Peptides from Different Carcass Elements of Organic and Conventional Pork—Potential Source of Antioxidant Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Meat Samples
- The pigs were crossbreed (wbp × pbz);
- The first group was reared in conventional stables, while the other group was reared in special stables, arranged according to the Commission Regulation (EC) No. 889/2008 of 5 September 2008 [31], following the detailed rules of Council Regulation (EC) No. 834/2007 [32] on organic production and labeling of organic products with regard to organic production, labeling, and control;
- The conventional rearing system comprised an indoor area of 1 m2, whereas the organic rearing system had an indoor area with organic saw dust of 1 m2 and an outdoor area with free ranges of 1 m2. The conventional stable had a climate control system, and the organic stable had only gravitation ventilation.
- The organic feed consisted exclusively of raw materials from organic farms and was produced directly on the farm. The second group of animals was fed with commercial feeds available for conventional producers. The composition and nutritional value of the compound feed were in line with the pig nutrition standards of the National Research Council [33]. Both conventionally and organically reared animals were given their diets and water ad libitum.
- One day before slaughter, the hogs (weighing approximately 110 kg) were transported to the abattoir, where they were rested overnight with free access to water. The animals were conventionally slaughtered after an electrical stunning.
2.2. Peptide Extraction and LC–MS/MS Identification
2.3. The Identification of Bioactive Peptides—In Silico Analysis
2.4. Evaluation of Bioactive (Antioxidant) Peptides—In Vitro Analysis
2.4.1. Ability to Scavenge 2,2′-Azino-bis-3-ethylbenzthiazoline-6-sulfonic Acid (ABTS)
2.4.2. Ability to Chelate Fe(II) Ions
2.4.3. Fe(III) Reduction Power (FRAP)
2.5. Statistical Analysis
3. Results
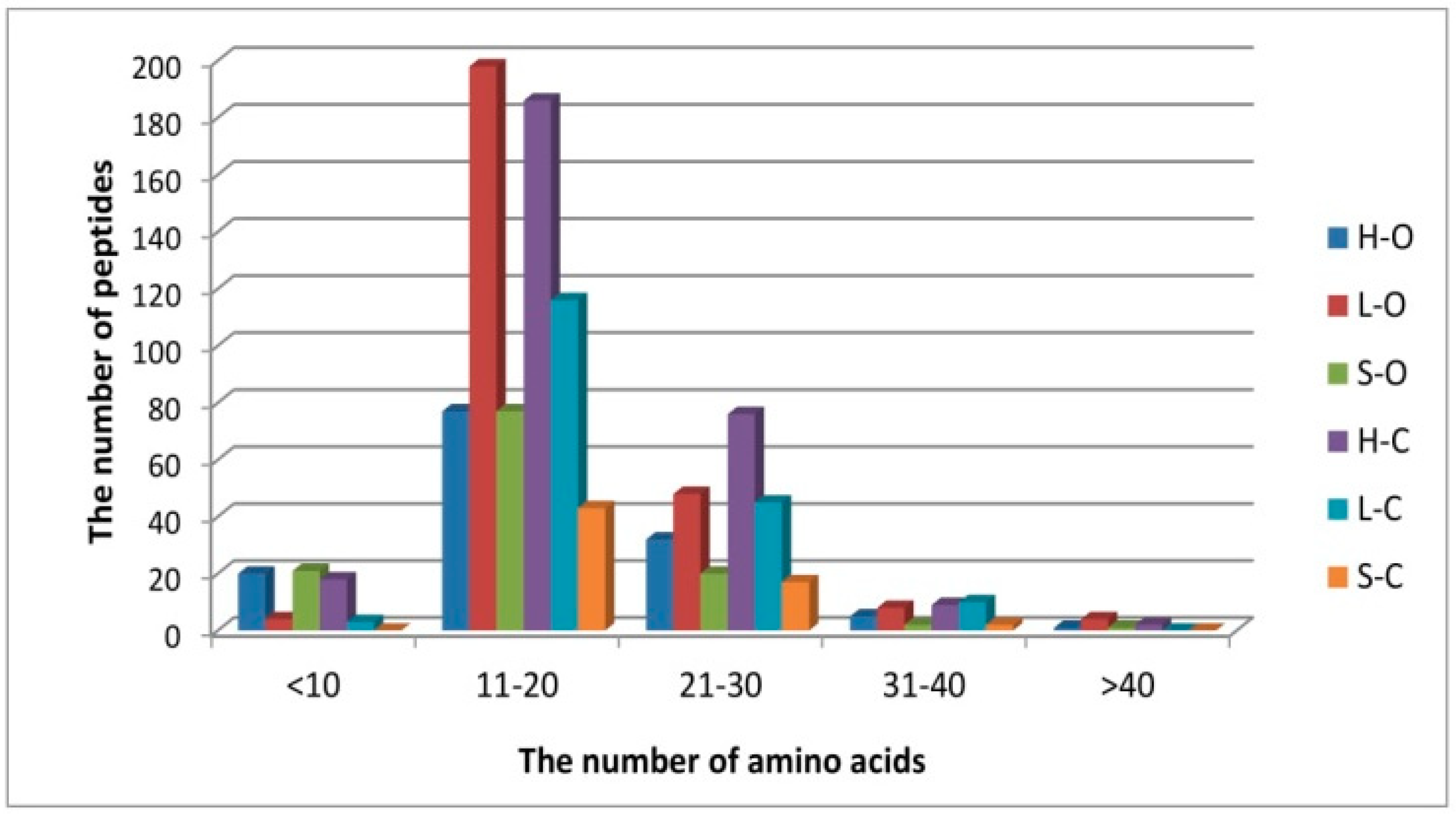
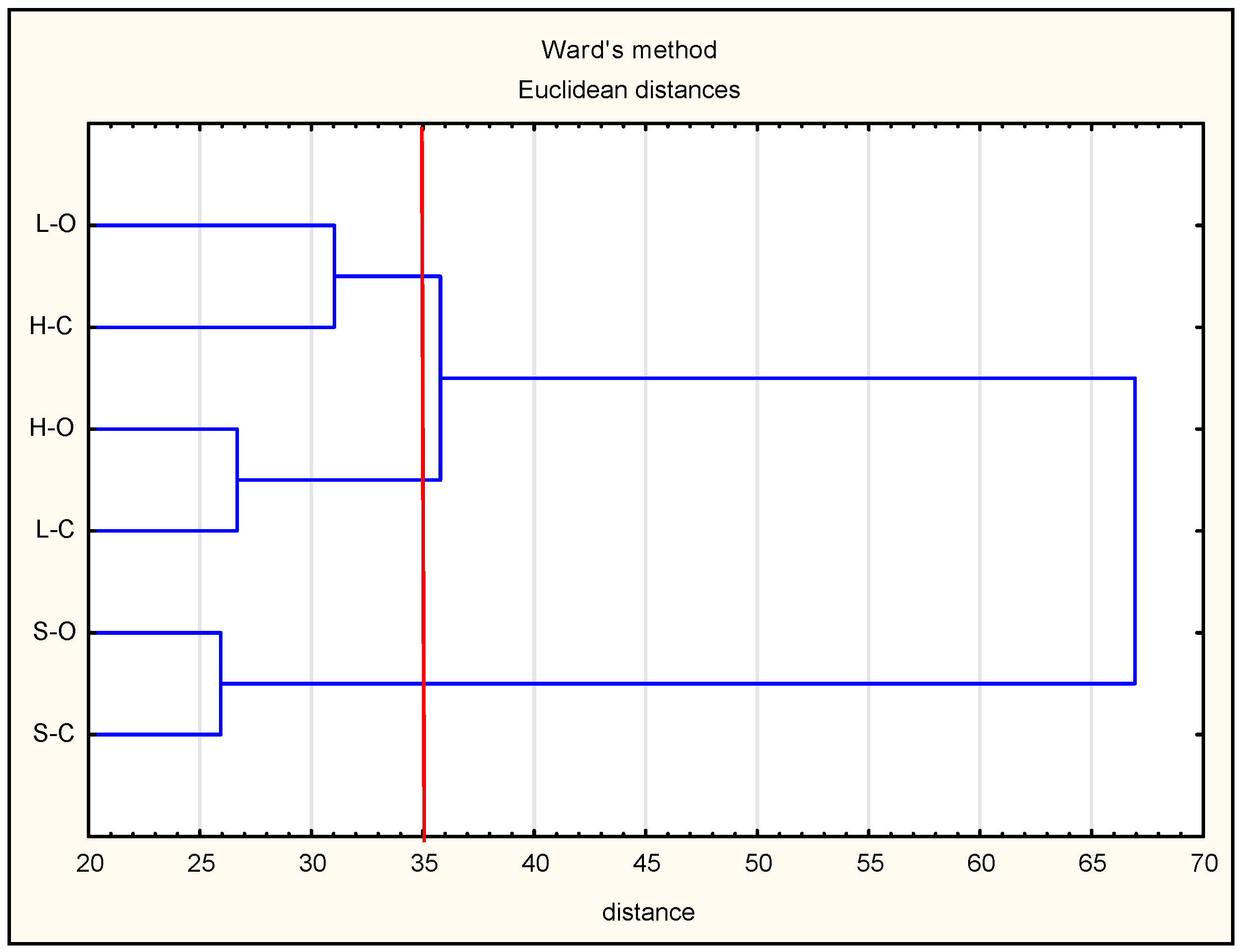
3.1. Protein Degradation and Peptide Formation
3.2. Antioxidant Properties of Peptides—In Silico Analysis
3.3. Antioxidant Properties of Peptides—In Vitro Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Halagarda, M. Decomposition analysis and consumer research as essential elements of the new food product development process. Br. Food J. 2017, 119, 511–526. [Google Scholar] [CrossRef]
- Halagarda, M.; Kędzior, W.; Pyrzyńska, E. Nutritional Value and Potential Chemical Food Safety Hazards of Selected Traditional and Conventional Pork Hams from Poland. J. Food Qual. 2017, 2017, 9037016. [Google Scholar] [CrossRef]
- Yadav, R.; Pathak, G.S. Intention to purchase organic food among young consumers: Evidences from a developing nation. Appetite 2016, 96, 122–128. [Google Scholar] [CrossRef] [PubMed]
