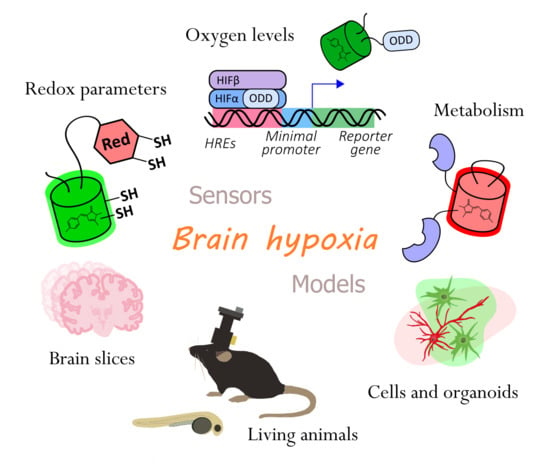
Genetically Encoded Tools for Research of Cell Signaling and Metabolism under Brain Hypoxia
Abstract
1. Introduction
2. Genetically Encoded Reporters for Hypoxia Investigation
2.1. Genetically Encoded Reporters for Oxygen Detection
2.1.1. Natural Oxygen-Sensing Platforms for the Development of Oxygen Reporters
2.1.2. Chromophore Maturation-Based Reporters
2.1.3. HRE-Based Reporters
2.1.4. ODD-Based Degradation Reporters
2.1.5. Combined Reporters (Chromophore Maturation, HRE-Mediated Expression, ODD-Mediated Degradation)
2.1.6. ODD-Based Hydroxylation Reporters
2.1.7. General Limitations of HIF System-Based Reporters for Hypoxia Visualization
2.1.8. Anaerobic GFP Redding-Based Imaging
2.1.9. Haem Proteins-Based Reporters
2.2. Genetically Encoded Reporters of Redox and Metabolic Parameters
2.2.1. Redox-Sensitive Fluorescent Proteins
2.2.2. Genetically Encoded Fluorescent Indicators of ROS and RNS
2.2.3. Genetically Encoded Fluorescent Indicators of NAD(H) and NADP(H)
2.2.4. Genetically Encoded Indicators of Metabolic Parameters
2.2.5. Genetically Encoded pH Indicators
3. In Vitro Models for Real-Time Imaging of Cell Signaling and Metabolism during the Course of Hypoxia
4. Animal Models for Real-Time Imaging of Cell Signaling and Metabolism during the Course of Hypoxia
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Stamati, K.; Mudera, V.; Cheema, U. Evolution of oxygen utilization in multicellular organisms and implications for cell signalling in tissue engineering. J. Tissue Eng. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Samanta, D.; Prabhakar, N.R.; Semenza, G.L. Systems biology of oxygen homeostasis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9, e1382. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, N.R.; Semenza, G.L. Oxygen Sensing and Homeostasis. Physiology 2015, 30, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Dunwoodie, S.L. The Role of Hypoxia in Development of the Mammalian Embryo. Dev. Cell 2009, 17, 755–773. [Google Scholar] [CrossRef] [PubMed]
- Mohyeldin, A.; Garzón-Muvdi, T.; Quiñones-Hinojosa, A. Oxygen in Stem Cell Biology: A Critical Component of the Stem Cell Niche. Cell Stem Cell 2010, 7, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.X.; Hu, M.S.; Esquivel, M.; Liang, G.Y.; Rennert, R.C.; McArdle, A.; Paik, K.J.; Duscher, D.; Gurtner, G.C.; Lorenz, H.P.; et al. The Role of Hypoxia-Inducible Factor in Wound Healing. Adv. Wound Care 2014, 3, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Ruthenborg, R.J.; Ban, J.-J.; Wazir, A.; Takeda, N.; Kim, J.-W. Regulation of wound healing and fibrosis by hypoxia and hypoxia-inducible factor-1. Mol. Cells 2014, 37, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Michiels, C. Physiological and pathological responses to hypoxia. Am. J. Pathol. 2004, 164, 1875–1882. [Google Scholar] [CrossRef]
- Kulandavelu, S.; Balkan, W.; Hare, J.M. Regulation of oxygen delivery to the body via hypoxic vasodilation. Proc. Natl. Acad. Sci. USA 2015, 112, 6254–6255. [Google Scholar] [CrossRef]
- Dunham-Snary, K.J.; Wu, D.; Sykes, E.A.; Thakrar, A.; Parlow, L.R.G.; Mewburn, J.D.; Parlow, J.L.; Archer, S.L. Hypoxic Pulmonary Vasoconstriction: From Molecular Mechanisms to Medicine. Chest 2017, 151, 181–192. [Google Scholar] [CrossRef]
- Krock, B.L.; Skuli, N.; Simon, M.C. Hypoxia-induced angiogenesis: Good and evil. Genes Cancer 2011, 2, 1117–1133. [Google Scholar] [CrossRef] [PubMed]
- Haase, V.H. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 2013, 27, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Oxygen Sensing, Hypoxia-Inducible Factors, and Disease Pathophysiology. Annu. Rev. Pathol. Mech. Dis. 2014, 9, 47–71. [Google Scholar] [CrossRef] [PubMed]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia Auckl. NZ 2015, 3, 83–92. [Google Scholar] [CrossRef]
- Ni, R.; Vaas, M.; Ren, W.; Klohs, J. Noninvasive detection of acute cerebral hypoxia and subsequent matrix-metalloproteinase activity in a mouse model of cerebral ischemia using multispectral-optoacoustic-tomography. Neurophotonics 2018, 5, 015005. [Google Scholar] [CrossRef]
- Heiss, W.-D. Radionuclide Imaging in Ischemic Stroke. J. Nucl. Med. 2014, 55, 1831–1841. [Google Scholar] [CrossRef]
- Mandeville, E.T.; Ayata, C.; Zheng, Y.; Mandeville, J.B. Translational MR Neuroimaging of Stroke and Recovery. Transl. Stroke Res. 2017, 8, 22–32. [Google Scholar] [CrossRef]
- Pak, Y.L.; Swamy, K.M.K.; Yoon, J. Recent Progress in Fluorescent Imaging Probes. Sensors 2015, 15, 24374–24396. [Google Scholar] [CrossRef]
- Kozma, E.; Kele, P. Fluorogenic probes for super-resolution microscopy. Org. Biomol. Chem. 2019, 17, 215–233. [Google Scholar] [CrossRef]
- Depaoli, M.R.; Bischof, H.; Eroglu, E.; Burgstaller, S.; Ramadani-Muja, J.; Rauter, T.; Schinagl, M.; Waldeck-Weiermair, M.; Hay, J.C.; Graier, W.F.; et al. Live cell imaging of signaling and metabolic activities. Pharmacol. Ther. 2019, 202, 98–119. [Google Scholar] [CrossRef]
- Kostyuk, A.I.; Panova, A.S.; Bilan, D.S.; Belousov, V.V. Redox biosensors in a context of multiparameter imaging. Curr. Fluoresc. Chemilumin. Approaches Free Radic. Redox Biol. 2018, 128, 23–39. [Google Scholar] [CrossRef]
- Kostyuk, A.I.; Demidovich, A.D.; Kotova, D.A.; Belousov, V.V.; Bilan, D.S. Circularly Permuted Fluorescent Protein-Based Indicators: History, Principles, and Classification. Int. J. Mol. Sci. 2019, 20, 4200. [Google Scholar] [CrossRef]
- Michael, O. Breckwoldt; Christine Wittmann; Thomas Misgeld; Martin Kerschensteiner; Clemens Grabher Redox imaging using genetically encoded redox indicators in zebrafish and mice. Biol. Chem. 2015, 396, 511–522. [Google Scholar] [CrossRef]
- Bilan, D.S.; Belousov, V.V. In Vivo Imaging of Hydrogen Peroxide with HyPer Probes. Antioxid. Redox Signal. 2018, 29, 569–584. [Google Scholar] [CrossRef] [PubMed]
- Leopold, A.V.; Shcherbakova, D.M.; Verkhusha, V.V. Fluorescent Biosensors for Neurotransmission and Neuromodulation: Engineering and Applications. Front. Cell. Neurosci. 2019, 13, 474. [Google Scholar] [CrossRef] [PubMed]
- Newman, R.H.; Fosbrink, M.D.; Zhang, J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 2011, 111, 3614–3666. [Google Scholar] [CrossRef]
- Shirmanova, M.V.; Druzhkova, I.N.; Lukina, M.M.; Matlashov, M.E.; Belousov, V.V.; Snopova, L.B.; Prodanetz, N.N.; Dudenkova, V.V.; Lukyanov, S.A.; Zagaynova, E.V. Intracellular pH imaging in cancer cells in vitro and tumors in vivo using the new genetically encoded sensor SypHer2. Biochim. Biophys. Acta BBA-Gen. Subj. 2015, 1850, 1905–1911. [Google Scholar] [CrossRef]
- Wagener, K.C.; Kolbrink, B.; Dietrich, K.; Kizina, K.M.; Terwitte, L.S.; Kempkes, B.; Bao, G.; Müller, M. Redox Indicator Mice Stably Expressing Genetically Encoded Neuronal roGFP: Versatile Tools to Decipher Subcellular Redox Dynamics in Neuropathophysiology. Antioxid. Redox Signal. 2016, 25, 41–58. [Google Scholar] [CrossRef]
- Breckwoldt, M.O.; Pfister, F.M.J.; Bradley, P.M.; Marinković, P.; Williams, P.R.; Brill, M.S.; Plomer, B.; Schmalz, A.; St Clair, D.K.; Naumann, R.; et al. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat. Med. 2014, 20, 555–560. [Google Scholar] [CrossRef]
- Dana, H.; Novak, O.; Guardado-Montesino, M.; Fransen, J.W.; Hu, A.; Borghuis, B.G.; Guo, C.; Kim, D.S.; Svoboda, K. Thy1 transgenic mice expressing the red fluorescent calcium indicator jRGECO1a for neuronal population imaging in vivo. PLoS ONE 2018, 13, e0205444. [Google Scholar] [CrossRef]
- Al-Osta, I.; Mucha, M.; Pereda, D.; Piqué-Gili, M.; Okorocha, A.E.; Thomas, R.; Hartell, N.A. Imaging Calcium in Hippocampal Presynaptic Terminals With a Ratiometric Calcium Sensor in a Novel Transgenic Mouse. Front. Cell. Neurosci. 2018, 12, 209. [Google Scholar] [CrossRef] [PubMed]
- Sahl, S.J.; Hell, S.W.; Jakobs, S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 2017, 18, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Vu, C.Q.; Matsuda, T.; Nagai, T. Fluorescent Protein-Based Indicators for Functional Super-Resolution Imaging of Biomolecular Activities in Living Cells. Int. J. Mol. Sci. 2019, 20, 5784. [Google Scholar] [CrossRef] [PubMed]
- Mishina, N.M.; Mishin, A.S.; Belyaev, Y.; Bogdanova, E.A.; Lukyanov, S.; Schultz, C.; Belousov, V.V. Live-Cell STED Microscopy with Genetically Encoded Biosensor. Nano Lett. 2015, 15, 2928–2932. [Google Scholar] [CrossRef]
- Brahimi-Horn, M.C.; Pouysségur, J. HIF at a glance. J. Cell Sci. 2009, 122, 1055. [Google Scholar] [CrossRef] [PubMed]
- Larigot, L.; Juricek, L.; Dairou, J.; Coumoul, X. AhR signaling pathways and regulatory functions. Biochim. Open 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dengler, V.L.; Galbraith, M.; Espinosa, J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Su, H.; Soga, T.; Kranc, K.R.; Pollard, P.J. Prolyl hydroxylase domain enzymes: Important regulators of cancer metabolism. Hypoxia (Auckl. NZ) 2014, 2, 127–142. [Google Scholar] [CrossRef]
- Pugh, C.W.; Ratcliffe, P.J. The von Hippel–Lindau tumor suppressor, hypoxia-inducible factor-1 (HIF-1) degradation, and cancer pathogenesis. Semin. Cancer Biol. 2003, 13, 83–89. [Google Scholar] [CrossRef]
- Frew, I.J.; Krek, W. pVHL: A multipurpose adaptor protein. Sci. Signal. 2008, 1, pe30. [Google Scholar] [CrossRef]
- Craggs, T.D. Green fluorescent protein: Structure, folding and chromophore maturation. Chem. Soc. Rev. 2009, 38, 2865–2875. [Google Scholar] [CrossRef] [PubMed]
- Elowitz, M.B.; Surette, M.G.; Wolf, P.-E.; Stock, J.; Leibler, S. Photoactivation turns green fluorescent protein red. Curr. Biol. 1997, 7, 809–812. [Google Scholar] [CrossRef]
- Lidsky, P.V.; Lukyanov, K.A.; Misra, T.; Handke, B.; Mishin, A.S.; Lehner, C.F. A genetically encoded fluorescent probe for imaging of oxygenation gradients in living Drosophila. Development 2018, 145, dev156257. [Google Scholar] [CrossRef] [PubMed]
- Yanushevich, Y.G.; Staroverov, D.B.; Savitsky, A.P.; Fradkov, A.F.; Gurskaya, N.G.; Bulina, M.E.; Lukyanov, K.A.; Lukyanov, S.A. A strategy for the generation of non-aggregating mutants of Anthozoa fluorescent proteins. FEBS Lett. 2002, 511, 11–14. [Google Scholar] [CrossRef]
- Terskikh, A.; Fradkov, A.; Ermakova, G.; Zaraisky, A.; Tan, P.; Kajava, A.V.; Zhao, X.; Lukyanov, S.; Matz, M.; Kim, S.; et al. “Fluorescent Timer”: Protein That Changes Color with Time. Science 2000, 290, 1585. [Google Scholar] [CrossRef]
- Verkhusha, V.V.; Chudakov, D.M.; Gurskaya, N.G.; Lukyanov, S.; Lukyanov, K.A. Common pathway for the red chromophore formation in fluorescent proteins and chromoproteins. Chem. Biol. 2004, 11, 845–854. [Google Scholar] [CrossRef]
- Baird, G.S.; Zacharias, D.A.; Tsien, R.Y. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 2000, 97, 11984–11989. [Google Scholar] [CrossRef]
- Drepper, T.; Eggert, T.; Circolone, F.; Heck, A.; Krauss, U.; Guterl, J.-K.; Wendorff, M.; Losi, A.; Gartner, W.; Jaeger, K.-E. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 2007, 25, 443–445. [Google Scholar] [CrossRef]
- Potzkei, J.; Kunze, M.; Drepper, T.; Gensch, T.; Jaeger, K.-E.; Büchs, J. Real-time determination of intracellular oxygen in bacteria using a genetically encoded FRET-based biosensor. BMC Biol. 2012, 10, 28. [Google Scholar] [CrossRef]
- Eichhof, I.; Ernst, J.F. Oxygen-independent FbFP: Fluorescent sentinel and oxygen sensor component in Saccharomyces cerevisiae and Candida albicans. Fungal Genet. Biol. 2016, 92, 14–25. [Google Scholar] [CrossRef]
- Semenza, G.L.; Jiang, B.-H.; Leung, S.W.; Passantino, R.; Concordet, J.-P.; Maire, P.; Giallongo, A. Hypoxia Response Elements in the Aldolase A, Enolase 1, and Lactate Dehydrogenase A Gene Promoters Contain Essential Binding Sites for Hypoxia-inducible Factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef] [PubMed]
- Berra, E.; Benizri, E.; Ginouvès, A.; Volmat, V.; Roux, D.; Pouysségur, J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003, 22, 4082–4090. [Google Scholar] [CrossRef]
