxo-6′,7′-didehydro-5,5′,6,6′,7,8-hexahydro-5,6-epoxy-β,β-caroten-3′-yl acetate | Decreased Nfe2l2 hypermethylation | Reduced Dnmt activity | Mouse epidermal JB6 P+ cells | [85] |
| 7. | Luteolin | 2-(3,4dihydroxyphenyl)-5,7-dihydroxy-chromen-4-one | Decreased NFE2L2 hypermethylation | Decreased expression of DNMT1, DNMT3A and DNMT3B; decreased HDAC1, HDAC2, HDAC3, HDAC6, HDAC7; reduced activities of DNMTs and HDACs; increased ten-eleven translocation 1, 2 and 3 (TET1, TET2, and TET3) | Human colon cancer cells and SNU-407 cells | [86,87] |
| 8. | Pelargonidin | 3,5,7-Trihydroxy-2-(4hydroxyphenyl) chromenium | Decreased methylated CpGs in Nfe2l2 promoter | Decreased Dnmt1 and Dnmt3b expression; reduced levels of Hdacs 1–4 and Hdac7 | JB6 P+ cells | [88] |
| 9. | Polydatin | 3-Hydroxy-5-[(E)-2-(4-hydroxyphenyl)vinyl] phenyl β-d-glucopyranoside | Enhanced miR-200a targeting KEAP1 to activate NRF2 signaling | Increased miR-200a expression under high fructose induction; downregulated KEAP1 mRNA and protein | Buffalo rat liver (BRL-3A) and human HepG2 cells | [89] |
| 10. | Reserpine | Methyl 18β-hydroxy-11,17α-dimethoxy-3β,20α-yohimban-16βcarboxylate 3,4,5-trimethoxybenzoate | Decreased proportion of methylated CpG sites in the Nfe2l2 promoter | Concentration-dependent decreased mRNA and protein expression of Dnmt1, Dnmt3a, and Dnmt3b | JB6 P+ Cell | [90] |
| 11. | Resveratrol | 3,4′,5-trihydroxystilbene | Decreased methylation of the NFE2L2 promoter | Inhibited expression and activity of DNMT1, DNMT3a, and DNMT3b; miR93 implicated | HepG2 cells and estradiol-induced breast cancer | [91,92] |
| 12. | Sulforaphane | 1-Isothiocyanato-4-(methanesulfinyl)butane | CpG demethylation and histone acetylation at the Nfe2l2 promoter; lncRNA upregulation | Inhibition of Dnmt1, Dnmt3a, Dnmt3b, Hdacs 1–5, and Hdac7; upregulated functional pseudogene NMRAL2P | JB6 P+ cells; TRAMP C1 cells; human colon cancer cells | [93,94] |
| 13. | Tanshinone IIA | 1,6,6-trimethyl-8,9- dihydro-7H-naphtho [1,2-g] benzofuran-10,11-dione | Decreased methylated CpGs in Nfe2l2 promoter; increased recruitment of RNA polymerase complex II at the NFE2L2 transcription start site | Decreased mRNA and protein levels of HDAC1, HDAC3, and HDAC8, as well as DNMT1, DNMT3a, and DNMT3b; induced expression of TET2 | JB6 P+ cells, human normal hepatocyte and Hepa RG cells; rifampicin-induced liver injury in mice | [95,96] |
| 14. | Taxifolin | (2S,3S)-2-(3,4dihydroxy-phenyl)-3,5,7-trihydroxy-2,3dihydro-4H-chromen-4-one | Decreased proportion of methylated CpGs in the Nfe2l2 promoter | Reduced protein levels of Dnmt1, Dnmt3a and Dnmt3b as well as Hdacs 1, 3, 4, and 8 | JB6 P+ cells | [97] |
| 15. | Ursolic acid | (3β)-3-Hydroxyurs-12-en-28-oic acid | Nfe2l2 mouse promoter demethylation; increased acetylation and K4 monomethylation of histone H3 in human cells | Reduced DNMT1 and DNMT3a protein levels; inhibited expression of HDACs 1-3 and 8 (Class I) and HDAC 6 and 7 (Class II); induced Setd7 | JB6 P+ cells PC3 and LnCaP cells | [98,99] |
| 16. | γ Tocopherol–rich mixture of tocopherols (γ-TmT) | (2R)-2,5,7,8-tetramethyl-2-[ (4R,8R)-4,8,12-trimethyl-tridecyl]-6-chromanol | Reversed hyper-methylation in the Nfe2l2 promoter | Inhibited protein levels of Dnmt1, Dnmt3a, and Dnmt3b | Prostate tissues of C57BL/TGN TRAMP mice | [100] |
| 17. | Z-Ligustilide | (3E)-3-butylidene-4,5-dihydro-2-benzofuran-1-one | Decreased methylation of the first five CpGs of the Nfe2l2 promoter | Inhibited Dnmt activity | TRAMP C1 cells | [101] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhattacharjee, S.; Dashwood, R.H. Epigenetic Regulation of NRF2/KEAP1 by Phytochemicals. Antioxidants 2020, 9, 865. https://doi.org/10.3390/antiox9090865
Bhattacharjee S, Dashwood RH. Epigenetic Regulation of NRF2/KEAP1 by Phytochemicals. Antioxidants. 2020; 9(9):865. https://doi.org/10.3390/antiox9090865
Chicago/Turabian StyleBhattacharjee, Shamee, and Roderick H. Dashwood. 2020. "Epigenetic Regulation of NRF2/KEAP1 by Phytochemicals" Antioxidants 9, no. 9: 865. https://doi.org/10.3390/antiox9090865
APA StyleBhattacharjee, S., & Dashwood, R. H. (2020). Epigenetic Regulation of NRF2/KEAP1 by Phytochemicals. Antioxidants, 9(9), 865. https://doi.org/10.3390/antiox9090865