- IFOAM Organics International. IFOAM—Organics International. 2016. Available online: https://www.ifoam.bio/ (accessed on 4 November 2019).
- Dangour, A.D.; Allen, E.; Lock, K.; Uauy, R. Nutritional composition and health benefits of organic foods—Using systematic reviews to question the available evidence. Indian J. Med. Res. 2010, 131, 478–480. [Google Scholar] [PubMed]
- Dangour, A.D.; Lock, K.; Hayter, A.; Aikenhead, A.; Allen, E.; Uauy, R. Nutrition-related health effects of organic foods: A systematic review. Am. J. Clin. Nutr. 2010, 92, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Komprda, T. Comparison of quality and safety of organic and conventional foods. Chemickélisty 2009, 103, 729–732. [Google Scholar]
- Schulzová, V.; Hajšlová, J.; Botek, P.; Peroutka, R. Furanocoumarins in vegetables: Influence of farming system and other factors on levels of toxicants. J. Sci. Food Agric. 2007, 87, 2763–2767. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, D.; Yang, S. Effect of organic and conventional rearing system on the mineral content of pork. Meat Sci. 2016, 118, 103–107. [Google Scholar] [CrossRef]
- De Smet, S.; Vossen, E. Meat: The balance between nutrition and health. A review. Meat Sci. 2016, 120, 145–156. [Google Scholar] [CrossRef]
- Cicero, A.F.; Fogacci, F.; Colletti, A. Potential role of bioactive peptides in prevention and treatment of chronic diseases: A narrative review. Br. J. Pharm. 2017, 174, 1378–1394. [Google Scholar] [CrossRef]
- Falowo, A.B.; Fayemi, P.O.; Muchenje, V. Natural antioxidants against lipid–protein oxidative deterioration in meat and meat products: A review. Food Res. Int. 2014, 64, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Mora, L.; Sentandreu, M.A.; Toldrá, F. Identification of small troponin T peptides generated in dry-cured ham. Food Chem. 2010, 123, 691–697. [Google Scholar] [CrossRef]
- Kęska, P.; Stadnik, J. Stability of antiradical activity of protein extracts and hydrolysates from dry-cured pork loins with probiotic strains of LAB. Nutrients 2018, 10, 521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wójciak, K.M.; Kęska, P.; Okoń, A.; Solska, E.; Libera, J.; Dolatowski, Z.J. The influence of acid whey on the antioxidant peptides generated to reduce oxidation and improve colour stability in uncured roast beef. J. Sci. Food Agric. 2018, 98, 3728–3734. [Google Scholar] [CrossRef]
- Gallego, M.; Mora, L.; Toldrá, F. The relevance of dipeptides and tripeptides in the bioactivity and taste of dry-cured ham. Food Prod. Process. Nutr. 2019, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Xiong, Y.L. Natural antioxidants as food and feed additives to promote health benefits and quality of meat products: A review. Meat Sci. 2016, 120, 107–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohaib, M.; Anjum, F.M.; Sahar, A.; Arshad, M.S.; Rahman, U.U.; Imran, A.; Hussain, S. Antioxidant proteins and peptides to enhance the oxidative stability of meat and meat products: A comprehensive review. Int. J. Food Prop. 2017, 20, 2581–2593. [Google Scholar] [CrossRef]