- Emerling, B.M.; Weinberg, F.; Liu, J.-L.; Mak, T.W.; Chandel, N.S. PTEN regulates p300-dependent hypoxia-inducible factor 1 transcriptional activity through Forkhead transcription factor 3a (FOXO3a). Proc. Natl. Acad. Sci. USA 2008, 105, 2622. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Dosey, T.L.; Biechele, T.; Moon, R.T.; Horwitz, M.S.; Ruohola-Baker, H. Assessment of Hypoxia Inducible Factor Levels in Cancer Cell Lines upon Hypoxic Induction Using a Novel Reporter Construct. PLoS ONE 2011, 6, e27460. [Google Scholar] [CrossRef] [PubMed]
- Bagnall, J.; Leedale, J.; Taylor, S.E.; Spiller, D.G.; White, M.R.H.; Sharkey, K.J.; Bearon, R.N.; Sée, V. Tight Control of Hypoxia-inducible Factor-α Transient Dynamics Is Essential for Cell Survival in Hypoxia. J. Biol. Chem. 2014, 289, 5549–5564. [Google Scholar] [CrossRef]
- Phillips, M.I.; Tang, Y.; Schmidt-Ott, K.; Qian, K.; Kagiyama, S. Vigilant Vector: Heart-Specific Promoter in an Adeno-Associated Virus Vector for Cardioprotection. Hypertension 2002, 39, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Jackson, M.; Qian, K.; Phillips, M. Ian Hypoxia Inducible Double Plasmid System for Myocardial Ischemia Gene Therapy. Hypertension 2002, 39, 695–698. [Google Scholar] [CrossRef]
- Shibata, T.; Akiyama, N.; Noda, M.; Sasai, K.; Hiraoka, M. Enhancement of gene expression under hypoxic conditions using fragments of the human vascular endothelial growth factor and the erythropoietin genes. Int. J. Radiat. Oncol. Biol. Phys. 1998, 42, 913–916. [Google Scholar] [CrossRef]
- Shibata, T.; Giaccia, A.J.; Brown, J.M. Development of a hypoxia-responsive vector for tumor-specific gene therapy. Gene Ther. 2000, 7, 493–498. [Google Scholar] [CrossRef]
- Sun, X.; Xing, L.; Deng, X.; Hsiao, H.T.; Manami, A.; Koutcher, J.A.; Clifton Ling, C.; Li, G.C. Hypoxia targeted bifunctional suicide gene expression enhances radiotherapy in vitro and in vivo. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2012, 105, 57–63. [Google Scholar] [CrossRef]
- Hsiao, H.T.; Xing, L.; Deng, X.; Sun, X.; Ling, C.C.; Li, G.C. Hypoxia-targeted triple suicide gene therapy radiosensitizes human colorectal cancer cells. Oncol. Rep. 2014, 32, 723–729. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Post, D.; Van Meir, E. Generation of bidirectional hypoxia/HIF-responsive expression vectors to target gene expression to hypoxic cells. Gene Ther. 2001, 8, 1801–1807. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, H.; Mu, Z.; Jiang, Z.; Wang, Y.; Yang, G.-Y.; Zhang, Z. Hypoxia-controlled matrix metalloproteinase-9 hyperexpression promotes behavioral recovery after ischemia. Neurosci. Bull. 2015, 31, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Ma, Y.; Jiang, L.; Mu, Z.; Jiang, Z.; Chen, X.; Wang, Y.; Yang, G.-Y.; Zhang, Z. Hypoxia Response Element-Regulated MMP-9 Promotes Neurological Recovery via Glial Scar Degradation and Angiogenesis in Delayed Stroke. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 1448–1459. [Google Scholar] [CrossRef]
- Indovina, P.; Collini, M.; Chirico, G.; Santini, M.T. Three-dimensional cell organization leads to almost immediate HRE activity as demonstrated by molecular imaging of MG-63 spheroids using two-photon excitation microscopy. FEBS Lett. 2007, 581, 719–726. [Google Scholar] [CrossRef]
- Moeller, B.J.; Cao, Y.; Li, C.Y.; Dewhirst, M.W. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell 2004, 5, 429–441. [Google Scholar] [CrossRef]
- Cao, Y.; Li, C.-Y.; Moeller, B.J.; Yu, D.; Zhao, Y.; Dreher, M.R.; Shan, S.; Dewhirst, M.W. Observation of Incipient Tumor Angiogenesis That Is Independent of Hypoxia and Hypoxia Inducible Factor-1 Activation. Cancer Res. 2005, 65, 5498. [Google Scholar] [CrossRef][Green Version]
- Le, A.; Stine, Z.E.; Nguyen, C.; Afzal, J.; Sun, P.; Hamaker, M.; Siegel, N.M.; Gouw, A.M.; Kang, B.; Yu, S.-H.; et al. Tumorigenicity of hypoxic respiring cancer cells revealed by a hypoxia–cell cycle dual reporter. Proc. Natl. Acad. Sci. USA 2014, 111, 12486. [Google Scholar] [CrossRef]
- Lehmann, S.; Stiehl, D.P.; Honer, M.; Dominietto, M.; Keist, R.; Kotevic, I.; Wollenick, K.; Ametamey, S.; Wenger, R.H.; Rudin, M. Longitudinal and multimodal in vivo imaging of tumor hypoxia and its downstream molecular events. Proc. Natl. Acad. Sci. USA 2009, 106, 14004–14009. [Google Scholar] [CrossRef]
- He, F.; Deng, X.; Wen, B.; Liu, Y.; Sun, X.; Xing, L.; Minami, A.; Huang, Y.; Chen, Q.; Zanzonico, P.B.; et al. Noninvasive molecular imaging of hypoxia in human xenografts: Comparing hypoxia-induced gene expression with endogenous and exogenous hypoxia markers. Cancer Res. 2008, 68, 8597–8606. [Google Scholar] [CrossRef]
- Penet, M.-F.; Pathak, A.P.; Raman, V.; Ballesteros, P.; Artemov, D.; Bhujwalla, Z.M. Noninvasive Multiparametric Imaging of Metastasis-Permissive Microenvironments in a Human Prostate Cancer Xenograft. Cancer Res. 2009, 69, 8822. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Greenwood, T.R.; Artemov, D.; Raman, V.; Winnard, P.T.; Heeren, R.M.A.; Bhujwalla, Z.M.; Glunde, K. Localized Hypoxia Results in Spatially Heterogeneous Metabolic Signatures in Breast Tumor Models. Neoplasia 2012, 14, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.-W.; Feng, H.-L.; Wang, M.; Zhu, Q.-L.; Liu, Y.-Q.; Jiang, Y.-X. In vivo longitudinal and multimodal imaging of hypoxia-inducible factor 1α and angiogenesis in breast cancer. Chin. Med. J. (Engl.) 2020, 133. [Google Scholar] [CrossRef] [PubMed]
- Lavista-Llanos, S.; Centanin, L.; Irisarri, M.; Russo, D.M.; Gleadle, J.M.; Bocca, S.N.; Muzzopappa, M.; Ratcliffe, P.J.; Wappner, P. Control of the Hypoxic Response in Drosophila melanogaster by the Basic Helix-Loop-Helix PAS Protein Similar. Mol. Cell. Biol. 2002, 22, 6842. [Google Scholar] [CrossRef]
- Santhakumar, K.; Judson, E.C.; Elks, P.M.; McKee, S.; Elworthy, S.; van Rooijen, E.; Walmsley, S.S.; Renshaw, S.; Cross, S.S.; van Eeden, F.J. A zebrafish model to study and therapeutically manipulate hypoxia signaling in tumorigenesis. Cancer Res. 2012, 72, 4017–4027. [Google Scholar] [CrossRef]
- Boast, K.; Binley, K.; Iqball, S.; Price, T.; Spearman, H.; Kingsman, S.; Kingsman, A.; Naylor, S. Characterization of Physiologically Regulated Vectors for the Treatment of Ischemic Disease. Hum. Gene Ther. 1999, 10, 2197–2208. [Google Scholar] [CrossRef]
- Fomicheva, E.V.; Turner, I.I.; Edwards, T.G.; Hoff, J.; Arden, E.; D’Alecy, L.G.; Metzger, J.M. Double Oxygen–sensing Vector System for Robust Hypoxia/Ischemia-regulated Gene Induction in Cardiac Muscle In Vitro and In Vivo. Mol. Ther. 2008, 16, 1594–1601. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Inubushi, M.; Jin, Y.-N.; Murai, C.; Tsuji, A.B.; Hata, H.; Kitagawa, Y.; Saga, T. Detailed assessment of gene activation levels by multiple hypoxia-responsive elements under various hypoxic conditions. Ann. Nucl. Med. 2014, 28, 1011–1019. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Lee, Y.-S.; Kim, J.-M.; Kim, K.L.; Lee, J.-S.; Jang, H.-S.; Shin, I.-S.; Suh, W.; Jeon, E.-S.; Byun, J.; et al. A novel chimeric promoter that is highly responsive to hypoxia and metals. Gene Ther. 2006, 13, 857–868. [Google Scholar] [CrossRef]
- Iglesias, P.; Costoya, J.A. A novel BRET-based genetically encoded biosensor for functional imaging of hypoxia. Biosens. Bioelectron. 2009, 24, 3126–3130. [Google Scholar] [CrossRef]
- Morfoisse, F.; Renaud, E.; Hantelys, F.; Prats, A.-C.; Garmy-Susini, B. Role of hypoxia and vascular endothelial growth factors in lymphangiogenesis. Mol. Cell. Oncol. 2014, 1, e29907. [Google Scholar] [CrossRef][Green Version]
- Cavadas, M.A.S.; Cheong, A.; Taylor, C.T. The regulation of transcriptional repression in hypoxia. Exp. Cell Res. 2017, 356, 173–181. [Google Scholar] [CrossRef]
- Wen, B.; Burgman, P.; Zanzonico, P.; O’Donoghue, J.; Cai, S.; Finn, R.; Serganova, I.; Blasberg, R.; Gelovani, J.; Li, G.C.; et al. A preclinical model for noninvasive imaging of hypoxia-induced gene expression; comparison with an exogenous marker of tumor hypoxia. Eur. J. Nucl. Med. Mol. Imaging 2004, 31, 1530–1538. [Google Scholar] [CrossRef] [PubMed]
- Serganova, I.; Doubrovin, M.; Vider, J.; Ponomarev, V.; Soghomonyan, S.; Beresten, T.; Ageyeva, L.; Serganov, A.; Cai, S.; Balatoni, J.; et al. Molecular Imaging of Temporal Dynamics and Spatial Heterogeneity of Hypoxia-Inducible Factor-1 Signal Transduction Activity in Tumors in Living Mice. Cancer Res. 2004, 64, 6101. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Doran, D.M.; Kulkarni-Datar, K.; Cool, D.R.; Brown, T.L. Hypoxia activates constitutive luciferase reporter constructs. Biochimie 2011, 93, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-H.; Kuo, J.-W.; Lee, Y.-J.; Chang, C.-W.; Gelovani, J.G.; Liu, R.-S. Construction of mutant TKGFP for real-time imaging of temporal dynamics of HIF-1 signal transduction activity mediated by hypoxia and reoxygenation in tumors in living mice. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2009, 50, 2049–2057. [Google Scholar] [CrossRef]
- D’Angelo, G.; Duplan, E.; Vigne, P.; Frelin, C. Cyclosporin A Prevents the Hypoxic Adaptation by Activating Hypoxia-inducible Factor-1α Pro-564 Hydroxylation. J. Biol. Chem. 2003, 278, 15406–15411. [Google Scholar] [CrossRef]
- D’Angelo, G.; Duplan, E.; Boyer, N.; Vigne, P.; Frelin, C. Hypoxia Up-regulates Prolyl Hydroxylase Activity: A FEEDBACK MECHANSIM THAT LIMITS HIF-1 RESPONSES DURING REOXYGENATION. J. Biol. Chem. 2003, 278, 38183–38187. [Google Scholar] [CrossRef]
- Masson, N.; Willam, C.; Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001, 20, 5197–5206. [Google Scholar] [CrossRef]
- Chowdhury, R.; Leung, I.K.H.; Tian, Y.-M.; Abboud, M.I.; Ge, W.; Domene, C.; Cantrelle, F.-X.; Landrieu, I.; Hardy, A.P.; Pugh, C.W.; et al. Structural basis for oxygen degradation domain selectivity of the HIF prolyl hydroxylases. Nat. Commun. 2016, 7, 12673. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.-M.; Yeoh, K.K.; Lee, M.K.; Eriksson, T.; Kessler, B.M.; Kramer, H.B.; Edelmann, M.J.; Willam, C.; Pugh, C.W.; Schofield, C.J.; et al. Differential sensitivity of hypoxia inducible factor hydroxylation sites to hypoxia and hydroxylase inhibitors. J. Biol. Chem. 2011, 286, 13041–13051. [Google Scholar] [CrossRef] [PubMed]
- Hirsilä, M.; Koivunen, P.; Günzler, V.; Kivirikko, K.I.; Myllyharju, J. Characterization of the Human Prolyl 4-Hydroxylases That Modify the Hypoxia-inducible Factor. J. Biol. Chem. 2003, 278, 30772–30780. [Google Scholar] [CrossRef]
- Smirnova, N.A.; Rakhman, I.; Moroz, N.; Basso, M.; Payappilly, J.; Kazakov, S.; Hernandez-Guzman, F.; Gaisina, I.N.; Kozikowski, A.P.; Ratan, R.R.; et al. Utilization of an in vivo reporter for high throughput identification of branched small molecule regulators of hypoxic adaptation. Chem. Biol. 2010, 17, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Koivunen, P.; Hirsilä, M.; Günzler, V.; Kivirikko, K.I.; Myllyharju, J. Catalytic Properties of the Asparaginyl Hydroxylase (FIH) in the Oxygen Sensing Pathway Are Distinct from Those of Its Prolyl 4-Hydroxylases. J. Biol. Chem. 2004, 279, 9899–9904. [Google Scholar] [CrossRef] [PubMed]
- Appelhoff, R.J.; Tian, Y.-M.; Raval, R.R.; Turley, H.; Harris, A.L.; Pugh, C.W.; Ratcliffe, P.J.; Gleadle, J.M. Differential Function of the Prolyl Hydroxylases PHD1, PHD2, and PHD3 in the Regulation of Hypoxia-inducible Factor. J. Biol. Chem. 2004, 279, 38458–38465. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, N.A.; Osipyants, A.I.; Khristichenko, A.Y.; Hushpulian, D.M.; Nikulin, S.V.; Chubar, T.A.; Zakhariants, A.A.; Tishkov, V.I.; Gazaryan, I.G.; Poloznikov, A.A. HIF2 ODD-luciferase reporter: The most sensitive assay for HIF prolyl hydroxylase inhibitors. Russ. Chem. Bull. 2018, 67, 150–156. [Google Scholar] [CrossRef]
- Okamoto, M.; Asamura, A.; Tanaka, K.; Soeda, T.; Watanabe, K.; Mizuguchi, H.; Ikeda, T. Expression of HIF-1α ODD domain fused canine caspase 3 by EGFR promoter-driven adenovirus vector induces cytotoxicity in canine breast tumor cells under hypoxia. Vet. Res. Commun. 2016, 40, 131–139. [Google Scholar] [CrossRef]
- Arquier, N.; Vigne, P.; Duplan, E.; Hsu, T.; Therond, P.P.; Frelin, C.; D’Angelo, G. Analysis of the hypoxia-sensing pathway in Drosophila melanogaster. Biochem. J. 2005, 393, 471–480. [Google Scholar] [CrossRef][Green Version]