- Liu, R.; Xing, L.; Fu, Q.; Zhou, G.H.; Zhang, W.G. A review of antioxidant peptides derived from meat muscle and by-products. Antioxidants 2016, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Kęska, P.; Stadnik, J. Porcine myofibrillar proteins as potential precursors of bioactive peptides–An in silico study. Food Funct. 2016, 7, 2878–2885. [Google Scholar] [CrossRef]
- Kęska, P.; Wójciak, K.M.; Stadnik, J. Effect of Marination Time on the Antioxidant Properties of Peptides Extracted from Organic Dry-Fermented Beef. Biomolecules 2019, 9, 614. [Google Scholar] [CrossRef] [Green Version]
- Kęska, P.; Stadnik, J. Taste-active peptides and amino acids of pork meat as components of dry-cured meat products: An in-silico study. J. Sens. Stud. 2017, 32, e12301. [Google Scholar] [CrossRef]
- Willcox, J.K.; Ash, S.L.; Catignani, G.L. Antioxidants and prevention of chronic disease. Crit. Rev. Food Sci. Nutr. 2004, 44, 275–295. [Google Scholar] [CrossRef] [PubMed]
- Hernández, P.; Zomeno, L.; Ariño, B.; Blasco, A. Antioxidant, lipolytic and proteolytic enzyme activities in pork meat from different genotypes. Meat Sci. 2004, 66, 525–529. [Google Scholar] [CrossRef]
- Cabrera, M.C.; Saadoun, A. An overview of the nutritional value of beef and lamb meat from South America. Meat Sci. 2014, 98, 435–444. [Google Scholar] [CrossRef]
- Ntawubizi, M.; Raes, K.; De Smet, S. Genetic parameter estimates for plasma oxidative status traits in slaughter pigs. J. Anim. Sci. 2020, 98, skz378. [Google Scholar] [CrossRef]
- Gatellier, P.; Mercier, Y.; Renerre, M. Effect of diet finishing mode (pasture or mixed diet) on antioxidant status of Charolais bovine meat. Meat Sci. 2004, 67, 385–394. [Google Scholar] [CrossRef]
- Descalzo, A.M.; Rossetti, L.; Grigioni, G.; Irurueta, M.; Sancho, A.M.; Carrete, J.; Pensel, N.A. Antioxidant status and odour profile in fresh beef from pasture or grain-fed cattle. Meat Sci. 2007, 75, 299–307. [Google Scholar] [CrossRef]
- Nian, Y.; Allen, P.; Harrison, S.M.; Brunton, N.P.; Prendiville, R.; Kerry, J.P. Fatty acid composition of young dairy bull beef as affected by breed type, production treatment, and relationship to sensory characteristics. Anim. Prod. Sci. 2019, 59, 1360–1372. [Google Scholar] [CrossRef]
- Xu, X.; Chen, X.; Chen, D.; Yu, B.; Yin, J.; Huang, Z. Effects of dietary apple polyphenol supplementation on carcass traits, meat quality, muscle amino acid and fatty acid composition in finishing pigs. Food Funct. 2019, 10, 7426–7434. [Google Scholar] [CrossRef]
- Commission Regulation (EC) No 889/2008 of 5 September 2008 Laying down Detailed Rules for the Implementation of Council Regulation (EC) No 834/2007 on Organic Production and Labelling of Organic Products with Regard to Organic Production, Labelling and Control. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32008R0889 (accessed on 7 December 2019).
- Council Regulation (EC) No 834/2007 of 28 June 2007 on organic production and labelling of organic products and repealing Regulation (EEC) No 2092/91. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32007R0834 (accessed on 7 December 2019).
- National Research Council. Nutrient Requirements of Swine; Tenth Revised Edition; The national Academies Press: Washington, DC, USA, 1998.
- BIOPEP-UWM Database. Available online: http://www.uwm.edu.pl/biochemia/index.php/pl/biopep (accessed on 14 December 2019).
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Decker, E.A.; Welch, B. Role of ferritin as a lipid oxidation catalyst in muscle food. J. Agric. Food Chem. 1990, 38, 674–677. [Google Scholar] [CrossRef]
- Oyaizu, M. Studies on products of browning reaction: Antioxidative activity of products of browning reaction. Jpn. J. Nutr. 1986, 44, 307–315. [Google Scholar] [CrossRef] [Green Version]
- Picard, B.; Gagaoua, M.; Al-Jammas, M.; De Koning, L.; Valais, A.; Bonnet, M. Beef tenderness and intramuscular fat proteomic biomarkers: Muscle type effect. Peer J. 2018, 6, e4891. [Google Scholar] [CrossRef] [PubMed]
- Picard, B.; Gagaoua, M. Meta-proteomics for the discovery of protein biomarkers of beef tenderness: An overview of integrated studies. Food Res. Int. 2019, 127, 108739. [Google Scholar] [CrossRef] [PubMed]
- Picard, B.; Gagaoua, M.; Al Jammas, M.; Bonnet, M. Beef tenderness and intramuscular fat proteomic biomarkers: Effect of gender and rearing practices. J. Proteom. 2019, 200, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Y.; Wang, C.; Tang, C.B.; Dai, C.; Bai, Y.; Yu, X.B.; Li, C.B.; Xu, X.L.; Zhou, G.H.; Cao, J.X. Label-free proteomics reveals the mechanism of bitterness and adhesiveness in Jinhua ham. Food Chem. 2019, 297, 125012. [Google Scholar] [CrossRef]
- Taylor, R.G.; Geesink, G.H.; Thompson, V.F.; Koohmaraie, M.; Goll, D.E. Is Z-disk degradation responsible for postmortem tenderization? J. Anim. Sci. 1995, 73, 1351–1367. [Google Scholar] [CrossRef] [Green Version]
- Huff-Lonergan, E.; Mitsuhashi, T.; Beekman, D.D.; Parrish, F.C., Jr.; Olson, D.G.; Robson, R.M. Proteolysis of specific muscle structural proteins by µ-calpain at low pH and temperature is similar to degradation in postmortem bovine muscle. J. Anim. Sci. 1996, 74, 993–1008. [Google Scholar] [CrossRef] [Green Version]
- Huff-Lonergan, E.; Mitsuhashi, T.; Parrish, F.C., Jr.; Robson, R.M. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blotting comparisons of purified myofibrils and whole muscle preparations for evaluating titin and nebulin in postmortem bovine muscle. J. Anim. Sci. 1996, 74, 779–785. [Google Scholar] [CrossRef] [Green Version]