- Misra, T.; Baccino-Calace, M.; Meyenhofer, F.; Rodriguez-Crespo, D.; Akarsu, H.; Armenta-Calderón, R.; Gorr, T.A.; Frei, C.; Cantera, R.; Egger, B.; et al. A genetically encoded biosensor for visualising hypoxia responses in vivo. Biol. Open 2017, 6, 296. [Google Scholar] [CrossRef]
- Baccino-Calace, M.; Prieto, D.; Cantera, R.; Egger, B. Compartment and cell type-specific hypoxia responses in the developing Drosophila brain. bioRxiv 2019. [Google Scholar] [CrossRef]
- Safran, M.; Kim, W.Y.; O’Connell, F.; Flippin, L.; Günzler, V.; Horner, J.W.; DePinho, R.A.; Kaelin, W.G. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: Assessment of an oral agent that stimulates erythropoietin production. Proc. Natl. Acad. Sci. USA 2006, 103, 105. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, A.; Pace, A.; Rockwell, C.E.; Roth, K.J.; Chow, A.; O’Brien, K.M.; Albee, R.; Kelly, K.; Towery, K.; Luyendyk, J.P.; et al. Hepatic Stellate Cells Orchestrate Clearance of Necrotic Cells in a Hypoxia-Inducible Factor-1α–Dependent Manner by Modulating Macrophage Phenotype in Mice. J. Immunol. 2014, 1303195. [Google Scholar] [CrossRef] [PubMed]
- Goldman, S.J.; Chen, E.; Taylor, R.; Zhang, S.; Petrosky, W.; Reiss, M.; Jin, S. Use of the ODD-Luciferase Transgene for the Non-Invasive Imaging of Spontaneous Tumors in Mice. PLoS ONE 2011, 6, e18269. [Google Scholar] [CrossRef] [PubMed]
- Goldman, S.J.; Jin, S. The bioluminescent imaging of spontaneously occurring tumors in immunocompetent. Methods Mol. Biol. (Clifton NJ) 2014, 1098, 129–143. [Google Scholar] [CrossRef]
- Harada, H.; Kizaka-Kondoh, S.; Itasaka, S.; Shibuya, K.; Morinibu, A.; Shinomiya, K.; Hiraoka, M. The combination of hypoxia-response enhancers and an oxygen-dependent proteolytic motif enables real-time imaging of absolute HIF-1 activity in tumor xenografts. Biochem. Biophys. Res. Commun. 2007, 360, 791–796. [Google Scholar] [CrossRef]
- Harada, H.; Itasaka, S.; Kizaka-Kondoh, S.; Shibuya, K.; Morinibu, A.; Shinomiya, K.; Hiraoka, M. The Akt/mTOR Pathway Assures the Synthesis of HIF-1α Protein in a Glucose- and Reoxygenation-dependent Manner in Irradiated Tumors. J. Biol. Chem. 2009, 284, 5332–5342. [Google Scholar] [CrossRef]
- Kadonosono, T.; Kuchimaru, T.; Yamada, S.; Takahashi, Y.; Murakami, A.; Tani, T.; Watanabe, H.; Tanaka, T.; Hirota, K.; Inoue, M.; et al. Detection of the Onset of Ischemia and Carcinogenesis by Hypoxia-Inducible Transcription Factor-Based In Vivo Bioluminescence Imaging. PLoS ONE 2011, 6, e26640. [Google Scholar] [CrossRef]
- Saha, D.; Dunn, H.; Zhou, H.; Harada, H.; Hiraoka, M.; Mason, R.P.; Zhao, D. In vivo bioluminescence imaging of tumor hypoxia dynamics of breast cancer brain metastasis in a mouse model. J. Vis. Exp. JoVE 2011, 3175. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Li, J.; Entenberg, D.; Xue, A.; Wang, W.; Condeelis, J. Direct visualization of the phenotype of hypoxic tumor cells at single cell resolution in vivo using a new hypoxia probe. IntraVital 2016, 5, e1187803. [Google Scholar] [CrossRef]
- Fluegen, G.; Avivar-Valderas, A.; Wang, Y.; Padgen, M.R.; Williams, J.K.; Nobre, A.R.; Calvo, V.; Cheung, J.F.; Bravo-Cordero, J.J.; Entenberg, D.; et al. Phenotypic heterogeneity of disseminated tumour cells is preset by primary tumour hypoxic microenvironments. Nat. Cell Biol. 2017, 19, 120–132. [Google Scholar] [CrossRef]
- Burr, S.P.; Costa, A.S.H.; Grice, G.L.; Timms, R.T.; Lobb, I.T.; Freisinger, P.; Dodd, R.B.; Dougan, G.; Lehner, P.J.; Frezza, C.; et al. Mitochondrial Protein Lipoylation and the 2-Oxoglutarate Dehydrogenase Complex Controls HIF1α Stability in Aerobic Conditions. Cell Metab. 2016, 24, 740–752. [Google Scholar] [CrossRef]
- Miles, A.L.; Burr, S.P.; Grice, G.L.; Nathan, J.A. The vacuolar-ATPase complex and assembly factors, TMEM199 and CCDC115, control HIF1α prolyl hydroxylation by regulating cellular iron levels. eLife 2017, 6, e22693. [Google Scholar] [CrossRef] [PubMed]
- Danhier, P.; Krishnamachary, B.; Bharti, S.; Kakkad, S.; Mironchik, Y.; Bhujwalla, Z.M. Combining Optical Reporter Proteins with Different Half-lives to Detect Temporal Evolution of Hypoxia and Reoxygenation in Tumors. Neoplasia (N. Y. NY) 2015, 17, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Erapaneedi, R.; Belousov, V.V.; Schäfers, M.; Kiefer, F. A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO J. 2016, 35, 102–113. [Google Scholar] [CrossRef]
- Kumagai, A.; Ando, R.; Miyatake, H.; Greimel, P.; Kobayashi, T.; Hirabayashi, Y.; Shimogori, T.; Miyawaki, A. A Bilirubin-Inducible Fluorescent Protein from Eel Muscle. Cell 2013, 153, 1602–1611. [Google Scholar] [CrossRef] [PubMed]
- Keyaerts, M.; Remory, I.; Caveliers, V.; Breckpot, K.; Bos, T.J.; Poelaert, J.; Bossuyt, A.; Lahoutte, T. Inhibition of Firefly Luciferase by General Anesthetics: Effect on In Vitro and In Vivo Bioluminescence Imaging. PLoS ONE 2012, 7, e30061. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, E.H.; Niedre, M.J.; Jarvi, M.T.; Mocanu, J.D.; Moriyama, Y.; Subarsky, P.; Li, B.; Lilge, L.D.; Wilson, B.C. The influence of hypoxia on bioluminescence in luciferase-transfected gliosarcoma tumor cells in vitro. Photochem. Photobiol. Sci. 2008, 7, 675–680. [Google Scholar] [CrossRef]
- Hagen, T.; Taylor, C.T.; Lam, F.; Moncada, S. Redistribution of Intracellular Oxygen in Hypoxia by Nitric Oxide: Effect on HIF1α. Science 2003, 302, 1975. [Google Scholar] [CrossRef]
- Feeney, K.A.; Putker, M.; Brancaccio, M.; O’Neill, J.S. In-depth Characterization of Firefly Luciferase as a Reporter of Circadian Gene Expression in Mammalian Cells. J. Biol. Rhythm. 2016, 31, 540–550. [Google Scholar] [CrossRef]
- Krishnamachary, B.; Penet, M.-F.; Nimmagadda, S.; Mironchik, Y.; Raman, V.; Solaiyappan, M.; Semenza, G.L.; Pomper, M.G.; Bhujwalla, Z.M. Hypoxia Regulates CD44 and Its Variant Isoforms through HIF-1α in Triple Negative Breast Cancer. PLoS ONE 2012, 7, e44078. [Google Scholar] [CrossRef]
- Doherty, G.P.; Bailey, K.; Lewis, P.J. Stage-specific fluorescence intensity of GFP and mCherry during sporulation In Bacillus Subtilis. BMC Res. Notes 2010, 3, 303. [Google Scholar] [CrossRef] [PubMed]
- Hebisch, E.; Knebel, J.; Landsberg, J.; Frey, E.; Leisner, M. High Variation of Fluorescence Protein Maturation Times in Closely Related Escherichia coli Strains. PLoS ONE 2013, 8, e75991. [Google Scholar] [CrossRef]
- Youssef, S.; Ren, W.; Ai, H.-W. A Genetically Encoded FRET Sensor for Hypoxia and Prolyl Hydroxylases. ACS Chem. Biol. 2016, 11, 2492–2498. [Google Scholar] [CrossRef]
- Mizukami, Y.; Jo, W.-S.; Duerr, E.-M.; Gala, M.; Li, J.; Zhang, X.; Zimmer, M.A.; Iliopoulos, O.; Zukerberg, L.R.; Kohgo, Y.; et al. Induction of interleukin-8 preserves the angiogenic response in HIF-1α–deficient colon cancer cells. Nat. Med. 2005, 11, 992–997. [Google Scholar] [CrossRef]
- Treins, C.; Giorgetti-Peraldi, S.; Murdaca, J.; Semenza, G.L.; Van Obberghen, E. Insulin Stimulates Hypoxia-inducible Factor 1 through a Phosphatidylinositol 3-Kinase/Target of Rapamycin-dependent Signaling Pathway. J. Biol. Chem. 2002, 277, 27975–27981. [Google Scholar] [CrossRef]
- Treins, C.; Giorgetti-Peraldi, S.; Murdaca, J.; Monthouël-Kartmann, M.-N.; Van Obberghen, E. Regulation of Hypoxia-Inducible Factor (HIF)-1 Activity and Expression of HIF Hydroxylases in Response to Insulin-Like Growth Factor I. Mol. Endocrinol. 2005, 19, 1304–1317. [Google Scholar] [CrossRef]
- Zhu, K.; Chan, W.; Heymach, J.; Wilkinson, M.; McConkey, D.J. Control of HIF-1alpha expression by eIF2 alpha phosphorylation-mediated translational repression. Cancer Res. 2009, 69, 1836–1843. [Google Scholar] [CrossRef]
- Galbán, S.; Kuwano, Y.; Pullmann, R., Jr.; Martindale, J.L.; Kim, H.H.; Lal, A.; Abdelmohsen, K.; Yang, X.; Dang, Y.; Liu, J.O.; et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 2008, 28, 93–107. [Google Scholar] [CrossRef]
- Isaacs, J.S.; Jung, Y.-J.; Mimnaugh, E.G.; Martinez, A.; Cuttitta, F.; Neckers, L.M. Hsp90 Regulates a von Hippel Lindau-independent Hypoxia-inducible Factor-1α-degradative Pathway. J. Biol. Chem. 2002, 277, 29936–29944. [Google Scholar] [CrossRef]
- Stiehl, D.P.; Wirthner, R.; Köditz, J.; Spielmann, P.; Camenisch, G.; Wenger, R.H. Increased Prolyl 4-Hydroxylase Domain Proteins Compensate for Decreased Oxygen Levels: EVIDENCE FOR AN AUTOREGULATORY OXYGEN-SENSING SYSTEM. J. Biol. Chem. 2006, 281, 23482–23491. [Google Scholar] [CrossRef]
- Selak, M.A.; Armour, S.M.; MacKenzie, E.D.; Boulahbel, H.; Watson, D.G.; Mansfield, K.D.; Pan, Y.; Simon, M.C.; Thompson, C.B.; Gottlieb, E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 2005, 7, 77–85. [Google Scholar] [CrossRef]
- Fong, G.-H.; Takeda, K. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 2008, 15, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Galanis, A.; Pappa, A.; Giannakakis, A.; Lanitis, E.; Dangaj, D.; Sandaltzopoulos, R. Reactive oxygen species and HIF-1 signalling in cancer. Oxidative Stress Carcinog. Spec. Issue 2008, 266, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Tafani, M.; Sansone, L.; Limana, F.; Arcangeli, T.; De Santis, E.; Polese, M.; Fini, M.; Russo, M.A. The Interplay of Reactive Oxygen Species, Hypoxia, Inflammation, and Sirtuins in Cancer Initiation and Progression. Oxid. Med. Cell. Longev. 2015, 2016, 3907147. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Lai, U.H.; Zhu, L.; Singh, A.; Ahmed, M.; Forsyth, N.R. Reactive Oxygen Species Formation in the Brain at Different Oxygen Levels: The Role of Hypoxia Inducible Factors. Front. Cell Dev. Biol. 2018, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Adryan, B.; Decker, H.-J.H.; Papas, T.S.; Hsu, T. Tracheal development and the von Hippel–Lindau tumor suppressor homolog in Drosophila. Oncogene 2000, 19, 2803–2811. [Google Scholar] [CrossRef] [PubMed]
- Southall, T.D.; Gold, K.S.; Egger, B.; Davidson, C.M.; Caygill, E.E.; Marshall, O.J.; Brand, A.H. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: Assaying RNA Pol II occupancy in neural stem cells. Dev. Cell 2013, 26, 101–112. [Google Scholar] [CrossRef]
- O’Hagan, K.A.; Cocchiglia, S.; Zhdanov, A.V.; Tambuwala, M.M.; Cummins, E.P.; Monfared, M.; Agbor, T.A.; Garvey, J.F.; Papkovsky, D.B.; Taylor, C.T.; et al. PGC-1α is coupled to HIF-1α-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2188. [Google Scholar] [CrossRef]
- Takahashi, E.; Takano, T.; Numata, A.; Hayashi, N.; Okano, S.; Nakajima, O.; Nomura, Y.; Sato, M. Genetic Oxygen Sensor. In Oxygen Transport to Tissue XXVI; Okunieff, P., Williams, J., Chen, Y., Eds.; Springer: Boston, MA, USA, 2005; pp. 39–44. [Google Scholar]
- Takahashi, E.; Sato, M. Imaging of oxygen gradients in monolayer cultured cells using green fluorescent protein. Am. J. Physiol.-Cell Physiol. 2010, 299, C1318–C1323. [Google Scholar] [CrossRef]
- Takahashi, E.; Takano, T.; Nomura, Y.; Okano, S.; Nakajima, O.; Sato, M. In vivo oxygen imaging using green fluorescent protein. Am. J. Physiol.-Cell Physiol. 2006, 291, C781–C787. [Google Scholar] [CrossRef]
- Tuckerman, J.R.; Gonzalez, G.; Sousa, E.H.S.; Wan, X.; Saito, J.A.; Alam, M.; Gilles-Gonzalez, M.-A. An Oxygen-Sensing Diguanylate Cyclase and Phosphodiesterase Couple for c-di-GMP Control. Biochemistry 2009, 48, 9764–9774. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Suquet, C.; Satterlee, J.-D.; Kang, C. Insights into Signal Transduction Involving PAS Domain Oxygen-Sensing Heme Proteins from the X-ray Crystal Structure of Escherichia Coli Dos Heme Domain (Ec DosH). Biochemistry 2004, 43, 2738–2746. [Google Scholar] [CrossRef] [PubMed]
- Ishitsuka, Y.; Araki, Y.; Tanaka, A.; Igarashi, J.; Ito, O.; Shimizu, T. Arg97 at the Heme-Distal Side of the Isolated Heme-Bound PAS Domain of a Heme-Based Oxygen Sensor from Escherichia coli (Ec DOS) Plays Critical Roles in Autoxidation and Binding to Gases, Particularly O2. Biochemistry 2008, 47, 8874–8884. [Google Scholar] [CrossRef] [PubMed]
- Nomata, J.; Hisabori, T. Development of heme protein based oxygen sensing indicators. Sci. Rep. 2018, 8, 11849. [Google Scholar] [CrossRef] [PubMed]