- Ladrat, C.; Verrez-Bagnis, V.; Noël, J.; Fleurence, J. In vitro proteolysis of myofibrillar and sarcoplasmic proteins of white muscle of sea bass (Dicentrarchus labrax L.): Effects of cathepsins B, D and L. Food Chem. 2003, 81, 517–525. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Li, Z.; Li, X.; Xin, J.; Wang, Y.; Li, G.; Wu, L.; Shen, Q.W.; Zhang, D. Comparative profiling of sarcoplasmic phosphoproteins in ovine muscle with different color stability. Food Chem. 2018, 240, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Sarah, S.A.; Faradalila, W.N.; Salwani, M.S.; Amin, I.; Karsani, S.A.; Sazili, A.Q. LC–QTOF-MS identification of porcine-specific peptide in heat treated pork identifies candidate markers for meat species determination. Food Chem. 2016, 199, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.D.; Seo, J.K.; Yum, H.W.; Jeong, J.Y.; Yang, H.S. Protein markers for discrimination of meat species in raw beef, pork and poultry and their mixtures. Food Chem. 2017, 217, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Li, X.; Zhang, C.H.; Liu, J.Q.; Huang, D.Q. Characterization and discrimination of Tibetan and Duroc×(Landrace× Yorkshire) pork using label-free quantitative proteomics analysis. Food Res. Int. 2019, 119, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Soulat, J.; Bons, J.; Léger, S.; De Koning, L.; Carapito, C.; Picard, B. Quantification of biomarkers for beef meat qualities using a combination of Parallel Reaction Monitoring-and antibody-based proteomics. Food Chem. 2020, 317, 126376. [Google Scholar] [CrossRef] [PubMed]
- Bazile, J.; Picard, B.; Chambon, C.; Valais, A.; Bonnet, M. Pathways and biomarkers of marbling and carcass fat deposition in bovine revealed by a combination of gel-based and gel-free proteomic analyses. Meat Sci. 2019, 156, 146–155. [Google Scholar] [CrossRef]
- Zhu, C.Z.; Zhang, W.G.; Zhou, G.H.; Xu, X.L.; Kang, Z.L.; Yin, Y. Isolation and identification of antioxidant peptides from Jinhua ham. J. Agric. Food Chem. 2013, 61, 1265–1271. [Google Scholar] [CrossRef]
- Khaket, T.P.; Redhu, D.; Dhanda, S.; Singh, J. In silico evaluation of potential DPP-III inhibitor precursors from dietary proteins. Int. J. Food Prop. 2015, 18, 499–507. [Google Scholar] [CrossRef] [Green Version]
- Descalzo, A.M.; Insani, E.M.; Biolatto, A.; Sancho, A.M.; Garcia, P.T.; Pensel, N.A.; Josifovich, J.A. Influence of pasture or grain-based diets supplemented with vitamin E on antioxidant/oxidative balance of Argentine beef. Meat Sci. 2005, 70, 35–44. [Google Scholar] [CrossRef]
- Manso, M.A.; Miguel, M.; Even, J.; Hernández, R.; Aleixandre, A.; López-Fandiño, R. Effect of the long-term intake of an egg white hydrolysate on the oxidative status and blood lipid profile of spontaneously hypertensive rats. Food Chem. 2008, 109, 361–367. [Google Scholar] [CrossRef]