- Penjweini, R.; Andreoni, A.; Rosales, T.; Kim, J.; Brenner, M.D.; Sackett, D.L.; Chung, J.H.; Knutson, J.R. Intracellular oxygen mapping using a myoglobin-mCherry probe with fluorescence lifetime imaging. J. Biomed. Opt. 2018, 23, 1–14. [Google Scholar] [CrossRef]
- Chandel, N.S.; McClintock, D.S.; Feliciano, C.E.; Wood, T.M.; Melendez, J.A.; Rodriguez, A.M.; Schumacker, P.T. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: A mechanism of O2 sensing. J. Biol. Chem. 2000, 275, 25130–25138. [Google Scholar] [CrossRef]
- Solaini, G.; Baracca, A.; Lenaz, G.; Sgarbi, G. Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta-Bioenerg. 2010, 1797, 1171–1177. [Google Scholar] [CrossRef]
- Smith, K.A.; Waypa, G.B.; Schumacker, P.T. Redox signaling during hypoxia in mammalian cells. Redox Biol. 2017, 13, 228–234. [Google Scholar] [CrossRef]
- Li, C.; Jackson, R.M. Reactive species mechanisms of cellular hypoxia-reoxygenation injury. Am. J. Physiol.-Cell Physiol. 2002, 282, C227–C241. [Google Scholar] [CrossRef]
- Hanson, G.T.; Aggeler, R.; Oglesbee, D.; Cannon, M.; Capaldi, R.A.; Tsien, R.Y.; Remington, S.J. Investigating Mitochondrial Redox Potential with Redox-sensitive Green Fluorescent Protein Indicators. J. Biol. Chem. 2004, 279, 13044–13053. [Google Scholar] [CrossRef]
- Dooley, C.T.; Dore, T.M.; Hanson, G.T.; Jackson, W.C.; Remington, S.J.; Tsien, R.Y. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 2004, 279, 22284–22293. [Google Scholar] [CrossRef]
- Cannon, M.B.; Remington, S.J. Re-engineering redox-sensitive green fluorescent protein for improved response rate. Protein Sci. Publ. Protein Soc. 2006, 15, 45–57. [Google Scholar] [CrossRef]
- Lohman, J.R.; Remington, S.J. Development of a family of redox-sensitive green fluorescent protein indicators for use in relatively oxidizing subcellular environments. Biochemistry 2008, 47, 8678–8688. [Google Scholar] [CrossRef]
- Gutscher, M.; Pauleau, A.-L.; Marty, L.; Brach, T.; Wabnitz, G.H.; Samstag, Y.; Meyer, A.J.; Dick, T.P. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 2008, 5, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.B.; Remington, S.J. Redox-sensitive green fluorescent protein: Probes for dynamic intracellular redox responses. A review. Methods Mol. Biol. (Clifton NJ) 2008, 476, 51–65. [Google Scholar] [CrossRef]
- Meyer, A.J.; Dick, T.P. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 2010, 13, 621–650. [Google Scholar] [CrossRef] [PubMed]
- Shokhina, A.G.; Kostyuk, A.I.; Ermakova, Y.G.; Panova, A.S.; Staroverov, D.B.; Egorov, E.S.; Baranov, M.S.; van Belle, G.J.; Katschinski, D.M.; Belousov, V.V.; et al. Red fluorescent redox-sensitive biosensor Grx1-roCherry. Redox Biol. 2019, 21, 101071. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Makar, M.; Wang, M.X.; Ai, H.-W. Monitoring thioredoxin redox with a genetically encoded red fluorescent biosensor. Nat. Chem. Biol. 2017, 13, 1045–1052. [Google Scholar] [CrossRef]
- Fan, Y.; Chen, Z.; Ai, H. Monitoring redox dynamics in living cells with a redox-sensitive red fluorescent protein. Anal. Chem. 2015, 87, 2802–2810. [Google Scholar] [CrossRef]
- Hu, H.; Wang, A.; Huang, L.; Zou, Y.; Gu, Y.; Chen, X.; Zhao, Y.; Yang, Y. Monitoring cellular redox state under hypoxia using a fluorescent sensor based on eel fluorescent protein. Free Radic. Biol. Med. 2018, 120, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, H.; Henriksen, A.; Hansen, F.G.; Winther, J.R. Shedding light on disulfide bond formation: Engineering a redox switch in green fluorescent protein. EMBO J. 2001, 20, 5853–5862. [Google Scholar] [CrossRef] [PubMed]
- Björnberg, O.; Østergaard, H.; Winther, J.R. Mechanistic insight provided by glutaredoxin within a fusion to redox-sensitive yellow fluorescent protein. Biochemistry 2006, 45, 2362–2371. [Google Scholar] [CrossRef] [PubMed]
- Piattoni, C.V.; Sardi, F.; Klein, F.; Pantano, S.; Bollati-Fogolin, M.; Comini, M. New red-shifted fluorescent biosensor for monitoring intracellular redox changes. Free Radic. Biol. Med. 2019, 134, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Bilan, D.S.; Belousov, V.V. HyPer Family Probes: State of the Art. Antioxid. Redox Signal. 2016, 24, 731–751. [Google Scholar] [CrossRef] [PubMed]
- Belousov, V.V.; Fradkov, A.F.; Lukyanov, K.A.; Staroverov, D.B.; Shakhbazov, K.S.; Terskikh, A.V.; Lukyanov, S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 2006, 3, 281–286. [Google Scholar] [CrossRef]
- Markvicheva, K.N.; Bilan, D.S.; Mishina, N.M.; Gorokhovatsky, A.Y.; Vinokurov, L.M.; Lukyanov, S.; Belousov, V.V. A genetically encoded sensor for H2O2 with expanded dynamic range. Bioorg. Med. Chem. 2011, 19, 1079–1084. [Google Scholar] [CrossRef]
- Bilan, D.S.; Pase, L.; Joosen, L.; Gorokhovatsky, A.Y.; Ermakova, Y.G.; Gadella, T.W.J.; Grabher, C.; Schultz, C.; Lukyanov, S.; Belousov, V.V. HyPer-3: A genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem. Biol. 2013, 8, 535–542. [Google Scholar] [CrossRef]
- Ermakova, Y.G.; Bilan, D.S.; Matlashov, M.E.; Mishina, N.M.; Markvicheva, K.N.; Subach, O.M.; Subach, F.V.; Bogeski, I.; Hoth, M.; Enikolopov, G.; et al. Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nat. Commun. 2014, 5, 5222. [Google Scholar] [CrossRef]
- Melo, E.P.; Lopes, C.; Gollwitzer, P.; Lortz, S.; Lenzen, S.; Mehmeti, I.; Kaminski, C.F.; Ron, D.; Avezov, E. TriPer, an optical probe tuned to the endoplasmic reticulum tracks changes in luminal H2O2. BMC Biol. 2017, 15, 24. [Google Scholar] [CrossRef]
- Poburko, D.; Santo-Domingo, J.; Demaurex, N. Dynamic Regulation of the Mitochondrial Proton Gradient during Cytosolic Calcium Elevations. J. Biol. Chem. 2011, 286, 11672–11684. [Google Scholar] [CrossRef]
- Ermakova, Y.G.; Pak, V.V.; Bogdanova, Y.A.; Kotlobay, A.A.; Yampolsky, I.V.; Shokhina, A.G.; Panova, A.S.; Marygin, R.A.; Staroverov, D.B.; Bilan, D.S.; et al. SypHer3s: A genetically encoded fluorescent ratiometric probe with enhanced brightness and an improved dynamic range. Chem. Commun. Camb. Engl. 2018, 54, 2898–2901. [Google Scholar] [CrossRef] [PubMed]
- Pak, V.V.; Ezeriņa, D.; Lyublinskaya, O.G.; Pedre, B.; Tyurin-Kuzmin, P.A.; Mishina, N.M.; Thauvin, M.; Young, D.; Wahni, K.; Gache, S.A.M.; et al. Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metab. 2020, 31, 642–653.e6. [Google Scholar] [CrossRef] [PubMed]
- Gutscher, M.; Sobotta, M.C.; Wabnitz, G.H.; Ballikaya, S.; Meyer, A.J.; Samstag, Y.; Dick, T.P. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009, 284, 31532–31540. [Google Scholar] [CrossRef]
- Morgan, B.; Van Laer, K.; Owusu, T.N.E.; Ezeriņa, D.; Pastor-Flores, D.; Amponsah, P.S.; Tursch, A.; Dick, T.P. Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat. Chem. Biol. 2016, 12, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.C.; Cooper, C.E. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Lett. 1994, 356, 295–298. [Google Scholar] [CrossRef]
- Jeffrey Man, H.S.; Tsui, A.K.Y.; Marsden, P.A. Nitric Oxide and Hypoxia Signaling, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2014; Volume 96, ISBN 978-0-12-800254-4. [Google Scholar]
- Eroglu, E.; Gottschalk, B.; Charoensin, S.; Blass, S.; Bischof, H.; Rost, R.; Madreiter-Sokolowski, C.T.; Pelzmann, B.; Bernhart, E.; Sattler, W.; et al. Development of novel FP-based probes for live-cell imaging of nitric oxide dynamics. Nat. Commun. 2016, 7, 10623. [Google Scholar] [CrossRef]
- Lin, S.J.; Guarente, L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr. Opin. Cell Biol. 2003, 15, 241–246. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, R.S.; Handy, D.E.; Loscalzo, J. NAD(H) and NADP(H) Redox Couples and Cellular Energy Metabolism. Antioxid. Redox Signal. 2018, 28, 251–272. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, Y. Real-time and high-throughput analysis of mitochondrial metabolic states in living cells using genetically encoded NAD+/NADH sensors. Free Radic. Biol. Med. 2016, 100, 43–52. [Google Scholar] [CrossRef]
- Bilan, D.S.; Belousov, V.V. Genetically encoded probes for NAD+/NADH monitoring. Free Radic. Biol. Med. 2016, 100, 32–42. [Google Scholar] [CrossRef]
- Hung, Y.P.; Albeck, J.G.; Tantama, M.; Yellen, G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011, 14, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Bilan, D.S.; Matlashov, M.E.; Gorokhovatsky, A.Y.; Schultz, C.; Enikolopov, G.; Belousov, V.V. Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments. Biochim. Biophys. Acta 2014, 1840, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, Q.; Cheng, F.; Su, N.; Wang, A.; Zou, Y.; Hu, H.; Chen, X.; Zhou, H.-M.; Huang, X.; et al. SoNar, a Highly Responsive NAD+/NADH Sensor, Allows High-Throughput Metabolic Screening of Anti-tumor Agents. Cell Metab. 2015, 21, 777–789. [Google Scholar] [CrossRef]
- Tao, R.; Zhao, Y.; Chu, H.; Wang, A.; Zhu, J.; Chen, X.; Zou, Y.; Shi, M.; Liu, R.; Su, N.; et al. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat. Methods 2017, 14, 720–728. [Google Scholar] [CrossRef]
- Zhao, Y.; Jin, J.; Hu, Q.; Zhou, H.-M.; Yi, J.; Yu, Z.; Xu, L.; Wang, X.; Yang, Y.; Loscalzo, J. Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab. 2011, 14, 555–566. [Google Scholar] [CrossRef]
- Cambronne, X.A.; Stewart, M.L.; Kim, D.; Jones-Brunette, A.M.; Morgan, R.K.; Farrens, D.L.; Cohen, M.S.; Goodman, R.H. Biosensor reveals multiple sources for mitochondrial NAD+. Science 2016, 352, 1474–1477. [Google Scholar] [CrossRef]
- Zou, Y.; Wang, A.; Huang, L.; Zhu, X.; Hu, Q.; Zhang, Y.; Chen, X.; Li, F.; Wang, Q.; Wang, H.; et al. Illuminating NAD+ Metabolism in Live Cells and In Vivo Using a Genetically Encoded Fluorescent Sensor. Dev. Cell 2020, 53, 240–252.e7. [Google Scholar] [CrossRef]
- Wheaton, W.W.; Chandel, N.S. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am. J. Physiol.-Cell Physiol. 2011, 300, C385–C393. [Google Scholar] [CrossRef]
- Yaginuma, H.; Kawai, S.; Tabata, K.V.; Tomiyama, K.; Kakizuka, A.; Komatsuzaki, T.; Noji, H.; Imamura, H. Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging. Sci. Rep. 2014, 4, 6522. [Google Scholar] [CrossRef]
- Lobas, M.A.; Tao, R.; Nagai, J.; Kronschläger, M.T.; Borden, P.M.; Marvin, J.S.; Looger, L.L.; Khakh, B.S. A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nat. Commun. 2019, 10, 711. [Google Scholar] [CrossRef]
- Arai, S.; Kriszt, R.; Harada, K.; Looi, L.-S.; Matsuda, S.; Wongso, D.; Suo, S.; Ishiura, S.; Tseng, Y.-H.; Raghunath, M.; et al. RGB-Color Intensiometric Indicators to Visualize Spatiotemporal Dynamics of ATP in Single Cells. Angew. Chem. Int. Ed. Engl. 2018, 57, 10873–10878. [Google Scholar] [CrossRef]
- Imamura, H.; Nhat, K.P.H.; Togawa, H.; Saito, K.; Iino, R.; Kato-Yamada, Y.; Nagai, T.; Noji, H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. USA 2009, 106, 15651–15656. [Google Scholar] [CrossRef]
- Nakano, M.; Imamura, H.; Nagai, T.; Noji, H. Ca2+ regulation of mitochondrial ATP synthesis visualized at the single cell level. ACS Chem. Biol. 2011, 6, 709–715. [Google Scholar] [CrossRef]
- Yoshida, T.; Kakizuka, A.; Imamura, H. BTeam, a Novel BRET-based Biosensor for the Accurate Quantification of ATP Concentration within Living Cells. Sci. Rep. 2016, 6, 39618. [Google Scholar] [CrossRef]
- Berg, J.; Hung, Y.P.; Yellen, G. A genetically encoded fluorescent reporter of ATP: ADP ratio. Nat. Methods 2009, 6, 161–166. [Google Scholar] [CrossRef]
- Tantama, M.; Martínez-François, J.R.; Mongeon, R.; Yellen, G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 2013, 4, 1–11. [Google Scholar] [CrossRef]
- Alberti, K.G.M.M. The biochemical consequences of hypoxia. J. Clin. Pathol. 1977, 3, 14–20. [Google Scholar] [CrossRef]
- San Martín, A.; Ceballo, S.; Ruminot, I.; Lerchundi, R.; Frommer, W.B.; Barros, L.F. A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PLoS ONE 2013, 8, e57712. [Google Scholar] [CrossRef]
- Mächler, P.; Wyss, M.T.; Elsayed, M.; Stobart, J.; Gutierrez, R.; Von Faber-Castell, A.; Kaelin, V.; Zuend, M.; San Martín, A.; Romero-Gómez, I.; et al. In Vivo Evidence for a Lactate Gradient from Astrocytes to Neurons. Cell Metab. 2016, 23, 94–102. [Google Scholar] [CrossRef]
- Martynov, V.I.; Pakhomov, A.A.; Deyev, I.E.; Petrenko, A.G. Genetically encoded fluorescent indicators for live cell pH imaging. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 2924–2939. [Google Scholar] [CrossRef]
- Shen, Y.; Rosendale, M.; Campbell, R.E.; Perrais, D. pHuji, a pH-sensitive red fluorescent protein for imaging of exo- and endocytosis. J. Cell Biol. 2014, 207, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tsien, R.W. pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat. Neurosci. 2012, 15, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Tantama, M.; Hung, Y.P.; Yellen, G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J. Am. Chem. Soc. 2011, 133, 10034–10037. [Google Scholar] [CrossRef]
- Johnson, D.E.; Ai, H.-W.; Wong, P.; Young, J.D.; Campbell, R.E.; Casey, J.R. Red fluorescent protein pH biosensor to detect concentrative nucleoside transport. J. Biol. Chem. 2009, 284, 20499–20511. [Google Scholar] [CrossRef] [PubMed]
- Kogure, T.; Karasawa, S.; Araki, T.; Saito, K.; Kinjo, M.; Miyawaki, A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat. Biotechnol. 2006, 24, 577–581. [Google Scholar] [CrossRef]
- Miesenböck, G.; De Angelis, D.A.; Rothman, J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998, 394, 192–195. [Google Scholar] [CrossRef]
- Mahon, M.J. pHluorin2: An enhanced, ratiometric, pH-sensitive green florescent protein. Adv. Biosci. Biotechnol. Print 2011, 2, 132–137. [Google Scholar] [CrossRef]
- Hanson, G.T.; McAnaney, T.B.; Park, E.S.; Rendell, M.E.P.; Yarbrough, D.K.; Chu, S.; Xi, L.; Boxer, S.G.; Montrose, M.H.; Remington, S.J. Green fluorescent protein variants as ratiometric dual emission pH sensors. 1. Structural characterization and preliminary application. Biochemistry 2002, 41, 15477–15488. [Google Scholar] [CrossRef]
- Llopis, J.; McCaffery, J.M.; Miyawaki, A.; Farquhar, M.G.; Tsien, R.Y. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 6803–6808. [Google Scholar] [CrossRef]
- Bizzarri, R.; Arcangeli, C.; Arosio, D.; Ricci, F.; Faraci, P.; Cardarelli, F.; Beltram, F. Development of a novel GFP-based ratiometric excitation and emission pH indicator for intracellular studies. Biophys. J. 2006, 90, 3300–3314. [Google Scholar] [CrossRef]
- Valkonen, M.; Mojzita, D.; Penttilä, M.; Benčina, M. Noninvasive High-Throughput Single-Cell Analysis of the Intracellular pH of Saccharomyces cerevisiae by Ratiometric Flow Cytometry. Appl. Environ. Microbiol. 2013, 79, 7179–7187. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Wood, A.; Bowlby, M.R. Brain slices as models for neurodegenerative disease and screening platforms to identify novel therapeutics. Curr. Neuropharmacol. 2007, 5, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Holopainen, I.E. Organotypic hippocampal slice cultures: A model system to study basic cellular and molecular mechanisms of neuronal cell death, neuroprotection, and synaptic plasticity. Neurochem. Res. 2005, 30, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Schnichels, S.; Blak, M.; Hurst, J.; Dorfi, T.; Bartz-Schmidt, K.U.; Ziemssen, F.; Spitzer, M.S.; Schultheiss, M. Establishment of a retinal hypoxia organ culture model. Biol. Open 2017, 6, 1056–1064. [Google Scholar] [CrossRef]
- Lee, H.K.; Keum, S.; Sheng, H.; Warner, D.S.; Lo, D.C.; Marchuk, D.A. Natural allelic variation of the IL-21 receptor modulates ischemic stroke infarct volume. J. Clin. Investig. 2016, 126, 2827–2838. [Google Scholar] [CrossRef][Green Version]
- Keum, S.; Lee, H.K.; Chu, P.-L.; Kan, M.J.; Huang, M.-N.; Gallione, C.J.; Gunn, M.D.; Lo, D.C.; Marchuk, D.A. Natural genetic variation of integrin alpha L (Itgal) modulates ischemic brain injury in stroke. PLoS Genet. 2013, 9, e1003807. [Google Scholar] [CrossRef]
- Mao, L. Genetic Background Specific Hypoxia Resistance in Rat is Correlated with Balanced Activation of a Cross-Chromosomal Genetic Network Centering on Physiological Homeostasis. Front. Genet. 2012, 3, 208. [Google Scholar] [CrossRef]
- Du, R.; Zhou, J.; Lorenzano, S.; Liu, W.; Charoenvimolphan, N.; Qian, B.; Xu, J.; Wang, J.; Zhang, X.; Wang, X.; et al. Integrative Mouse and Human Studies Implicate ANGPT1 and ZBTB7C as Susceptibility Genes to Ischemic Injury. Stroke 2015, 46, 3514–3522. [Google Scholar] [CrossRef]
- Byrne, M.B.; Leslie, M.T.; Patel, H.S.; Gaskins, H.R.; Kenis, P.J.A. Design considerations for open-well microfluidic platforms for hypoxic cell studies. Biomicrofluidics 2017, 11, 054116. [Google Scholar] [CrossRef]
- Funamoto, K.; Zervantonakis, I.K.; Liu, Y.; Ochs, C.J.; Kim, C.; Kamm, R.D. A novel microfluidic platform for high-resolution imaging of a three-dimensional cell culture under a controlled hypoxic environment. Lab. Chip 2012, 12, 4855–4863. [Google Scholar] [CrossRef]
- Martewicz, S.; Michielin, F.; Serena, E.; Zambon, A.; Mongillo, M.; Elvassore, N. Reversible alteration of calcium dynamics in cardiomyocytes during acute hypoxia transient in a microfluidic platform. Integr. Biol. Quant. Biosci. Nano Macro 2012, 4, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Hellström Erkenstam, N.; Smith, P.L.P.; Fleiss, B.; Nair, S.; Svedin, P.; Wang, W.; Boström, M.; Gressens, P.; Hagberg, H.; Brown, K.L.; et al. Temporal Characterization of Microglia/Macrophage Phenotypes in a Mouse Model of Neonatal Hypoxic-Ischemic Brain Injury. Front. Cell. Neurosci. 2016, 10, 286. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Wang, J.; Gao, W.; Mohammadreza, A.; Kelbauskas, L.; Zhang, W.; Johnson, R.H.; Meldrum, D.R. Quantitative single-cell gene expression measurements of multiple genes in response to hypoxia treatment. Anal. Bioanal. Chem. 2011, 401, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Gropper, Y.; Feferman, T.; Shalit, T.; Salame, T.-M.; Porat, Z.; Shakhar, G. Culturing CTLs under Hypoxic Conditions Enhances Their Cytolysis and Improves Their Anti-tumor Function. Cell Rep. 2017, 20, 2547–2555. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wan, W.-W.; Xiong, S.-L.; Feng, H.; Wu, N. Cancer stem-like cells can be induced through dedifferentiation under hypoxic conditions in glioma, hepatoma and lung cancer. Cell Death Discov. 2017, 3, 16105. [Google Scholar] [CrossRef] [PubMed]
- Fenno, L.; Yizhar, O.; Deisseroth, K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011, 34, 389–412. [Google Scholar] [CrossRef]
- Roth, B.L. DREADDs for Neuroscientists. Neuron 2016, 89, 683–694. [Google Scholar] [CrossRef]
- Steinhorn, B.; Sorrentino, A.; Badole, S.; Bogdanova, Y.; Belousov, V.; Michel, T. Chemogenetic generation of hydrogen peroxide in the heart induces severe cardiac dysfunction. Nat. Commun. 2018, 9, 4044. [Google Scholar] [CrossRef]
- Ermakova, Y.G.; Lanin, A.A.; Fedotov, I.V.; Roshchin, M.; Kelmanson, I.V.; Kulik, D.; Bogdanova, Y.A.; Shokhina, A.G.; Bilan, D.S.; Staroverov, D.B.; et al. Thermogenetic neurostimulation with single-cell resolution. Nat. Commun. 2017, 8, 15362. [Google Scholar] [CrossRef]
- Roshchin, M.; Ermakova, Y.G.; Lanin, A.A.; Chebotarev, A.S.; Kelmanson, I.V.; Balaban, P.M.; Zheltikov, A.M.; Belousov, V.V.; Nikitin, E.S. Thermogenetic stimulation of single neocortical pyramidal neurons transfected with TRPV1-L channels. Neurosci. Lett. 2018, 687, 153–157. [Google Scholar] [CrossRef]
- Imamura, K.; Sahara, N.; Kanaan, N.M.; Tsukita, K.; Kondo, T.; Kutoku, Y.; Ohsawa, Y.; Sunada, Y.; Kawakami, K.; Hotta, A.; et al. Calcium dysregulation contributes to neurodegeneration in FTLD patient iPSC-derived neurons. Sci. Rep. 2016, 6, 34904. [Google Scholar] [CrossRef]
- Heman-Ackah, S.M.; Bassett, A.R.; Wood, M.J.A. Precision Modulation of Neurodegenerative Disease-Related Gene Expression in Human iPSC-Derived Neurons. Sci. Rep. 2016, 6, 28420. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Virumbrales, M.; Moreno, C.L.; Kruglikov, I.; Marazuela, P.; Sproul, A.; Jacob, S.; Zimmer, M.; Paull, D.; Zhang, B.; Schadt, E.E.; et al. CRISPR/Cas9-Correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer’s PSEN2 N141I neurons. Acta Neuropathol. Commun. 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Paquet, D.; Kwart, D.; Chen, A.; Sproul, A.; Jacob, S.; Teo, S.; Olsen, K.M.; Gregg, A.; Noggle, S.; Tessier-Lavigne, M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 2016, 533, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Pașca, S.P. The rise of three-dimensional human brain cultures. Nature 2018, 553, 437–445. [Google Scholar] [CrossRef]
- Yoon, S.-J.; Elahi, L.S.; Pașca, A.M.; Marton, R.M.; Gordon, A.; Revah, O.; Miura, Y.; Walczak, E.M.; Holdgate, G.M.; Fan, H.C.; et al. Reliability of human cortical organoid generation. Nat. Methods 2019, 16, 75–78. [Google Scholar] [CrossRef]
- Pașca, A.M.; Park, J.-Y.; Shin, H.-W.; Qi, Q.; Revah, O.; Krasnoff, R.; O’Hara, R.; Willsey, A.J.; Palmer, T.D.; Pașca, S.P. Human 3D cellular model of hypoxic brain injury of prematurity. Nat. Med. 2019, 25, 784–791. [Google Scholar] [CrossRef]
- Daviaud, N.; Chevalier, C.; Friedel, R.H.; Zou, H. Distinct Vulnerability and Resilience of Human Neuroprogenitor Subtypes in Cerebral Organoid Model of Prenatal Hypoxic Injury. Front. Cell. Neurosci. 2019, 13, 336. [Google Scholar] [CrossRef]
- Cakir, B.; Xiang, Y.; Tanaka, Y.; Kural, M.H.; Parent, M.; Kang, Y.-J.; Chapeton, K.; Patterson, B.; Yuan, Y.; He, C.-S.; et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 2019, 16, 1169–1175. [Google Scholar] [CrossRef]
- Rybachuk, O.; Kopach, O.; Krotov, V.; Voitenko, N.; Pivneva, T. Optimized Model of Cerebral Ischemia In situ for the Long-Lasting Assessment of Hippocampal Cell Death. Front. Neurosci. 2017, 11, 388. [Google Scholar] [CrossRef]
- Randić, M.; Jiang, M.C.; Cerne, R. Long-term potentiation and long-term depression of primary afferent neurotransmission in the rat spinal cord. J. Neurosci. Off. J. Soc. Neurosci. 1993, 13, 5228–5241. [Google Scholar] [CrossRef]
- Hutter-Schmid, B.; Kniewallner, K.M.; Humpel, C. Organotypic brain slice cultures as a model to study angiogenesis of brain vessels. Front. Cell Dev. Biol. 2015, 3, 52. [Google Scholar] [CrossRef] [PubMed]
- Raineteau, O.; Rietschin, L.; Gradwohl, G.; Guillemot, F.; Gähwiler, B.H. Neurogenesis in hippocampal slice cultures. Mol. Cell. Neurosci. 2004, 26, 241–250. [Google Scholar] [CrossRef]
- Haydar, T.F.; Bambrick, L.L.; Krueger, B.K.; Rakic, P. Organotypic slice cultures for analysis of proliferation, cell death, and migration in the embryonic neocortex. Brain Res. Brain Res. Protoc. 1999, 4, 425–437. [Google Scholar] [CrossRef]
- Kovács, R.; Papageorgiou, I.; Heinemann, U. Slice cultures as a model to study neurovascular coupling and blood brain barrier in vitro. Cardiovasc. Psychiatry Neurol. 2011, 2011, 646958. [Google Scholar] [CrossRef]
- Karra, D.; Dahm, R. Transfection techniques for neuronal cells. J. Neurosci. Off. J. Soc. Neurosci. 2010, 30, 6171–6177. [Google Scholar] [CrossRef]
- Murphy, R.C.; Messer, A. Gene transfer methods for CNS organotypic cultures: A comparison of three nonviral methods. Mol. Ther. J. Am. Soc. Gene Ther. 2001, 3, 113–121. [Google Scholar] [CrossRef]
- Sandbichler, A.M.; Jansen, B.; Peer, B.A.; Paulitsch, M.; Pelster, B.; Egg, M. Metabolic Plasticity Enables Circadian Adaptation to Acute Hypoxia in Zebrafish Cells. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 46, 1159–1174. [Google Scholar] [CrossRef]
- Simon, M.C.; Keith, B. The role of oxygen availability in embryonic development and stem cell function. Nat. Rev. Mol. Cell Biol. 2008, 9, 285–296. [Google Scholar] [CrossRef]
- Zheng, L.; Kelly, C.J.; Colgan, S.P. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am. J. Physiol. Cell Physiol. 2015, 309, C350–C360. [Google Scholar] [CrossRef]
- Rankin, E.B.; Giaccia, A.J. Hypoxic control of metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Rankin, E.B.; Nam, J.-M.; Giaccia, A.J. Hypoxia: Signaling the Metastatic Cascade. Trends Cancer 2016, 2, 295–304. [Google Scholar] [CrossRef]
- Lewis, D.M.; Park, K.M.; Tang, V.; Xu, Y.; Pak, K.; Eisinger-Mathason, T.S.K.; Simon, M.C.; Gerecht, S. Intratumoral oxygen gradients mediate sarcoma cell invasion. Proc. Natl. Acad. Sci. USA 2016, 113, 9292–9297. [Google Scholar] [CrossRef]