- Ebaid, H.; Salem, A.; Sayed, A.; Metwalli, A. Whey protein enhances normal inflammatory responses during cutaneous wound healing in diabetic rats. Lipids Health Dis. 2011, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katayama, S.; Xu, X.; Fan, M.Z.; Mine, Y. Antioxidative stress activity of oligophosphopeptides derived from hen egg yolk phosvitin in Caco-2 cells. J. Agric. Food Chem. 2006, 54, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Mendis, E.; Rajapakse, N.; Kim, S.K. Antioxidant properties of a radical-scavenging peptide purified from enzymatically prepared fish skin gelatin hydrolysate. J. Agric. Food Chem. 2005, 53, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Picariello, G.; Iacomino, G.; Mamone, G.; Ferranti, P.; Fierro, O.; Gianfrani, C.; Di Luccia, A.; Addeo, F. Transport across Caco-2 monolayers of peptides arising from in vitro digestion of bovine milk proteins. Food Chem. 2013, 139, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Liu, S.; Wang, C.; Li, B. Stability of casein antioxidant peptide fractions during in vitro digestion/Caco-2 cell model: Characteristics of the resistant peptides. Eur. Food Res. Technol. 2014, 239, 577–586. [Google Scholar] [CrossRef]
- Xie, N.; Wang, B.; Jiang, L.; Liu, C.; Li, B. Hydrophobicity exerts different effects on bioavailability and stability of antioxidant peptide fractions from casein during simulated gastrointestinal digestion and Caco-2 cell absorption. Food Res. Int. 2015, 76, 518–526. [Google Scholar] [CrossRef]
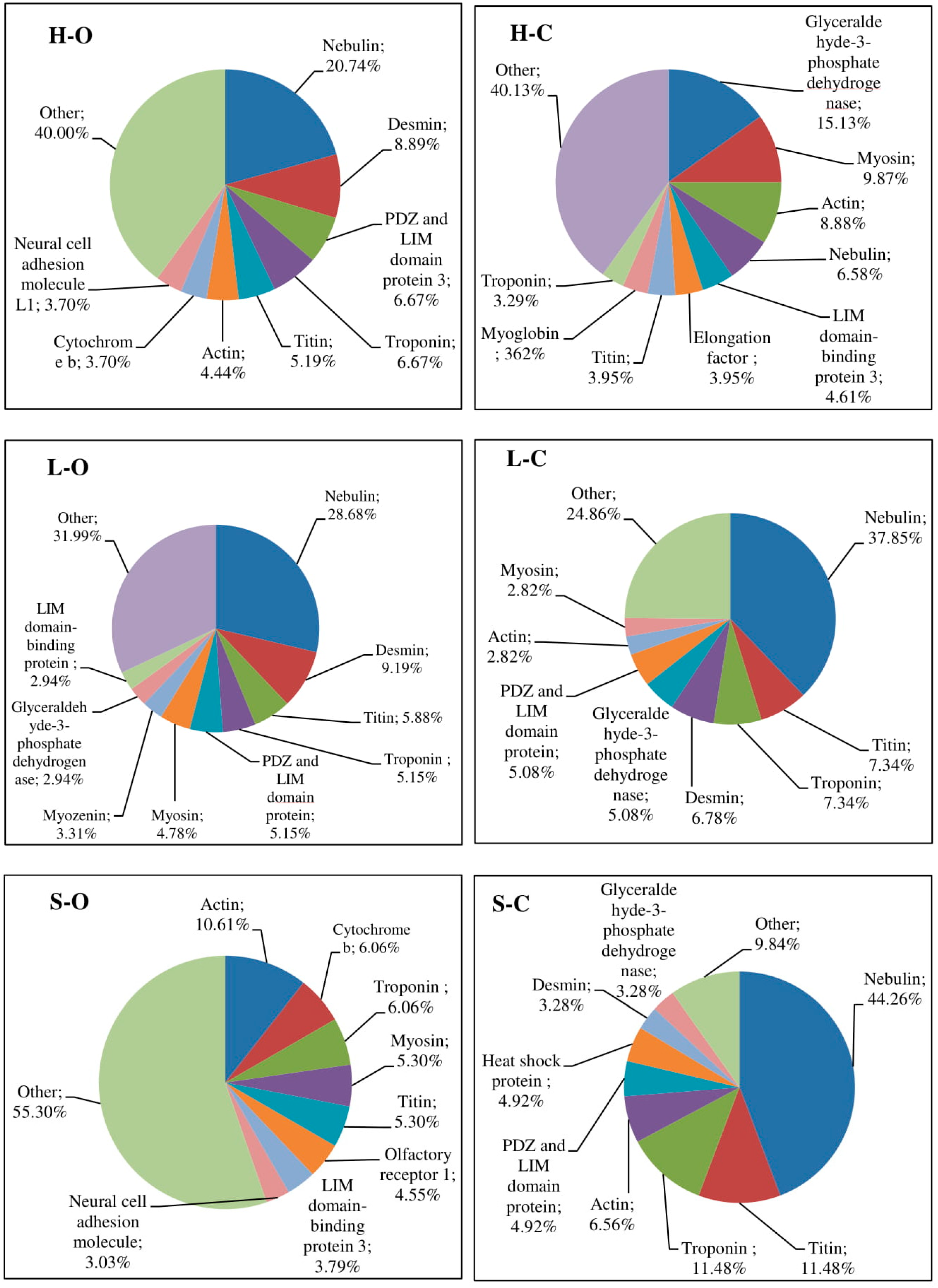
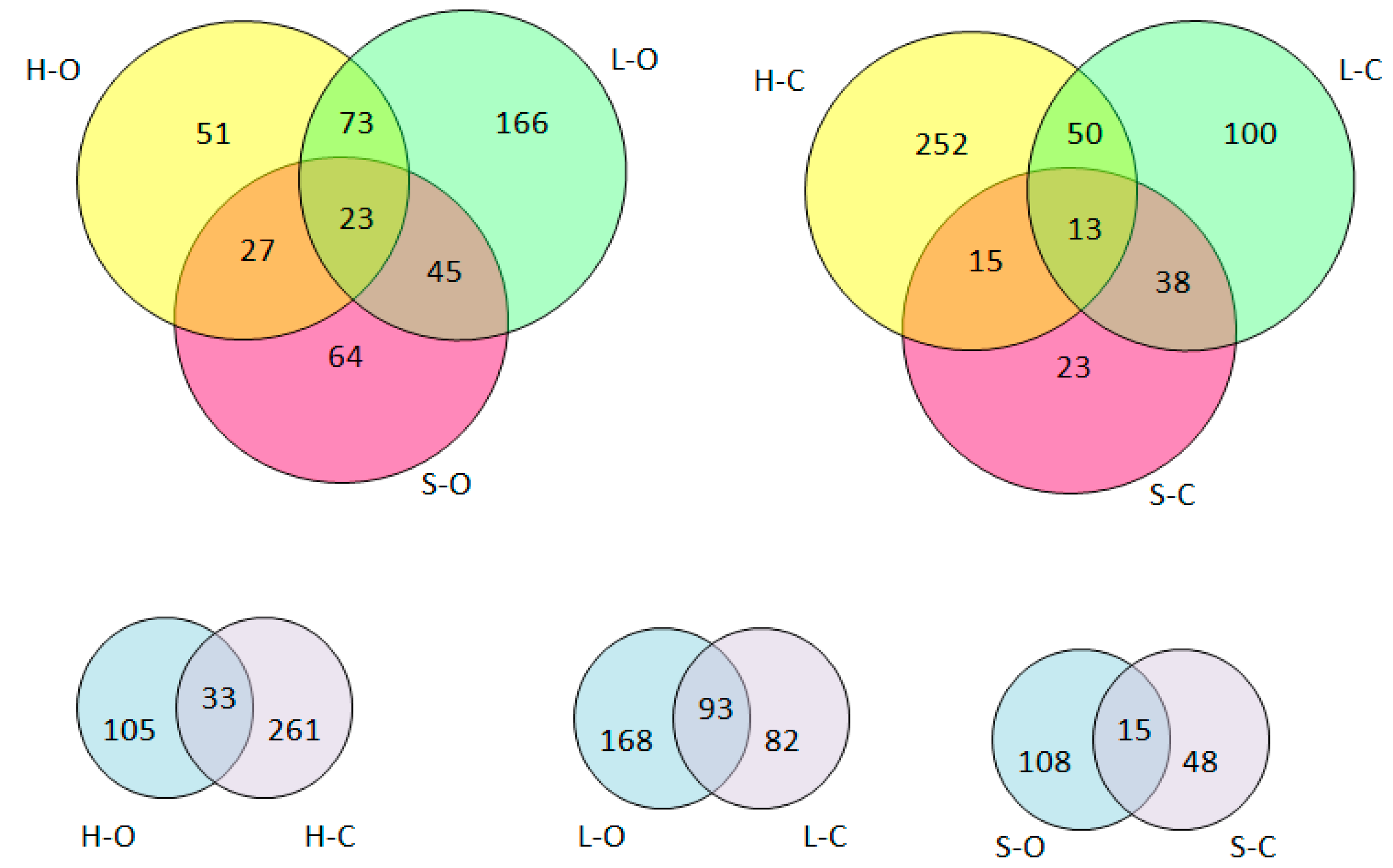
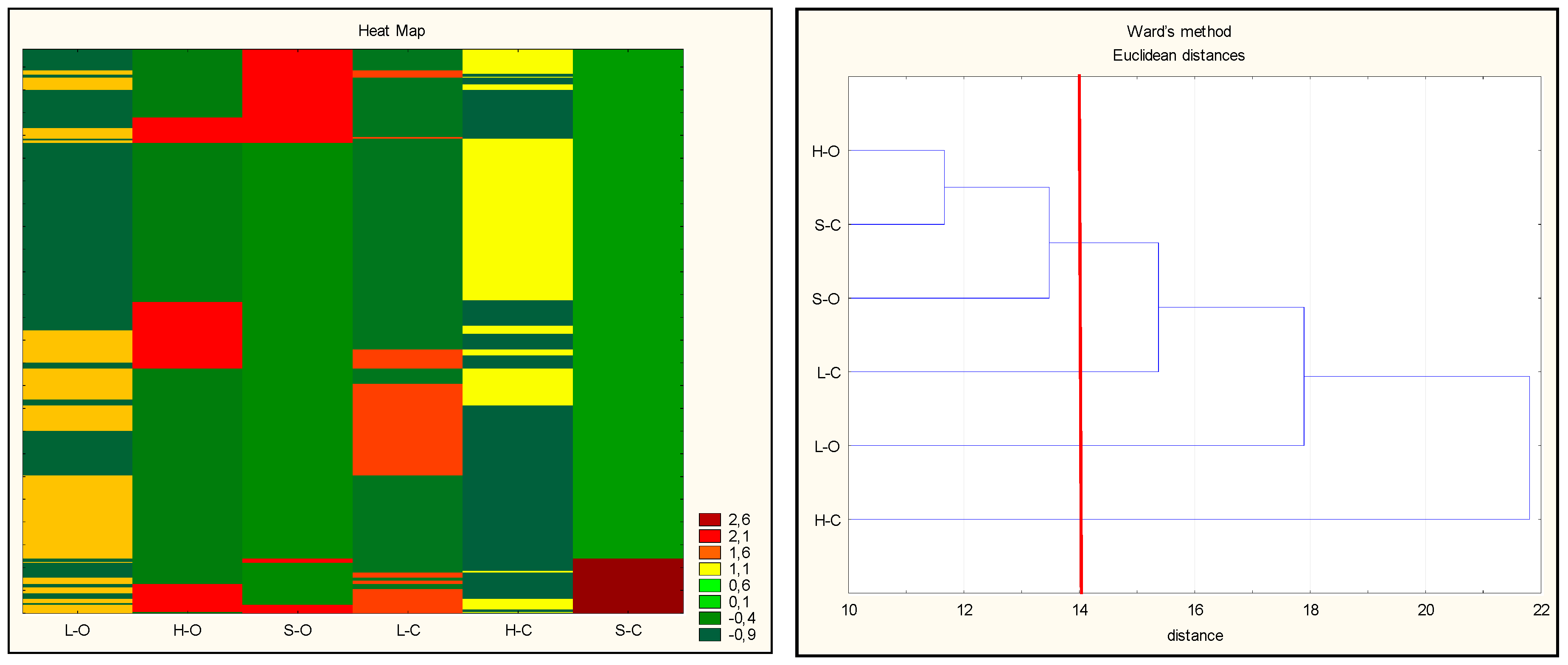
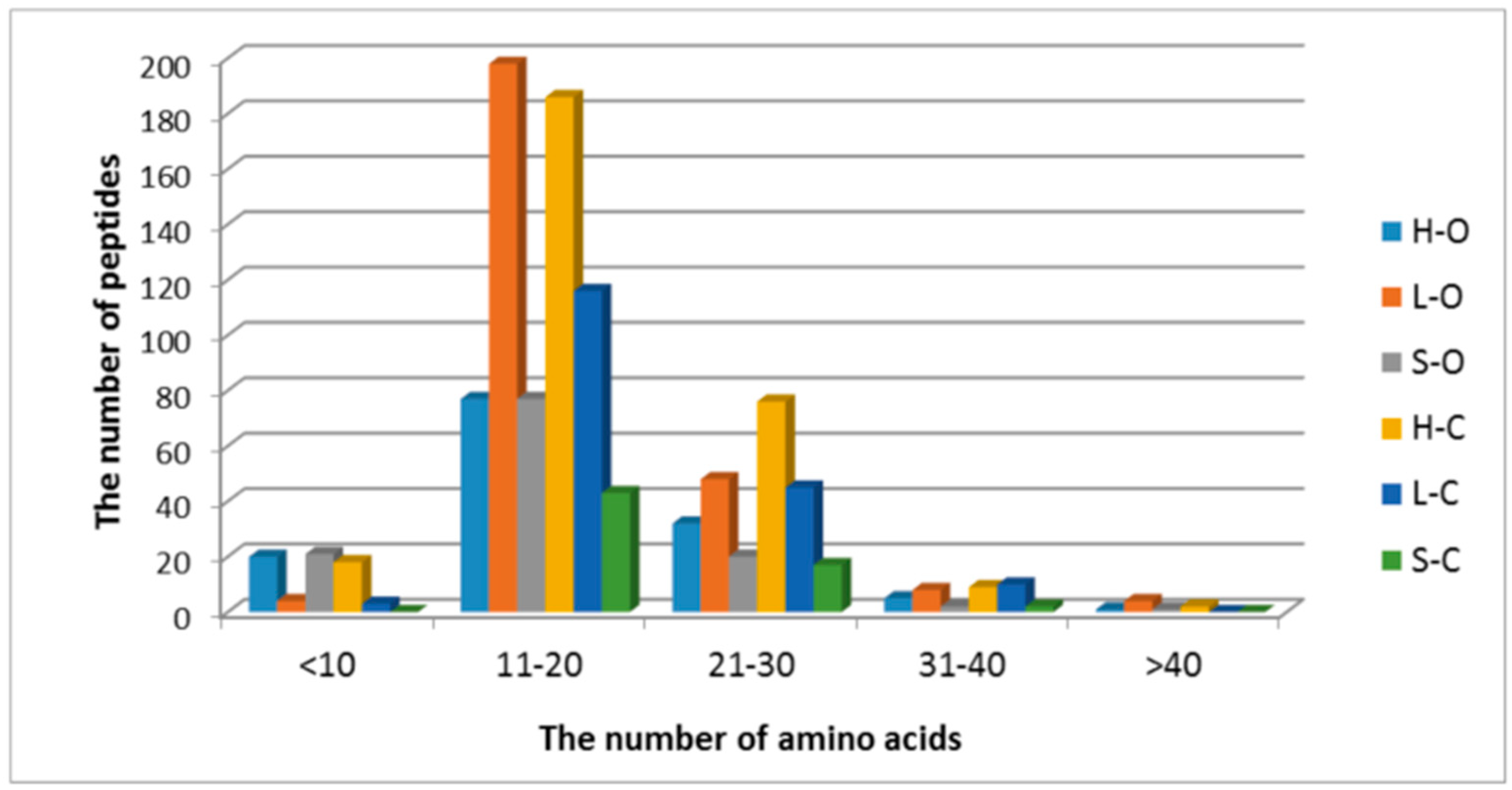
| Lp | Sequence 1 | Protein | Position 2 |
|---|---|---|---|
| 1 | R.VREPVISAVEQTAQR.T | Titin | [438–452] |
| 2 | T.IEPDAVHIKAAKDAYK.V | Nebulin | [5535–5550] |
| 3 | E.EAPPPPAEVHEVHEEVH.E | Troponin T, fast skeletal muscle | [21–38] |
| 4 | W.ITKQEYDEAGPSIVHRK.C | Actin, alpha-skeletal muscle | [358–375] |
| 5 | M.WITKQEYDEAGPSIVHRK.C | [357–375] |
| Sequence 1 | Protein | Position 2 | ACE Inhibitor | DPP-IV Inhibitor | Stimulating 3 | Antioxidant | |
|---|---|---|---|---|---|---|---|
| 1 | R.VREPVISAVEQTAQR.T | Titin | [438–452] | VR, VE, AV | EP, TA, VR, AV, PV, QT, VE, VI | - | - |
| 2 | S.VVDTPEIIHAQQVKN.L 4 | Nebulin | [6063–6077] | VK, EI, TP | VV, HA, TP, EI, IH, II, QQ, QV, VD, VK, | II | - |
| 3 | T.IEPDAVHIKAAKDAYK.V 5 | Nebulin | [5525–5540] | AY, AA, DA(2) 6, YK, KA, IE, AV, IEP | KA, EP, AA, AV, AY, HI, VH, YK | - | AY, KD |
| 4 | Q.IRKETEKAFVPKVVIS.A 7 | Titin | [512–527] | IR, AF, VP, KA, TE, EK, KE, VPK, FVP | KA, VP, VV, EK, AF, ET, IR, KE, KV, PK, RK, TE, VI | - | IR |
| 5 | E.EAPPPPAEVHEVHEEVH.E 8 | Troponin T, fast skeletal muscle | [22–38] | AP, EA, EV(3), EAP, PPP(2) | PPPP, PP(3), AP, PA, AE, EV(3), HE(2), VH(2) | EE | - |
| 6 | M.ARVREPVISAVEQTAQR.T 9 | Titin | [436–452] | VR, AR, VE, AV, RVR | EP, TA, VR, AV, PV, QT, VE, VI | - | - |
| 7 | T.IETRDGEVVSEATQQQH.E 10 | Desmin | [452–468] | GE, EA, DG, IE, EV, TQ, | VV, AT, ET, EV, GE, QH, QQ(2), TQ, TR, VS, | SE | - |