- Deygas, M.; Gadet, R.; Gillet, G.; Rimokh, R.; Gonzalo, P.; Mikaelian, I. Redox regulation of EGFR steers migration of hypoxic mammary cells towards oxygen. Nat. Commun. 2018, 9, 4545. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Satomi, T.; Abiru, A.; Miyamoto, K.; Nagasawa, K.; Maruyama, M.; Yamamoto, S.; Kikuchi, K.; Fuse, H.; Noda, M.; et al. Antihypertrophic Effects of Small Molecules that Maintain Mitochondrial ATP Levels Under Hypoxia. EBioMedicine 2017, 24, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Steinbeck, J.; Fuchs, P.; Lichtenauer, S.; Elsässer, M.; Schippers, J.H.M.; Nietzel, T.; Ruberti, C.; Van Aken, O.; Meyer, A.J.; et al. Multiparametric real-time sensing of cytosolic physiology links hypoxia responses to mitochondrial electron transport. New Phytol. 2019, 224, 1668–1684. [Google Scholar] [CrossRef] [PubMed]
- De Col, V.; Fuchs, P.; Nietzel, T.; Elsässer, M.; Voon, C.P.; Candeo, A.; Seeliger, I.; Fricker, M.D.; Grefen, C.; Møller, I.M.; et al. ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. eLife 2017, 6. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, C.; Lischinsky, J.E.; Jing, M.; Zhou, J.; Wang, H.; Zhang, Y.; Dong, A.; Wu, Z.; Wu, H.; et al. A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 2019, 102, 745–761.e8. [Google Scholar] [CrossRef]
- Marvin, J.S.; Shimoda, Y.; Magloire, V.; Leite, M.; Kawashima, T.; Jensen, T.P.; Kolb, I.; Knott, E.L.; Novak, O.; Podgorski, K.; et al. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods 2019, 16, 763–770. [Google Scholar] [CrossRef]
- Marvin, J.S.; Scholl, B.; Wilson, D.E.; Podgorski, K.; Kazemipour, A.; Müller, J.A.; Schoch, S.; Quiroz, F.J.U.; Rebola, N.; Bao, H.; et al. Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat. Methods 2018, 15, 936–939. [Google Scholar] [CrossRef]
- Dunn, T.A.; Wang, C.-T.; Colicos, M.A.; Zaccolo, M.; DiPilato, L.M.; Zhang, J.; Tsien, R.Y.; Feller, M.B. Imaging of cAMP levels and protein kinase A activity reveals that retinal waves drive oscillations in second-messenger cascades. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 12807–12815. [Google Scholar] [CrossRef] [PubMed]
- Weitz, A.C.; Behrend, M.R.; Lee, N.S.; Klein, R.L.; Chiodo, V.A.; Hauswirth, W.W.; Humayun, M.S.; Weiland, J.D.; Chow, R.H. Imaging the response of the retina to electrical stimulation with genetically encoded calcium indicators. J. Neurophysiol. 2013, 109, 1979–1988. [Google Scholar] [CrossRef] [PubMed]
- Dana, H.; Mohar, B.; Sun, Y.; Narayan, S.; Gordus, A.; Hasseman, J.P.; Tsegaye, G.; Holt, G.T.; Hu, A.; Walpita, D.; et al. Sensitive red protein calcium indicators for imaging neural activity. eLife 2016, 5. [Google Scholar] [CrossRef]
- Fernández-Moncada, I.; Ruminot, I.; Robles-Maldonado, D.; Alegría, K.; Deitmer, J.W.; Barros, L.F. Neuronal control of astrocytic respiration through a variant of the Crabtree effect. Proc. Natl. Acad. Sci. USA 2018, 115, 1623–1628. [Google Scholar] [CrossRef] [PubMed]
- Guzman, J.N.; Sanchez-Padilla, J.; Wokosin, D.; Kondapalli, J.; Ilijic, E.; Schumacker, P.T.; Surmeier, D.J. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature 2010, 468, 696–700. [Google Scholar] [CrossRef]
- Graves, S.M.; Xie, Z.; Stout, K.A.; Zampese, E.; Burbulla, L.F.; Shih, J.C.; Kondapalli, J.; Patriarchi, T.; Tian, L.; Brichta, L.; et al. Dopamine metabolism by a monoamine oxidase mitochondrial shuttle activates the electron transport chain. Nat. Neurosci. 2020, 23, 15–20. [Google Scholar] [CrossRef]
- Miller, N.; Shi, H.; Zelikovich, A.S.; Ma, Y.-C. Motor neuron mitochondrial dysfunction in spinal muscular atrophy. Hum. Mol. Genet. 2016, 25, 3395–3406. [Google Scholar] [CrossRef]
- Bebensee, D.F.; Can, K.; Müller, M. Increased Mitochondrial Mass and Cytosolic Redox Imbalance in Hippocampal Astrocytes of a Mouse Model of Rett Syndrome: Subcellular Changes Revealed by Ratiometric Imaging of JC-1 and roGFP1 Fluorescence. Oxid. Med. Cell. Longev. 2017, 2017, 3064016. [Google Scholar] [CrossRef]
- Díaz-García, C.M.; Mongeon, R.; Lahmann, C.; Koveal, D.; Zucker, H.; Yellen, G. Neuronal Stimulation Triggers Neuronal Glycolysis and Not Lactate Uptake. Cell Metab. 2017, 26, 361–374.e4. [Google Scholar] [CrossRef]
- Mongeon, R.; Venkatachalam, V.; Yellen, G. Cytosolic NADH-NAD(+) Redox Visualized in Brain Slices by Two-Photon Fluorescence Lifetime Biosensor Imaging. Antioxid. Redox Signal. 2016, 25, 553–563. [Google Scholar] [CrossRef]
- Hasel, P.; Mckay, S.; Qiu, J.; Hardingham, G.E. Selective dendritic susceptibility to bioenergetic, excitotoxic and redox perturbations in cortical neurons. Biochim. Biophys. Acta 2015, 1853, 2066–2076. [Google Scholar] [CrossRef]
- Can, K.; Menzfeld, C.; Rinne, L.; Rehling, P.; Kügler, S.; Golubiani, G.; Dudek, J.; Müller, M. Neuronal Redox-Imbalance in Rett Syndrome Affects Mitochondria as Well as Cytosol, and Is Accompanied by Intensified Mitochondrial O 2 Consumption and ROS Release. Front. Physiol. 2019, 10, 479. [Google Scholar] [CrossRef]
- Can, K.; Kügler, S.; Müller, M. Live Imaging of Mitochondrial ROS Production and Dynamic Redox Balance in Neurons. In Techniques to Investigate Mitochondrial Function in Neurons; Strack, S., Usachev, Y.M., Eds.; Neuromethods; Springer: New York, NY, USA, 2017; pp. 179–197. ISBN 978-1-4939-6890-9. [Google Scholar]
- Yin, B.; Barrionuevo, G.; Weber, S.G. Optimized real-time monitoring of glutathione redox status in single pyramidal neurons in organotypic hippocampal slices during oxygen-glucose deprivation and reperfusion. ACS Chem. Neurosci. 2015, 6, 1838–1848. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.; Barrionuevo, G.; Batinic-Haberle, I.; Sandberg, M.; Weber, S.G. Differences in Reperfusion-Induced Mitochondrial Oxidative Stress and Cell Death Between Hippocampal CA1 and CA3 Subfields Are Due to the Mitochondrial Thioredoxin System. Antioxid. Redox Signal. 2017, 27, 534–549. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.; Barrionuevo, G.; Weber, S.G. Mitochondrial GSH Systems in CA1 Pyramidal Cells and Astrocytes React Differently during Oxygen-Glucose Deprivation and Reperfusion. ACS Chem. Neurosci. 2018, 9, 738–748. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.C.; Jackson, J.G.; Robinson, M.B. Transient Oxygen/Glucose Deprivation Causes a Delayed Loss of Mitochondria and Increases Spontaneous Calcium Signaling in Astrocytic Processes. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 7109–7127. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef]
- Palmer, A.E.; Giacomello, M.; Kortemme, T.; Hires, S.A.; Lev-Ram, V.; Baker, D.; Tsien, R.Y. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem. Biol. 2006, 13, 521–530. [Google Scholar] [CrossRef]
- Gouriou, Y.; Bijlenga, P.; Demaurex, N. Mitochondrial Ca2+ uptake from plasma membrane Cav3.2 protein channels contributes to ischemic toxicity in PC12 cells. J. Biol. Chem. 2013, 288, 12459–12468. [Google Scholar] [CrossRef]
- Marina, N.; Ang, R.; Machhada, A.; Kasymov, V.; Karagiannis, A.; Hosford, P.S.; Mosienko, V.; Teschemacher, A.G.; Vihko, P.; Paton, J.F.R.; et al. Brainstem hypoxia contributes to the development of hypertension in the spontaneously hypertensive rat. Hypertension 2015, 65, 775–783. [Google Scholar] [CrossRef]
- Mank, M.; Santos, A.F.; Direnberger, S.; Mrsic-Flogel, T.D.; Hofer, S.B.; Stein, V.; Hendel, T.; Reiff, D.F.; Levelt, C.; Borst, A.; et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat. Methods 2008, 5, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.C.V.; De Silva, D.A.; Macleod, M.R.; Coutts, S.B.; Schwamm, L.H.; Davis, S.M.; Donnan, G.A. Ischaemic stroke. Nat. Rev. Dis. Primer 2019, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Millar, L.J.; Shi, L.; Hoerder-Suabedissen, A.; Molnár, Z. Neonatal Hypoxia Ischaemia: Mechanisms, Models, and Therapeutic Challenges. Front. Cell. Neurosci. 2017, 11, 78. [Google Scholar] [CrossRef]
- Gunn, A.J.; Thoresen, M. Neonatal encephalopathy and hypoxic–ischemic encephalopathy. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 162, pp. 217–237. ISBN 978-0-444-64029-1. [Google Scholar]
- Ferini-Strambi, L.; Lombardi, G.E.; Marelli, S.; Galbiati, A. Neurological Deficits in Obstructive Sleep Apnea. Curr. Treat. Options Neurol. 2017, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Gerez, E.; Fleming, I.N.; Parson, S.H. A role for spinal cord hypoxia in neurodegeneration. Cell Death Dis. 2019, 10, 861. [Google Scholar] [CrossRef]
- Lall, R.; Mohammed, R.; Ojha, U. What are the links between hypoxia and Alzheimer’s disease? Neuropsychiatr. Dis. Treat. 2019, 15, 1343–1354. [Google Scholar] [CrossRef]
- Antinucci, P.; Hindges, R. A crystal-clear zebrafish for in vivo imaging. Sci. Rep. 2016, 6, 29490. [Google Scholar] [CrossRef]
- Rafferty, S.A.; Quinn, T.A. A beginner’s guide to understanding and implementing the genetic modification of zebrafish. Prog. Biophys. Mol. Biol. 2018, 138, 3–19. [Google Scholar] [CrossRef]
- Chen, Z.; Truong, T.M.; Ai, H.-W. Illuminating Brain Activities with Fluorescent Protein-Based Biosensors. Chemosensors 2017, 5, 32. [Google Scholar] [CrossRef]
- Turrini, L.; Fornetto, C.; Marchetto, G.; Müllenbroich, M.C.; Tiso, N.; Vettori, A.; Resta, F.; Masi, A.; Mannaioni, G.; Pavone, F.S.; et al. Optical mapping of neuronal activity during seizures in zebrafish. Sci. Rep. 2017, 7, 3025. [Google Scholar] [CrossRef]
- Miyazawa, H.; Okumura, K.; Hiyoshi, K.; Maruyama, K.; Kakinuma, H.; Amo, R.; Okamoto, H.; Yamasu, K.; Tsuda, S. Optical interrogation of neuronal circuitry in zebrafish using genetically encoded voltage indicators. Sci. Rep. 2018, 8, 6048. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zeng, J.; Jing, M.; Zhou, J.; Feng, J.; Owen, S.F.; Luo, Y.; Li, F.; Wang, H.; Yamaguchi, T.; et al. A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 2018, 174, 481–496.e19. [Google Scholar] [CrossRef] [PubMed]
- Beckwith-Cohen, B.; Holzhausen, L.C.; Wang, T.-M.; Rajappa, R.; Kramer, R.H. Localizing Proton-Mediated Inhibitory Feedback at the Retinal Horizontal Cell–Cone Synapse with Genetically-Encoded pH Probes. J. Neurosci. 2019, 39, 651–662. [Google Scholar] [CrossRef]
- Kardash, E. Current Methods in Zebrafish Research. Mater. Methods 2012, 2, 109. [Google Scholar] [CrossRef]
- Horb, M.; Wlizla, M.; Abu-Daya, A.; McNamara, S.; Gajdasik, D.; Igawa, T.; Suzuki, A.; Ogino, H.; Noble, A. Centre de Ressource Biologique Xenope team in France Xenopus Resources: Transgenic, Inbred and Mutant Animals, Training Opportunities, and Web-Based Support. Front. Physiol. 2019, 10, 387. [Google Scholar] [CrossRef] [PubMed]
- Sagasti, A.; O’Brien, G.S.; Rieger, S.; Martin, S.M.; Cavanaugh, A.M.; Portera-Cailliau, C. Two-photon axotomy and time-lapse confocal imaging in live zebrafish embryos. J. Vis. Exp. 2009, 24, 1129. [Google Scholar] [CrossRef]
- Kioka, H.; Kato, H.; Fujita, T.; Asano, Y.; Shintani, Y.; Yamazaki, S.; Tsukamoto, O.; Imamura, H.; Kogo, M.; Kitakaze, M.; et al. In vivo real-time ATP imaging in zebrafish hearts reveals G0s2 induces ischemic tolerance. FASEB J. 2020, 34, 2041–2054. [Google Scholar] [CrossRef] [PubMed]
- Erickstad, M.; Hale, L.A.; Chalasani, S.H.; Groisman, A. A microfluidic system for studying the behavior of zebrafish larvae under acute hypoxia. Lab Chip 2015, 15, 857–866. [Google Scholar] [CrossRef]
- Niethammer, P.; Grabher, C.; Look, A.T.; Mitchison, T.J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 2009, 459, 996–999. [Google Scholar] [CrossRef]
- Yoo, S.K.; Starnes, T.W.; Deng, Q.; Huttenlocher, A. Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature 2011, 480, 109–112. [Google Scholar] [CrossRef]
- Pase, L.; Layton, J.E.; Wittmann, C.; Ellett, F.; Nowell, C.J.; Reyes-Aldasoro, C.C.; Varma, S.; Rogers, K.L.; Hall, C.J.; Keightley, M.C.; et al. Neutrophil-Delivered Myeloperoxidase Dampens the Hydrogen Peroxide Burst after Tissue Wounding in Zebrafish. Curr. Biol. 2012, 22, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Rieger, S.; Sagasti, A. Hydrogen Peroxide Promotes Injury-Induced Peripheral Sensory Axon Regeneration in the Zebrafish Skin. PLoS Biol. 2011, 9, e1000621. [Google Scholar] [CrossRef] [PubMed]