| 8 | W.ITKQEYDEAGPSIVHRK.C 11 | Actin, alpha-skeletal muscle | [359–375] | GP, AG, EA, EY, AGP | GP, AG, EY, HR, PS, QE, RK, SI, TK, VH, YD | IV | - |
| 9 | N.FTSVVDTPEIIHAQQVKN.L 12 | Nebulin | [6060–6077] | VK, EI, TP | VV, HA, TP, EI, HI, II, QQ, QV, SV, TS, VD, VK | II | - |
| 10 | N.YKADLKDLSKKGYDLRTD.A 13 | Nebulin | [1052–1069] | GY, KG, YK, KA, LR | KA, AD, GY, KG, KK, SK, TD, YD, YK | - | KD, LK |
| 11 | M.WITKQEYDEAGPSIVHRK.C 14 | Actin, alpha-skeletal muscle | [358–375] | GP, AG, EA, EY, AGP | GP, WI, AG, EY, HR, PS, QE, RK, SI, TK, VH, YD | IV | - |
| 12 | A.AVDMARVREPVISAVEQTAQR.T 15 | Titin | [432–452] | VR, AR, VE, AV(2), DM, RVR | MA, EP, TA, VR, AV(2), PV, QT, VD, VE, VI | - | - |
| 13 | L.YKEDVSPGTAIGKTPEMMRVKQTQDH.I 16 | Nebulin | [6154–6179] | VK, IG, GK, GT, PG, YK, TQ, KE, AI, TP, MM, VSP | TP, SP, TA, KE, KT, MM, MR, PG, QD, QT, TQ, VK, VS, YK | - | MM |
| Sequence 1 | Protein | Position 2 | ACE Inhibitor | DPP-IV Inhibitor | Stimulating 4 | Antioxidant | |
|---|---|---|---|---|---|---|---|
| 1 | V.IIIIIIIIII.I | Killer cell immunoglobulin-like receptor | [341–349] | - | II (9) 3 | II (9) | - |
| 2 | A.IILLLLILLI.L | Neural cell adhesion molecule | [1132–1142] | IL(2) | LL(4), II, IL(2), LI(2) | LLL(2), IL(2), LI(2), II, LL(4) | - |
| 3 | I.ILLLLILLIL.C | Neural cell adhesion molecule | [1133–1143] | IL(3) | LL(4), IL(3), LI(2) | LLL(2), IL(3), LI(2), LL(4) | - |
| 4 | S.ILILLIILLL.H | Cytochrome b | [297–307] | IL(3) | LL(3), II, IL(3), LI(2) | LLL(1), IL(3), LI(2), II, LL(3) | - |
| 5 | V.ILLLLLLLLL.F | Leukocyte immunoglobulin-like receptor subfamily B | [469–479] | IL(2) | LL(5), IL(2), LI(2) | LLL(4), IL(2), LI(2), LL(5) | - |
| 6 | F.LILILLLLLL.V | Cadherin-1 | [731–741] | IL(2) | LL(5), IL(2), LI(2) | LLL(4), IL(2), LI(2), LL(5) | - |
| 7 | G.LLILILLLLL.L | Cytochrome b | [232–241] | IL(2) | LL(5), IL(2), LI(2) | LLL(3), IL(2), LI(2), LL(5) | - |
| 8 | A.LLLILILLLL.V | Cytochrome b | [233–242] | IL(2) | LL(5), IL(2), LI(2) | LLL(3), IL(2), LI(2), LL(5) | - |
| 9 | H.LLLLLLIIIL.T | Myeloma-overexpressed gene protein | [302–311] | IL(2) | LL(5), II(2), IL, LI | LLL(4), IL, LI, II(2), LL(5) | - |
| 10 | C.LLLLLLLLIL.R | Ephrin | [186–195] | IL | LLL(6), IL, LI, LL(7) | LL(7), IL, LI | - |
| 11 | P.PPPAEVHEVHEEVH.E 5 | Troponin T, fast skeletal muscle | [25–38] | EV(3), PP(2), PPP | PP(2), PA, AE, EV(3), HE(2), VH(3) | EE | - |
| 12 | R.VREPVISAVEQTAQR.T | Titin | [437–452] | VR, VE, AV | EP, TA, VR, AV, PV, QT, VE, VI | - | - |
| 13 | S.VNVDYSKLKKEGPDF 6 | Cytochrome c oxidase subunit NDUFA4 | [68–82] | GP, EG, KL, KE, DY, DF | GP, EG, KE, KK, NV, SK, VD, VN, YS | - | LK |
| 14 | T.IEPDAVHIKAAKDAYK.V 7 | Nebulin | [5525–5540] | AY, AA, DA(2), YK, KA, IE, IEP, AV | KA, KD, AA, AV, AY, HI, VH, YK, | - | AY, KD |
| 15 | E.APPPPAEVHEVHEEVH.E 8 | Troponin T, fast skeletal muscle | [23–38] | AP, EV(3), PP(3) | PPPP, PPP(3), AP, PA, AE, EV(3), HE(2), VH(3) | EE | - |