- Meda, F.; Gauron, C.; Rampon, C.; Teillon, J.; Volovitch, M.; Vriz, S. Nerves Control Redox Levels in Mature Tissues Through Schwann Cells and Hedgehog Signaling. Antioxid. Redox Signal. 2016, 24, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Love, N.R.; Chen, Y.; Ishibashi, S.; Kritsiligkou, P.; Lea, R.; Koh, Y.; Gallop, J.L.; Dorey, K.; Amaya, E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat. Cell Biol. 2013, 15, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Zhou, X.-H.; Chang, N.; Xiao, C.-L.; Yan, S.; Ren, H.; Yang, X.-Z.; Zhang, M.-L.; Wu, Q.; Tang, B.; et al. Hydrogen peroxide primes heart regeneration with a derepression mechanism. Cell Res. 2014, 24, 1091–1107. [Google Scholar] [CrossRef]
- Gauron, C.; Meda, F.; Dupont, E.; Albadri, S.; Quenech’Du, N.; Ipendey, E.; Volovitch, M.; Del Bene, F.; Joliot, A.; Rampon, C.; et al. Hydrogen peroxide (H2O2) controls axon pathfinding during zebrafish development. Dev. Biol. 2016, 414, 133–141. [Google Scholar] [CrossRef]
- Han, Y.; Ishibashi, S.; Iglesias-Gonzalez, J.; Chen, Y.; Love, N.R.; Amaya, E. Ca2+ -Induced Mitochondrial ROS Regulate the Early Embryonic Cell Cycle. Cell Rep. 2018, 22, 218–231. [Google Scholar] [CrossRef]
- Rampon, C.; Volovitch, M.; Joliot, A.; Vriz, S. Hydrogen Peroxide and Redox Regulation of Developments. Antioxidants 2018, 7, 159. [Google Scholar] [CrossRef]
- Siemkowicz, E.; Hansen, A.J. Brain extracellular ion composition and EEG activity following 10 min ischemia in normo- and hyperglycemic rats. Stroke 1981, 12, 236–240. [Google Scholar] [CrossRef]
- Meyer, F.B.; Anderson, R.E.; Sundt, T.M.; Yaksh, T.L. Intracellular Brain pH, Indicator Tissue Perfusion, Electroencephalography, and Histology in Severe and Moderate Focal Cortical Ischemia in the Rabbit. J. Cereb. Blood Flow Metab. 1986, 6, 71–78. [Google Scholar] [CrossRef]
- Khacho, M.; Tarabay, M.; Patten, D.; Khacho, P.; MacLaurin, J.G.; Guadagno, J.; Bergeron, R.; Cregan, S.P.; Harper, M.-E.; Park, D.S.; et al. Acidosis overrides oxygen deprivation to maintain mitochondrial function and cell survival. Nat. Commun. 2014, 5, 3550. [Google Scholar] [CrossRef] [PubMed]
- Nakai, J.; Ohkura, M.; Imoto, K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001, 19, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, M.B.; Orger, M.B.; Robson, D.N.; Li, J.M.; Keller, P.J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 2013, 10, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Vladimirov, N.; Mu, Y.; Kawashima, T.; Bennett, D.V.; Yang, C.-T.; Looger, L.L.; Keller, P.J.; Freeman, J.; Ahrens, M.B. Light-sheet functional imaging in fictively behaving zebrafish. Nat. Methods 2014, 11, 883–884. [Google Scholar] [CrossRef]
- Wolf, S.; Supatto, W.; Debrégeas, G.; Mahou, P.; Kruglik, S.G.; Sintes, J.-M.; Beaurepaire, E.; Candelier, R. Whole-brain functional imaging with two-photon light-sheet microscopy. Nat. Methods 2015, 12, 379–380. [Google Scholar] [CrossRef]
- Akerboom, J.; Chen, T.-W.; Wardill, T.J.; Tian, L.; Marvin, J.S.; Mutlu, S.; Calderon, N.C.; Esposti, F.; Borghuis, B.G.; Sun, X.R.; et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci. 2012, 32, 13819–13840. [Google Scholar] [CrossRef] [PubMed]
- Muto, A.; Ohkura, M.; Abe, G.; Nakai, J.; Kawakami, K. Real-Time Visualization of Neuronal Activity during Perception. Curr. Biol. 2013, 23, 307–311. [Google Scholar] [CrossRef]
- Muto, A.; Kawakami, K. Calcium Imaging of Neuronal Activity in Free-Swimming Larval Zebrafish. In Zebrafish; Kawakami, K., Patton, E.E., Orger, M., Eds.; Springer: New York, NY, USA, 2016; Volume 1451, pp. 333–341. ISBN 978-1-4939-3769-1. [Google Scholar]
- Cong, L.; Wang, Z.; Chai, Y.; Hang, W.; Shang, C.; Yang, W.; Bai, L.; Du, J.; Wang, K.; Wen, Q. Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio). eLife 2017, 6, e28158. [Google Scholar] [CrossRef]
- Thouvenin, O.; Wyart, C. Tracking microscopy enables whole-brain imaging in freely moving zebrafish. Nat. Methods 2017, 14, 1041–1042. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, J.; Marques, J.C.; Grama, A.; Hildebrand, D.G.C.; Gu, W.; Li, J.M.; Robson, D.N. Pan-neuronal calcium imaging with cellular resolution in freely swimming zebrafish. Nat. Methods 2017, 14, 1107–1114. [Google Scholar] [CrossRef]
- Förster, D.; Dal Maschio, M.; Laurell, E.; Baier, H. An optogenetic toolbox for unbiased discovery of functionally connected cells in neural circuits. Nat. Commun. 2017, 8, 116. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, M.B.; Li, J.M.; Orger, M.B.; Robson, D.N.; Schier, A.F.; Engert, F.; Portugues, R. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 2012, 485, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, M.B.; Huang, K.H.; Narayan, S.; Mensh, B.D.; Engert, F. Two-photon calcium imaging during fictive navigation in virtual environments. Front. Neural Circuits 2013, 7. [Google Scholar] [CrossRef]
- Semmelhack, J.L.; Donovan, J.C.; Thiele, T.R.; Kuehn, E.; Laurell, E.; Baier, H. A dedicated visual pathway for prey detection in larval zebrafish. eLife 2014, 3, e04878. [Google Scholar] [CrossRef] [PubMed]
- Dunn, T.W.; Mu, Y.; Narayan, S.; Randlett, O.; Naumann, E.A.; Yang, C.-T.; Schier, A.F.; Freeman, J.; Engert, F.; Ahrens, M.B. Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion. eLife 2016, 5, e12741. [Google Scholar] [CrossRef] [PubMed]
- Migault, G.; van der Plas, T.L.; Trentesaux, H.; Panier, T.; Candelier, R.; Proville, R.; Englitz, B.; Debrégeas, G.; Bormuth, V. Whole-Brain Calcium Imaging during Physiological Vestibular Stimulation in Larval Zebrafish. Curr. Biol. 2018, 28, 3723–3735.e6. [Google Scholar] [CrossRef]
- Andalman, A.S.; Burns, V.M.; Lovett-Barron, M.; Broxton, M.; Poole, B.; Yang, S.J.; Grosenick, L.; Lerner, T.N.; Chen, R.; Benster, T.; et al. Neuronal Dynamics Regulating Brain and Behavioral State Transitions. Cell 2019, 177, 970–985.e20. [Google Scholar] [CrossRef]
- Rosch, R.; Hunter, P.; Baldeweg, T.; Friston, K.; Meyer, M. Calcium-imaging and dynamic causal modelling reveal brain-wide changes in effective connectivity and synaptic dynamics during epileptic seizures. PLoS Comput. Biol. 2018, 14, e1006375. [Google Scholar] [CrossRef]
- Liu, J.; Baraban, S.C. Network Properties Revealed during Multi-Scale Calcium Imaging of Seizure Activity in Zebrafish. Eneuro 2019, 6. [Google Scholar] [CrossRef]
- Burrows, D.R.W.; Samarut, É.; Liu, J.; Baraban, S.C.; Richardson, M.P.; Meyer, M.P.; Rosch, R.E. Imaging epilepsy in larval zebrafish. Eur. J. Paediatr. Neurol. 2020, 24, 70–80. [Google Scholar] [CrossRef]
- Ghannad-Rezaie, M.; Eimon, P.M.; Wu, Y.; Yanik, M.F. Engineering brain activity patterns by neuromodulator polytherapy for treatment of disorders. Nat. Commun. 2019, 10, 2620. [Google Scholar] [CrossRef]
- Durukan, A.; Tatlisumak, T. Preconditioning-induced ischemic tolerance: A window into endogenous gearing for cerebroprotection. Exp. Transl. Stroke Med. 2010, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Armstrong, E.A.; Yager, J.Y. Unilateral Common Carotid Artery Ligation as a Model of Perinatal Asphyxia: The Original Rice–Vannucci Model. In Animal Models of Neurodevelopmental Disorders; Yager, J.Y., Ed.; Neuromethods; Springer: New York, NY, USA, 2015; pp. 1–13. ISBN 978-1-4939-2709-8. [Google Scholar]
- Chiang, T.; Messing, R.O.; Chou, W.-H. Mouse Model of Middle Cerebral Artery Occlusion. J. Vis. Exp. JoVE 2011, 48, e2761. [Google Scholar] [CrossRef] [PubMed]
- Rumajogee, P.; Bregman, T.; Miller, S.P.; Yager, J.Y.; Fehlings, M.G. Rodent Hypoxia–Ischemia Models for Cerebral Palsy Research: A Systematic Review. Front. Neurol. 2016, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; McCullough, L.D. Middle Cerebral Artery Occlusion Model in Rodents: Methods and Potential Pitfalls. Available online: https://www.hindawi.com/journals/bmri/2011/464701/ (accessed on 22 February 2020).
- Durukan, A.; Tatlisumak, T. Acute ischemic stroke: Overview of major experimental rodent models, pathophysiology, and therapy of focal cerebral ischemia. Pharmacol. Biochem. Behav. 2007, 87, 179–197. [Google Scholar] [CrossRef]
- Silasi, G.; She, J.; Boyd, J.D.; Xue, S.; Murphy, T.H. A Mouse Model of Small-Vessel Disease that Produces Brain-Wide-Identified Microocclusions and Regionally Selective Neuronal Injury. J. Cereb. Blood Flow Metab. 2015, 35, 734–738. [Google Scholar] [CrossRef]
- Shih, A.Y.; Blinder, P.; Tsai, P.S.; Friedman, B.; Stanley, G.; Lyden, P.D.; Kleinfeld, D. The smallest stroke: Occlusion of one penetrating vessel leads to infarction and a cognitive deficit. Nat. Neurosci. 2013, 16, 55–63. [Google Scholar] [CrossRef]
- Uzdensky, A.B. Photothrombotic Stroke as a Model of Ischemic Stroke. Transl. Stroke Res. 2018, 9, 437–451. [Google Scholar] [CrossRef]
- Wright, J.L.; Chu, H.X.; Kagan, B.J.; Ermine, C.M.; Kauhausen, J.A.; Parish, C.L.; Sobey, C.G.; Thompson, L.H. Local Injection of Endothelin-1 in the Early Neonatal Rat Brain Models Ischemic Damage Associated with Motor Impairment and Diffuse Loss in Brain Volume. Neuroscience 2018, 393, 110–122. [Google Scholar] [CrossRef]
- Shibata, M.; Ohtani, R.; Ihara, M.; Tomimoto, H. White matter lesions and glial activation in a novel mouse model of chronic cerebral hypoperfusion. Stroke 2004, 35, 2598–2603. [Google Scholar] [CrossRef]
- Traystman, R.J. Animal Models of Focal and Global Cerebral Ischemia. ILAR J. 2003, 44, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, S.T. Rodent Models of Focal Stroke: Size, Mechanism, and Purpose. NeuroRx 2005, 2, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Braeuninger, S.; Kleinschnitz, C. Rodent models of focal cerebral ischemia: Procedural pitfalls and translational problems. Exp. Transl. Stroke Med. 2009, 1, 8. [Google Scholar] [CrossRef]
- Gennaro, M.; Mattiello, A.; Pizzorusso, T. Rodent Models of Developmental Ischemic Stroke for Translational Research: Strengths and Weaknesses. Available online: https://www.hindawi.com/journals/np/2019/5089321/ (accessed on 22 February 2020).
- Canazza, A.; Minati, L.; Boffano, C.; Parati, E.; Binks, S. Experimental Models of Brain Ischemia: A Review of Techniques, Magnetic Resonance Imaging, and Investigational Cell-Based Therapies. Front. Neurol. 2014, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.; Cho, A.; Limaye, A.; Cho, K.; Khillan, J.; Kulkarni, A.B. Genome Editing in Mice Using CRISPR/Cas9 Technology. Curr. Protoc. Cell Biol. 2018, 81, e57. [Google Scholar] [CrossRef]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Huijbers, I.J. Generating Genetically Modified Mice: A Decision Guide. In Site-Specific Recombinases: Methods and Protocols; Methods in Molecular Biology; Eroshenko, N., Ed.; Springer: New York, NY, USA, 2017; pp. 1–19. ISBN 978-1-4939-7169-5. [Google Scholar]
- Dana, H.; Chen, T.-W.; Hu, A.; Shields, B.C.; Guo, C.; Looger, L.L.; Kim, D.S.; Svoboda, K. Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS ONE 2014, 9, e108697. [Google Scholar] [CrossRef]
- Lou, S.; Adam, Y.; Weinstein, E.N.; Williams, E.; Williams, K.; Parot, V.; Kavokine, N.; Liberles, S.; Madisen, L.; Zeng, H.; et al. Genetically Targeted All-Optical Electrophysiology with a Transgenic Cre-Dependent Optopatch Mouse. J. Neurosci. 2016, 36, 11059–11073. [Google Scholar] [CrossRef]
- Song, C.; Do, Q.B.; Antic, S.D.; Knöpfel, T. Transgenic Strategies for Sparse but Strong Expression of Genetically Encoded Voltage and Calcium Indicators. Int. J. Mol. Sci. 2017, 18, 1461. [Google Scholar] [CrossRef]
- Wang, D.; McMahon, S.; Zhang, Z.; Jackson, M.B. Hybrid voltage sensor imaging of electrical activity from neurons in hippocampal slices from transgenic mice. J. Neurophysiol. 2012, 108, 3147–3160. [Google Scholar] [CrossRef][Green Version]
- Song, C.; Piscopo, D.M.; Niell, C.M.; Knöpfel, T. Cortical signatures of wakeful somatosensory processing. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Diuba, A.V.; Samigullin, D.V.; Kaszas, A.; Zonfrillo, F.; Malkov, A.; Petukhova, E.; Casini, A.; Arosio, D.; Esclapez, M.; Gross, C.T.; et al. CLARITY analysis of the Cl/pH sensor expression in the brain of transgenic mice. Neuroscience 2019. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Shinotsuka, N.; Nonomura, K.; Takemoto, K.; Kuida, K.; Yosida, H.; Miura, M. Live imaging of apoptosis in a novel transgenic mouse highlights its role in neural tube closure. J. Cell Biol. 2011, 195, 1047–1060. [Google Scholar] [CrossRef]