| 16 | I.TKQEYDEAGPSIVHRK.C 9 | Actin, alpha-skeletal muscle | [360–375] | GP, AG, EA, EY, AGP | GP, AG, EY, HR, PS, QE, RK, SI, TK, VH, YD | IV | - |
| 17 | L.KVSILAAIDEASKKLNAQ 10 | Apolipoprotein A-I | [248–265] | LAA, LA, AA, EA, KL, LN, AI, IL | LA, AA, AS, IL, KK, KV, LN, NA, SI, SK, VS | IL | - |
| 18 | E.EAPPPPAEVHEVHEEVH.E 11 | Troponin T, fast skeletal muscle | [22–38] | AP, EA, EV(3), PP(3), EAP, PPP(2) | PPPP, PP(3), AP, PA, AE, EV, HE, VH | EE | - |
| 19 | E.KAKDIEHAKKVSQQVSK.V 12 | Nebulin | [153–169] | AKK, KA, IE | KA, HA, EH, KK, KV, QQ, QV, SK, VS(2), KA | - | KD |
| 20 | W.ITKQEYDEAGPSIVHRK.C 13 | Actin, alpha-skeletal muscle | [359–375] | GP, AG, EA, EY, AGP | GP, AG, EY, HR, PS, QE, RK, SI, TK, VH, YD | IV | - |
| 21 | ISKQEYDESGPSIVHRK 14 | POTE ankyrin domain family member F | GP, SG, EY, SGP | GP, ES, EY, HR, PS, QE, RK, SI, SK, VH, YD | IV | - | |
| 22 | M.WITKQEYDEAGPSIVHRK.C 15 | Actin, alpha-skeletal muscle | [358–375] | GP, AG, EA, EY, AGP | GP, WI, AG, EY, HR, PS, QE, RK, SI, TK, VH, YD | IV | - |
| 23 | L.KPRPPPPPPAPPKEDVKEKIFQ.L 16 | Titin | [11804–11836] | PR, VK, RP, AP, IF, PAP, PPK, KP, PP(6), EK, KE(2), PAPPK, RPP, FQ, PPP(4) | PPPP(3), PP(6), AP, PA, RP, KP, EK, FQ, KE(2), KI, PK, VK, PR, | - | KP |
| Antioxidant Properties | Ham | Loin | Shoulder | Rearing System (A) | Meat Element (B) | A × B | |
|---|---|---|---|---|---|---|---|
| ABTS assay [%] | O | 39.80 ± 3.11 Aa | 41.89 ± 3.48 Aa | 33.92 ± 3.97 Ab | NS | *** | *** |
| C | 41.51 ± 3.21 Ba | 38.06 ± 2.69 Bb | 34.19 ± 1.71 Ac | ||||
| Fe(II) assay [%] | O | 12.91 ± 2.01 Aa | 13.49 ± 1.60 Aa | 13.24 ± 2.50 Aa | *** | * | ** |
| C | 17.37 ± 1.41 Ba | 14.77 ± 1.73 Ab | 14.72 ± 1.37 Ab | ||||
| FRAP assay | O | 0.567 ± 0.024 Aa | 0.653 ± 0.028 Ab | 0.606 ± 0.011 Ac | *** | *** | ** |
| C | 0.742 ± 0.058 Ba | 0.767 ± 0.036 Ba | 0.767 ± 0.039 Ba | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kęska, P.; Rohn, S.; Halagarda, M.; M. Wójciak, K. Peptides from Different Carcass Elements of Organic and Conventional Pork—Potential Source of Antioxidant Activity. Antioxidants 2020, 9, 835. https://doi.org/10.3390/antiox9090835
Kęska P, Rohn S, Halagarda M, M. Wójciak K. Peptides from Different Carcass Elements of Organic and Conventional Pork—Potential Source of Antioxidant Activity. Antioxidants. 2020; 9(9):835. https://doi.org/10.3390/antiox9090835
Chicago/Turabian StyleKęska, Paulina, Sascha Rohn, Michał Halagarda, and Karolina M. Wójciak. 2020. "Peptides from Different Carcass Elements of Organic and Conventional Pork—Potential Source of Antioxidant Activity" Antioxidants 9, no. 9: 835. https://doi.org/10.3390/antiox9090835
APA StyleKęska, P., Rohn, S., Halagarda, M., & M. Wójciak, K. (2020). Peptides from Different Carcass Elements of Organic and Conventional Pork—Potential Source of Antioxidant Activity. Antioxidants, 9(9), 835. https://doi.org/10.3390/antiox9090835