- Daigle, T.L.; Madisen, L.; Hage, T.A.; Valley, M.T.; Knoblich, U.; Larsen, R.S.; Takeno, M.M.; Huang, L.; Gu, H.; Larsen, R.; et al. A suite of transgenic driver and reporter mouse lines with enhanced brain cell type targeting and functionality. Cell 2018, 174, 465–480.e22. [Google Scholar] [CrossRef] [PubMed]
- Gee, J.M.; Gibbons, M.B.; Taheri, M.; Palumbos, S.; Morris, S.C.; Smeal, R.M.; Flynn, K.F.; Economo, M.N.; Cizek, C.G.; Capecchi, M.R.; et al. Imaging activity in astrocytes and neurons with genetically encoded calcium indicators following in utero electroporation. Front. Mol. Neurosci. 2015, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Z.; Schnitzer, M.J. Genetically encoded indicators of neuronal activity. Nat. Neurosci. 2016, 19, 1142–1153. [Google Scholar] [CrossRef]
- Broussard, G.J.; Liang, R.; Tian, L. Monitoring activity in neural circuits with genetically encoded indicators. Front. Mol. Neurosci. 2014, 7, 97. [Google Scholar] [CrossRef]
- Holtmaat, A.; Bonhoeffer, T.; Chow, D.K.; Chuckowree, J.; De Paola, V.; Hofer, S.B.; Hübener, M.; Keck, T.; Knott, G.; Lee, W.-C.A.; et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protoc. 2009, 4, 1128–1144. [Google Scholar] [CrossRef]
- Silasi, G.; Xiao, D.; Vanni, M.P.; Chen, A.C.N.; Murphy, T.H. Intact skull chronic windows for mesoscopic wide-field imaging in awake mice. J. Neurosci. Methods 2016, 267, 141–149. [Google Scholar] [CrossRef]
- Helmchen, F.; Denk, W. Deep tissue two-photon microscopy. Nat. Methods 2005, 2, 932–940. [Google Scholar] [CrossRef]
- Kondo, M.; Kobayashi, K.; Ohkura, M.; Nakai, J.; Matsuzaki, M. Two-photon calcium imaging of the medial prefrontal cortex and hippocampus without cortical invasion. eLife 2017, 6, e26839. [Google Scholar] [CrossRef] [PubMed]
- Horton, N.G.; Wang, K.; Kobat, D.; Clark, C.G.; Wise, F.W.; Schaffer, C.B.; Xu, C. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photonics 2013, 7, 205–209. [Google Scholar] [CrossRef]
- Sych, Y.; Chernysheva, M.; Sumanovski, L.T.; Helmchen, F. High-density multi-fiber photometry for studying large-scale brain circuit dynamics. Nat. Methods 2019, 16, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Barbera, G.; Liang, B.; Zhang, L.; Li, Y.; Lin, D.-T. A wireless miniScope for deep brain imaging in freely moving mice. J. Neurosci. Methods 2019, 323, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Jacob, A.D.; Ramsaran, A.I.; Mocle, A.J.; Tran, L.M.; Yan, C.; Frankland, P.W.; Josselyn, S.A. A Compact Head-Mounted Endoscope for In Vivo Calcium Imaging in Freely Behaving Mice. Curr. Protoc. Neurosci. 2018, 84, e51. [Google Scholar] [CrossRef]
- Feng, G.; Mellor, R.H.; Bernstein, M.; Keller-Peck, C.; Nguyen, Q.T.; Wallace, M.; Nerbonne, J.M.; Lichtman, J.W.; Sanes, J.R. Imaging Neuronal Subsets in Transgenic Mice Expressing Multiple Spectral Variants of GFP. Neuron 2000, 28, 41–51. [Google Scholar] [CrossRef]
- Zhang, Z.G.; Zhang, L.; Ding, G.; Jiang, Q.; Zhang, R.L.; Zhang, X.; Gan, W.-B.; Chopp, M. A Model of Mini-Embolic Stroke Offers Measurements of the Neurovascular Unit Response in the Living Mouse. Stroke 2005, 36, 2701–2704. [Google Scholar] [CrossRef]
- Zhang, S.; Boyd, J.; Delaney, K.; Murphy, T.H. Rapid Reversible Changes in Dendritic Spine Structure In Vivo Gated by the Degree of Ischemia. J. Neurosci. 2005, 25, 5333–5338. [Google Scholar] [CrossRef]
- Brown, C.E.; Li, P.; Boyd, J.D.; Delaney, K.R.; Murphy, T.H. Extensive Turnover of Dendritic Spines and Vascular Remodeling in Cortical Tissues Recovering from Stroke. J. Neurosci. 2007, 27, 4101–4109. [Google Scholar] [CrossRef]
- Zhang, S.; Murphy, T.H. Imaging the Impact of Cortical Microcirculation on Synaptic Structure and Sensory-Evoked Hemodynamic Responses In Vivo. PLoS Biol. 2007, 5, e119. [Google Scholar] [CrossRef]
- Enright, L.E.; Zhang, S.; Murphy, T.H. Fine Mapping of the Spatial Relationship between Acute Ischemia and Dendritic Structure Indicates Selective Vulnerability of Layer V Neuron Dendritic Tufts within Single Neurons in Vivo. J. Cereb. Blood Flow Metab. 2007, 27, 1185–1200. [Google Scholar] [CrossRef]
- Brown, C.E.; Murphy, T.H. Livin’ on the Edge: Imaging Dendritic Spine Turnover in the Peri-Infarct Zone during Ischemic Stroke and Recovery. Neuroscientist 2008, 14, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.E.; Boyd, J.D.; Murphy, T.H. Longitudinal in vivo Imaging Reveals Balanced and Branch-Specific Remodeling of Mature Cortical Pyramidal Dendritic Arbors after Stroke. J. Cereb. Blood Flow Metab. 2010, 30, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.H. Two-Photon Imaging of Neuronal Structural Plasticity in Mice during and after Ischemia. Cold Spring Harb. Protoc. 2015, 2015, pdb.prot087486. [Google Scholar] [CrossRef][Green Version]
- Ikonomidou, C.; Price, M.; Mosinger, J.; Frierdich, G.; Labruyere, J.; Salles, K.; Olney, J. Hypobaric-ischemic conditions produce glutamate-like cytopathology in infant rat brain. J. Neurosci. 1989, 9, 1693–1700. [Google Scholar] [CrossRef]
- Park, J.S.; Bateman, M.C.; Goldberg, M.P. Rapid Alterations in Dendrite Morphology during Sublethal Hypoxia or Glutamate Receptor Activation. Neurobiol. Dis. 1996, 3, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Lihavainen, E.; Kislin, M.; Toptunov, D.; Khiroug, L.; Ribeiro, A.S. Automatic quantification of mitochondrial fragmentation from two-photon microscope images of mouse brain tissue: Automatic quantification of mitochondrial fragmentation. J. Microsc. 2015, 260, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Kislin, M.; Sword, J.; Fomitcheva, I.V.; Croom, D.; Pryazhnikov, E.; Lihavainen, E.; Toptunov, D.; Rauvala, H.; Ribeiro, A.S.; Khiroug, L.; et al. Reversible Disruption of Neuronal Mitochondria by Ischemic and Traumatic Injury Revealed by Quantitative Two-Photon Imaging in the Neocortex of Anesthetized Mice. J. Neurosci. 2017, 37, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Risher, W.C.; Ard, D.; Yuan, J.; Kirov, S.A. Recurrent Spontaneous Spreading Depolarizations Facilitate Acute Dendritic Injury in the Ischemic Penumbra. J. Neurosci. 2010, 30, 9859–9868. [Google Scholar] [CrossRef]
- Risher, W.C.; Andrew, R.D.; Kirov, S.A. Real-time passive volume responses of astrocytes to acute osmotic and ischemic stress in cortical slices and in vivo revealed by two-photon microscopy. Glia 2009, 57, 207–221. [Google Scholar] [CrossRef]
- Risher, W.C.; Croom, D.; Kirov, S.A. Persistent astroglial swelling accompanies rapid reversible dendritic injury during stroke-induced spreading depolarizations. Glia 2012, 60, 1709–1720. [Google Scholar] [CrossRef]
- Balbi, M.; Vanni, M.P.; Silasi, G.; Sekino, Y.; Bolanos, L.; LeDue, J.M.; Murphy, T.H. Targeted ischemic stroke induction and mesoscopic imaging assessment of blood flow and ischemic depolarization in awake mice. Neurophotonics 2017, 4, 035001. [Google Scholar] [CrossRef]
- Balbi, M.; Vanni, M.P.; Vega, M.J.; Silasi, G.; Sekino, Y.; Boyd, J.D.; LeDue, J.M.; Murphy, T.H. Longitudinal monitoring of mesoscopic cortical activity in a mouse model of microinfarcts reveals dissociations with behavioral and motor function. J. Cereb. Blood Flow Metab. 2019, 39, 1486–1500. [Google Scholar] [CrossRef]
- Tvrdik, P.; Kearns, K.N.; Sharifi, K.A.; Sluzewski, M.F.; Acton, S.T.; Kalani, M.Y.S. Calcium Imaging of Microglial Network Activity in Stroke. In Microglia; Garaschuk, O., Verkhratsky, A., Eds.; Springer: New York, NY, USA, 2019; Volume 2034, pp. 267–279. ISBN 978-1-4939-9657-5. [Google Scholar]
- Steiger, H.-J.; Hänggi, D. Ischaemic preconditioning of the brain, mechanisms and applications. Acta Neurochir. (Wien) 2007, 149, 1–10. [Google Scholar] [CrossRef]
- Zhou, G.; Li, M.H.; Tudor, G.; Lu, H.T.; Kadirvel, R.; Kallmes, D. Remote Ischemic Conditioning in Cerebral Diseases and Neurointerventional Procedures: Recent Research Progress. Front. Neurol. 2018, 9, 339. [Google Scholar] [CrossRef]
- Gidday, J.M. Cerebral preconditioning and ischaemic tolerance. Nat. Rev. Neurosci. 2006, 7, 437–448. [Google Scholar] [CrossRef]
- Pietrogrande, G.; Zalewska, K.; Zhao, Z.; Abdolhoseini, M.; Chow, W.Z.; Sanchez-Bezanilla, S.; Ong, L.K.; Johnson, S.J.; Nilsson, M.; Walker, F.R. Low oxygen post conditioning prevents thalamic secondary neuronal loss caused by excitotoxicity after cortical stroke. Sci. Rep. 2019, 9, 4841. [Google Scholar] [CrossRef]
- Zhao, H. Ischemic postconditioning as a novel avenue to protect against brain injury after stroke. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2009, 29, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ren, C.; Chen, X.; Shen, J. From rapid to delayed and remote postconditioning: The evolving concept of ischemic postconditioning in brain ischemia. Curr. Drug Targets 2012, 13, 173–187. [Google Scholar] [CrossRef]
- Anttila, V.; Haapanen, H.; Yannopoulos, F.; Herajärvi, J.; Anttila, T.; Juvonen, T. Review of remote ischemic preconditioning: From laboratory studies to clinical trials. Scand. Cardiovasc. J. SCJ 2016, 50, 355–361. [Google Scholar] [CrossRef]
- Dezfulian, C.; Garrett, M.; Gonzalez, N.R. Clinical application of preconditioning and postconditioning to achieve neuroprotection. Transl. Stroke Res. 2013, 4, 19–24. [Google Scholar] [CrossRef]
- McCafferty, K.; Forbes, S.; Thiemermann, C.; Yaqoob, M.M. The challenge of translating ischemic conditioning from animal models to humans: The role of comorbidities. Dis. Model. Mech. 2014, 7, 1321–1333. [Google Scholar] [CrossRef]
- Vinciguerra, A.; Cuomo, O.; Cepparulo, P.; Anzilotti, S.; Brancaccio, P.; Sirabella, R.; Guida, N.; Annunziato, L.; Pignataro, G. Models and methods for conditioning the ischemic brain. J. Neurosci. Methods 2018, 310, 63–74. [Google Scholar] [CrossRef]
- Meller, R.; Simon, R.P. A critical review of mechanisms regulating remote preconditioning-induced brain protection. J. Appl. Physiol. Bethesda Md 1985 2015, 119, 1135–1142. [Google Scholar] [CrossRef]
- Chen, G.; Thakkar, M.; Robinson, C.; Doré, S. Limb Remote Ischemic Conditioning: Mechanisms, Anesthetics, and the Potential for Expanding Therapeutic Options. Front. Neurol. 2018, 9, 40. [Google Scholar] [CrossRef]
- Fan, Y.-Y.; Hu, W.-W.; Nan, F.; Chen, Z. Postconditioning-induced neuroprotection, mechanisms and applications in cerebral ischemia. Neurochem. Int. 2017, 107, 43–56. [Google Scholar] [CrossRef]
- Niedzielska, E.; Smaga, I.; Gawlik, M.; Moniczewski, A.; Stankowicz, P.; Pera, J.; Filip, M. Oxidative Stress in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 4094–4125. [Google Scholar] [CrossRef]
- Uttara, B.; Singh, A.V.; Zamboni, P.; Mahajan, R.T. Oxidative stress and neurodegenerative diseases: A review of upstream and downstream antioxidant therapeutic options. Curr. Neuropharmacol. 2009, 7, 65–74. [Google Scholar] [CrossRef]
- Mecocci, P.; Boccardi, V.; Cecchetti, R.; Bastiani, P.; Scamosci, M.; Ruggiero, C.; Baroni, M. A Long Journey into Aging, Brain Aging, and Alzheimer’s Disease Following the Oxidative Stress Tracks. J. Alzheimers Dis. JAD 2018, 62, 1319–1335. [Google Scholar] [CrossRef]
- Kumar, A.; Yegla, B.; Foster, T.C. Redox Signaling in Neurotransmission and Cognition during Aging. Antioxid. Redox Signal. 2018, 28, 1724–1745. [Google Scholar] [CrossRef]
- Pignataro, G.; Scorziello, A.; Di Renzo, G.; Annunziato, L. Post-ischemic brain damage: Effect of ischemic preconditioning and postconditioning and identification of potential candidates for stroke therapy. FEBS J. 2009, 276, 46–57. [Google Scholar] [CrossRef] [PubMed]
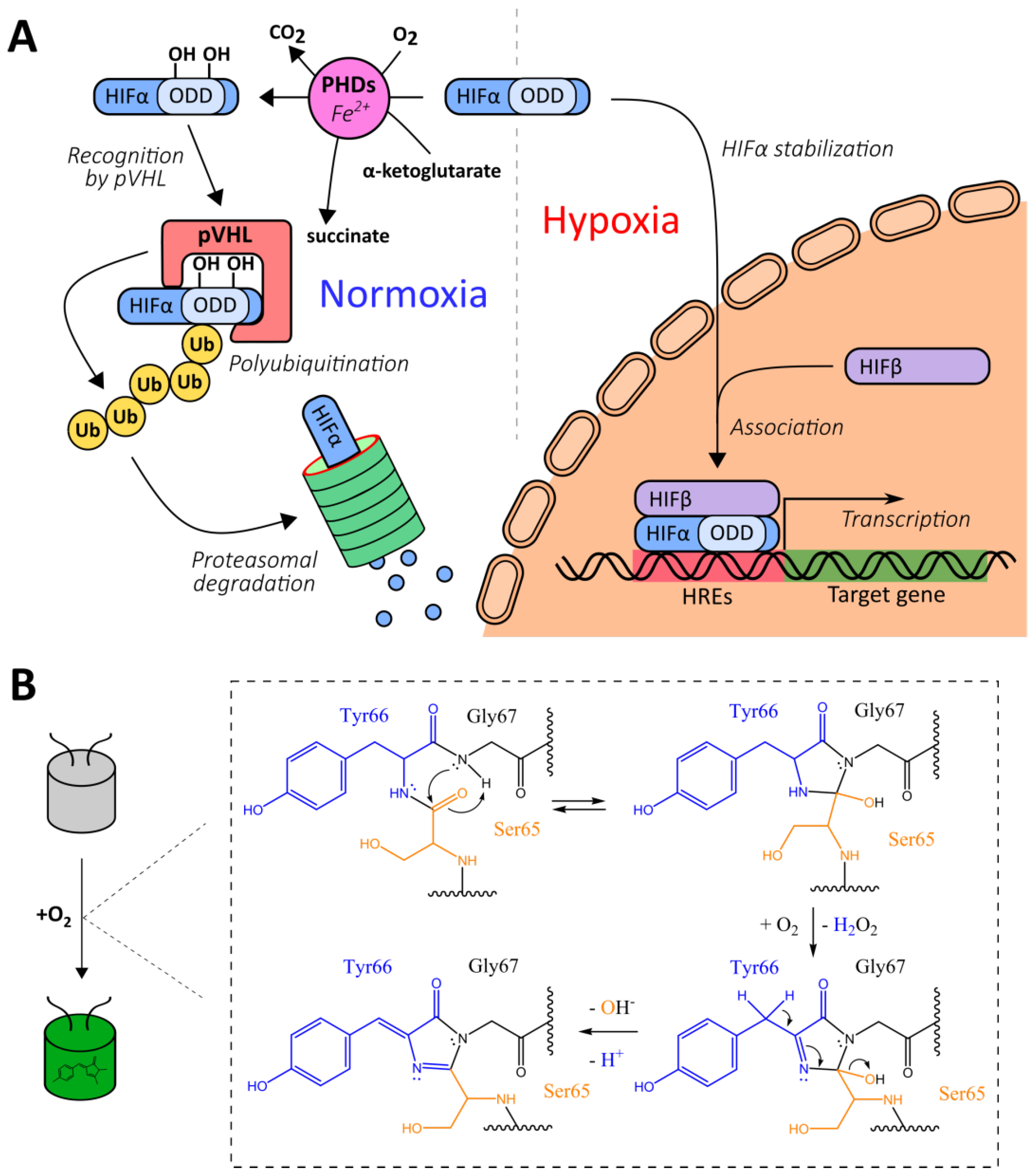

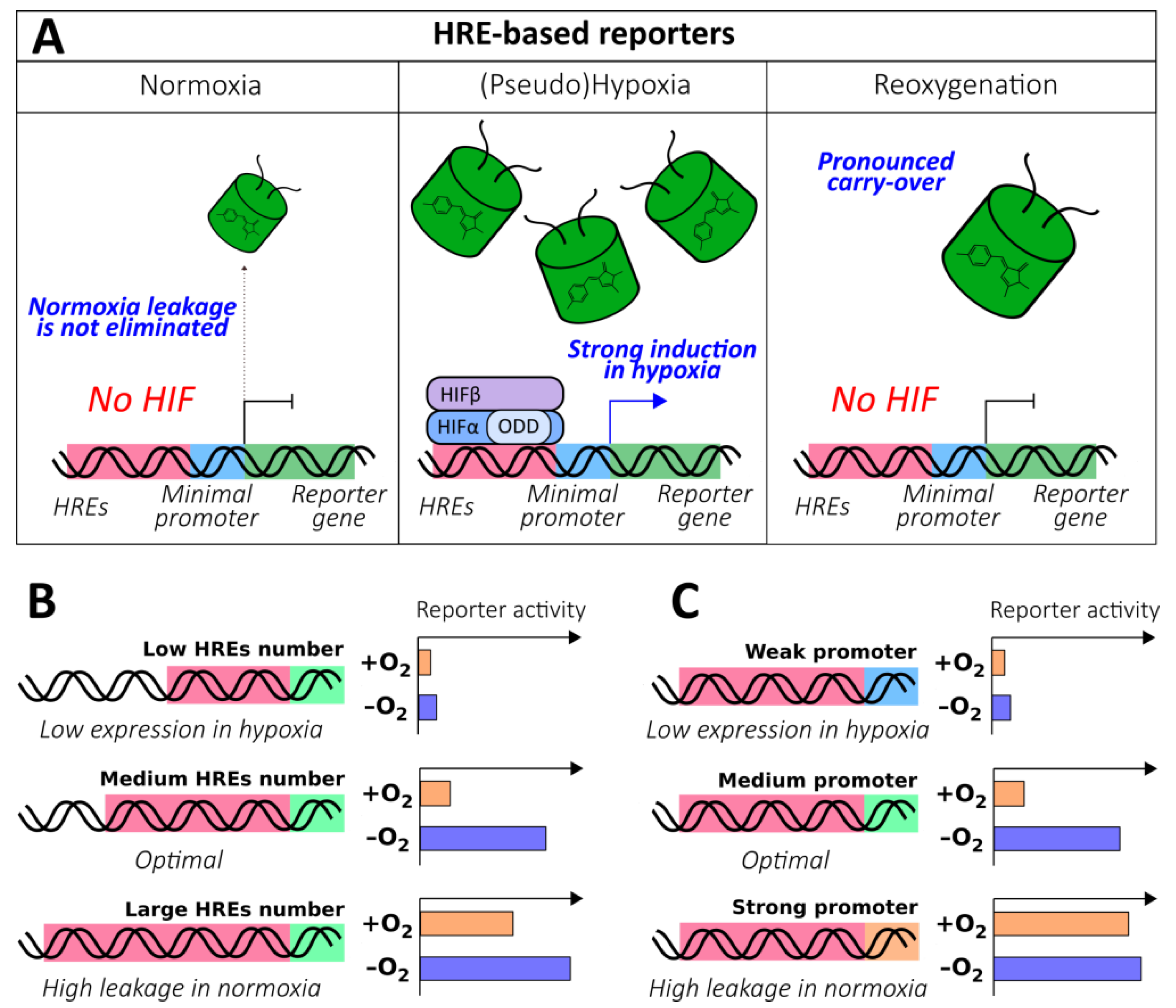
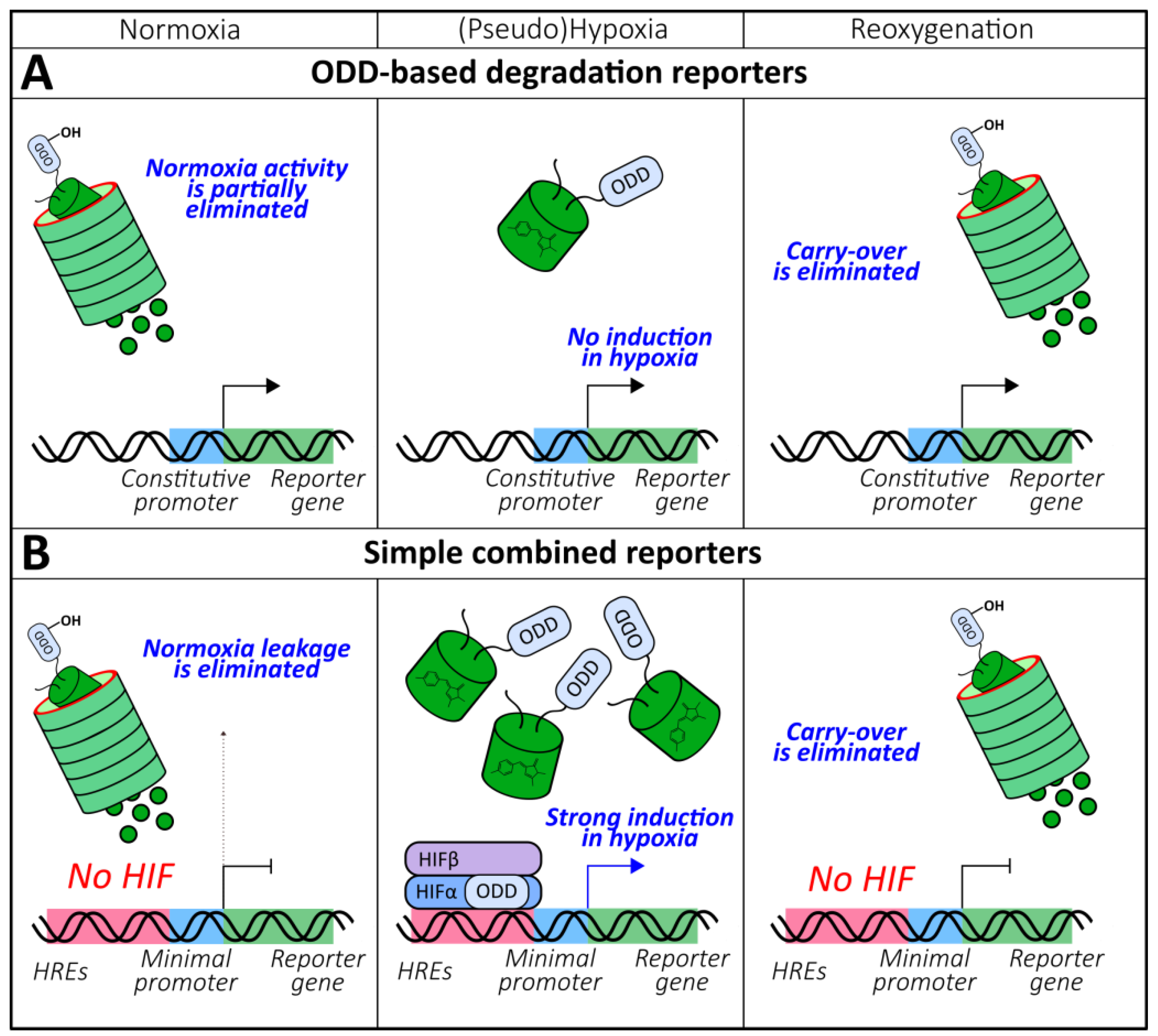

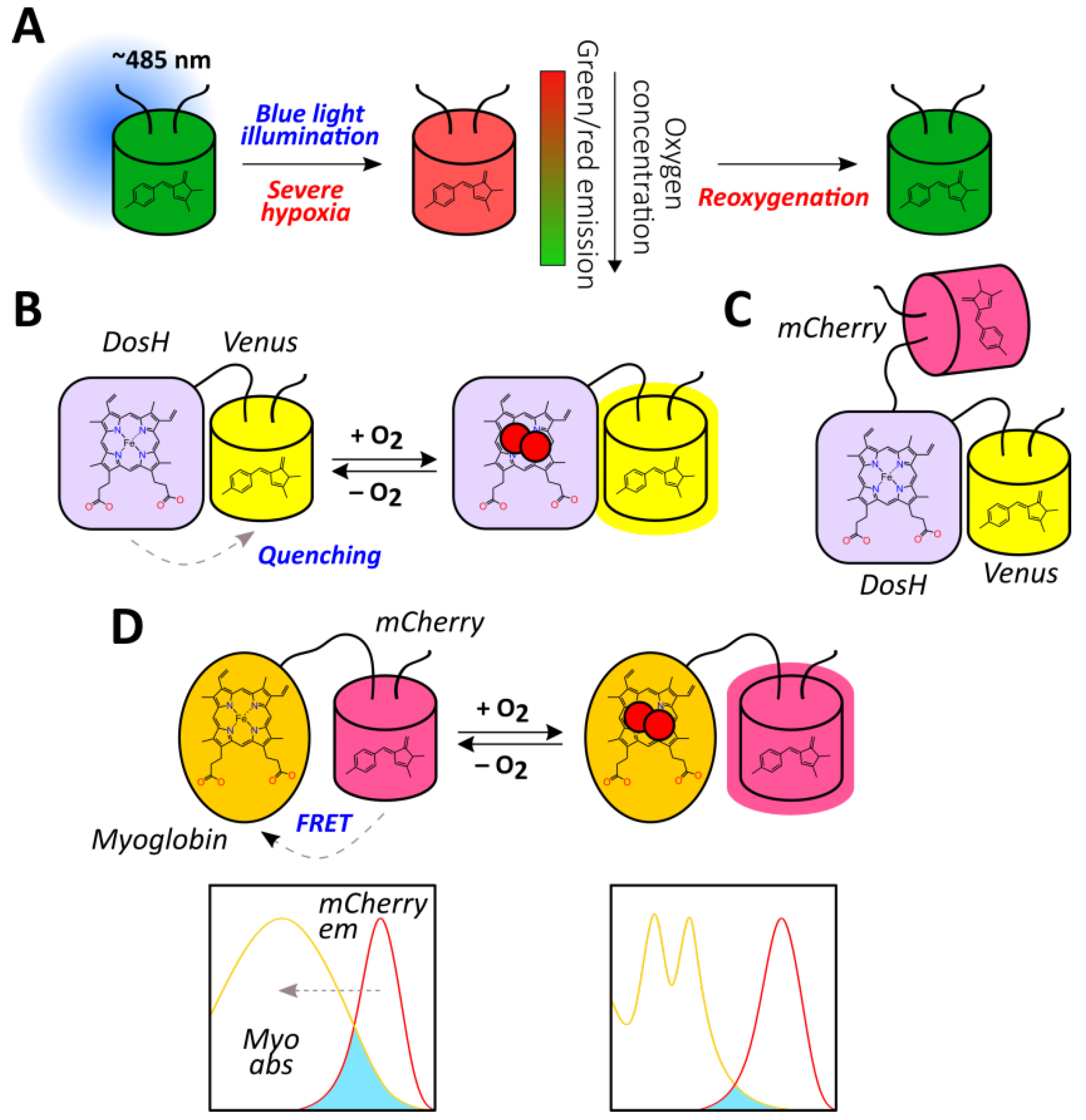
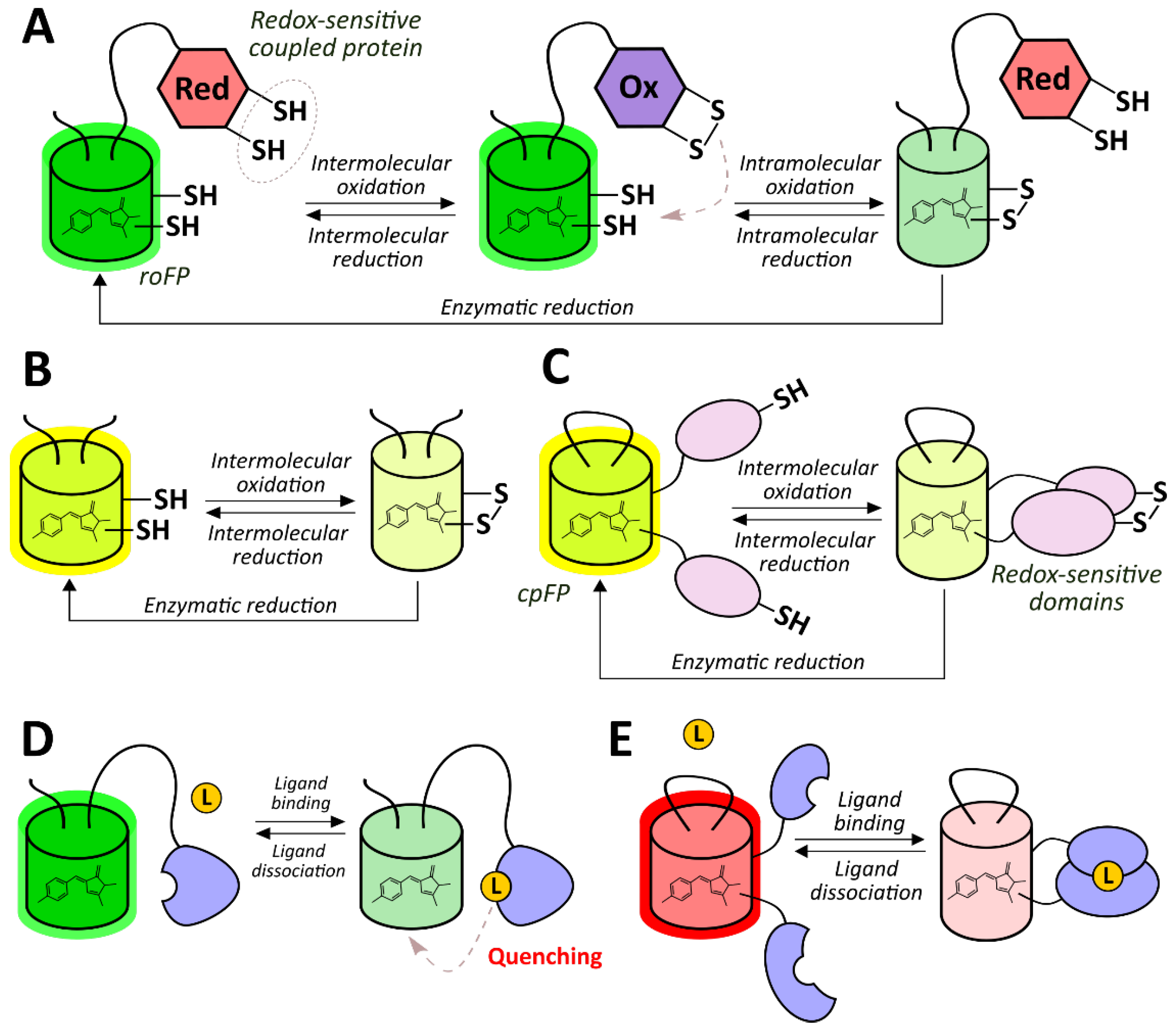
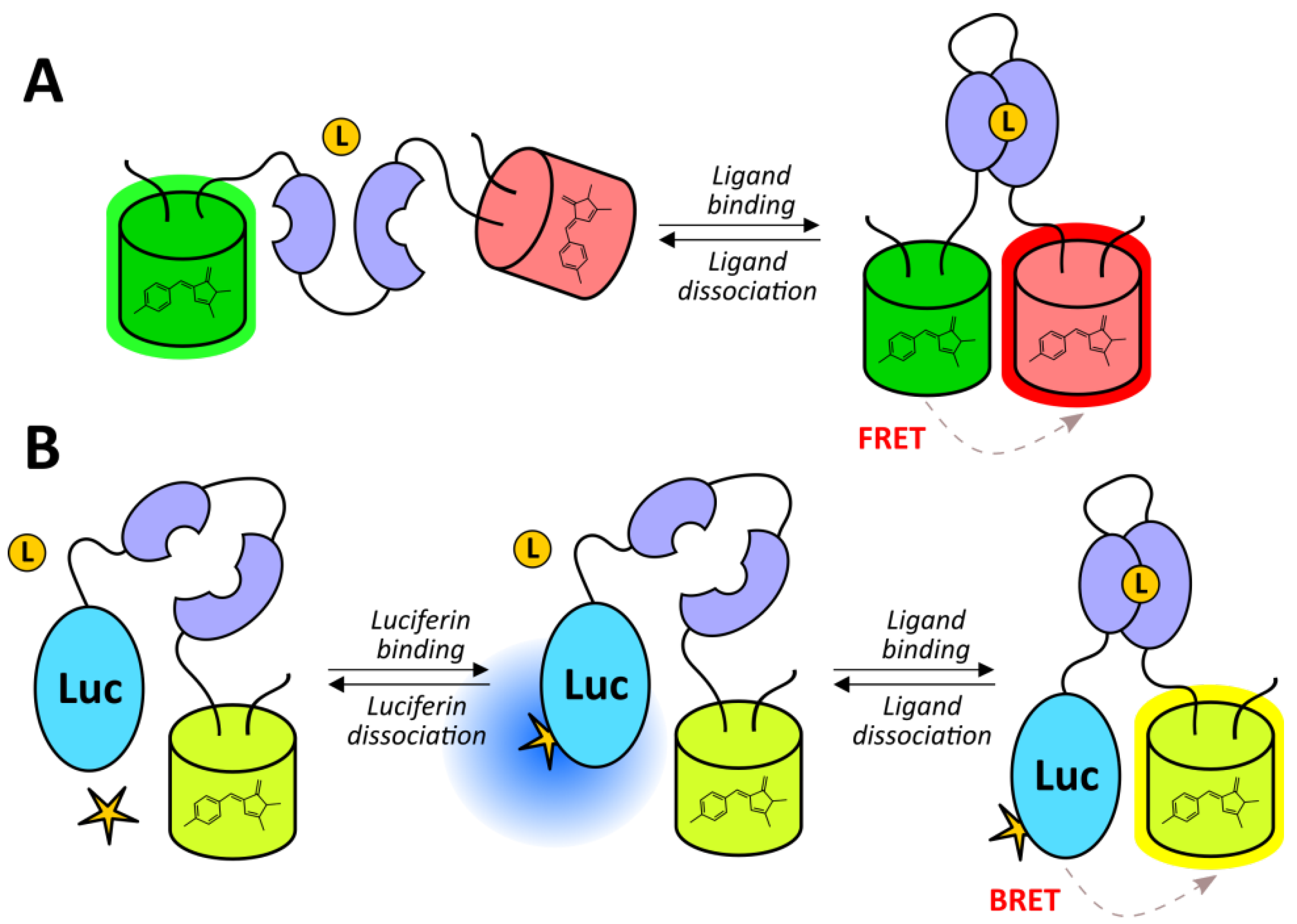
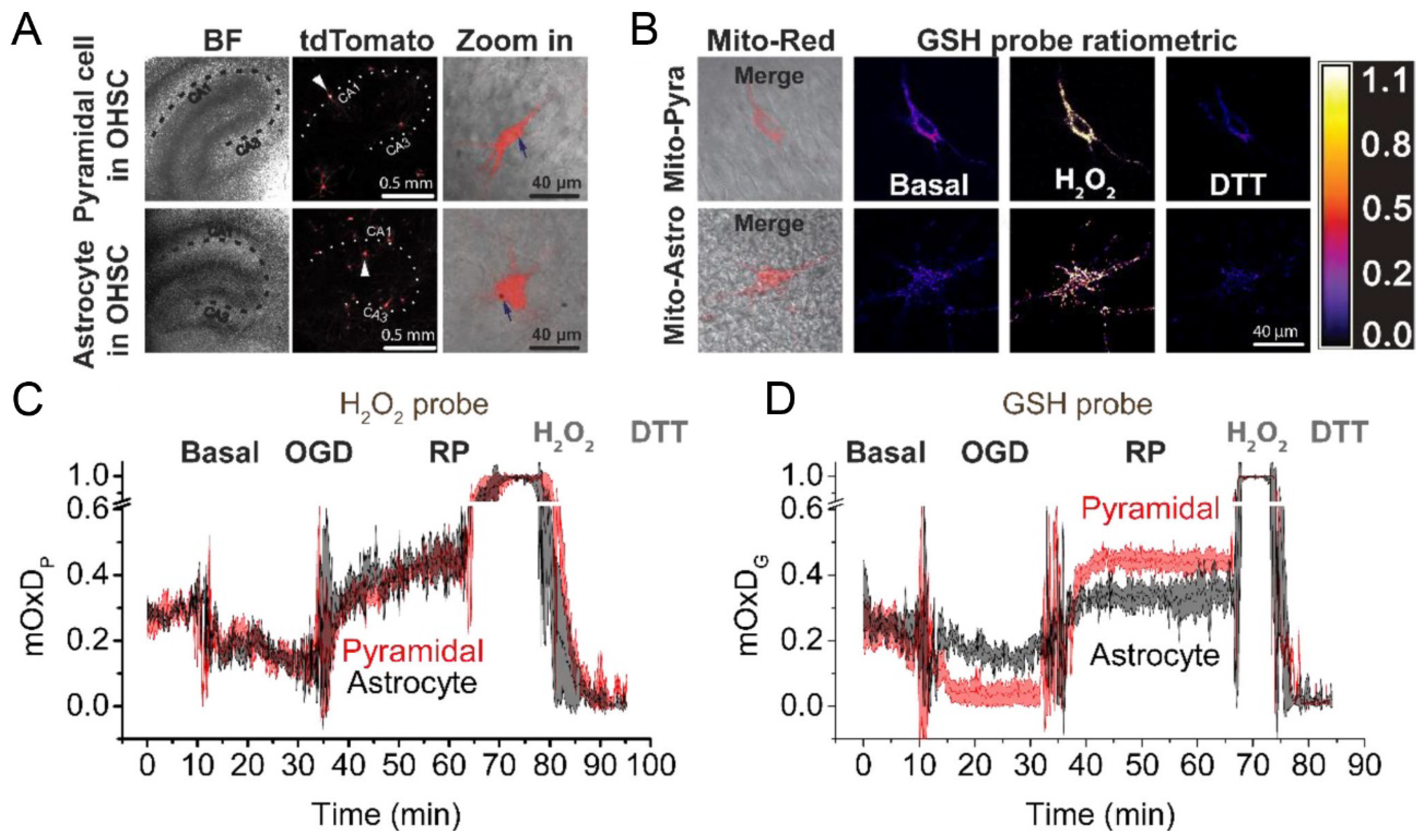
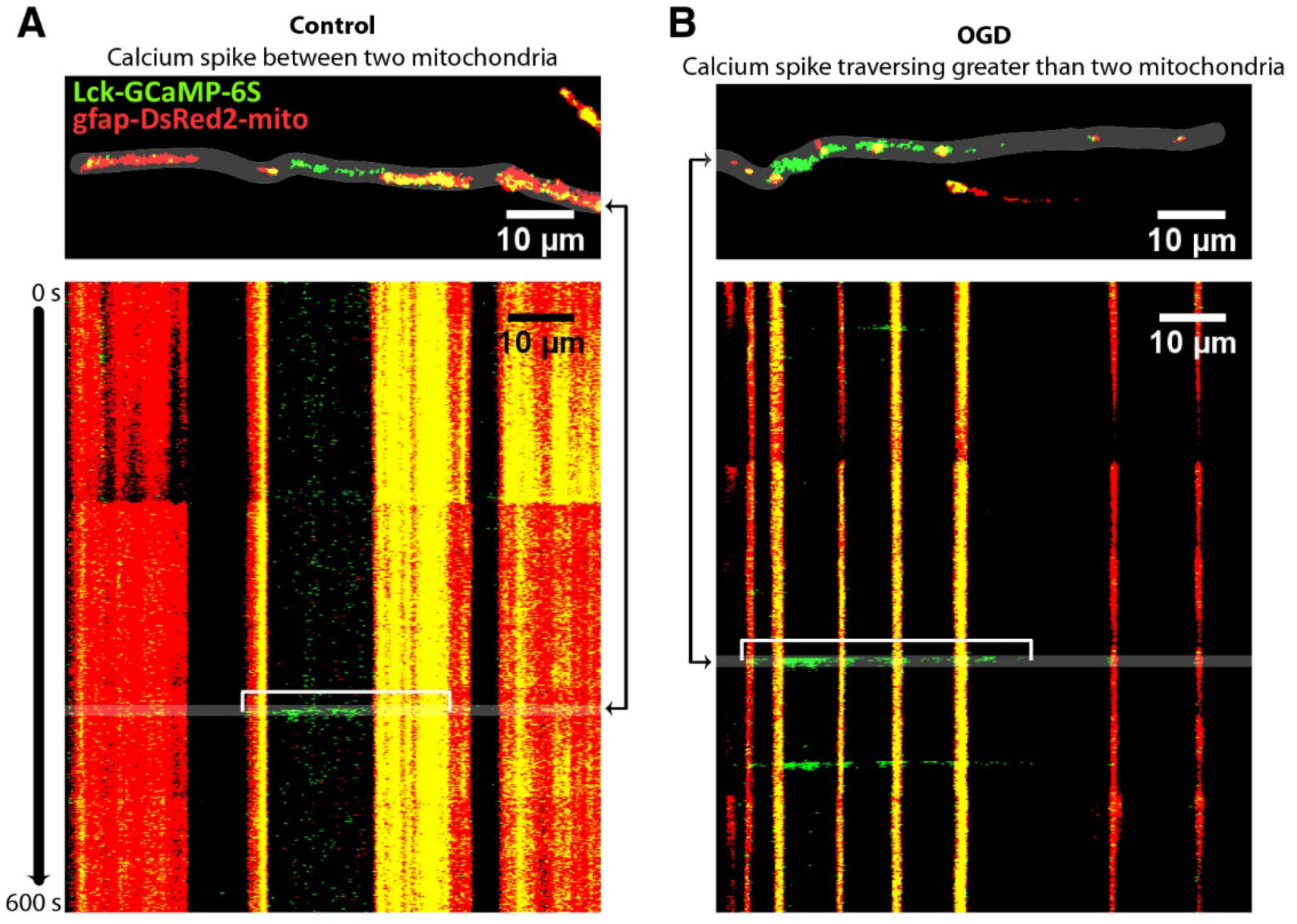
| Name | Range Max | Midpoint Potential/EC50 | Reference |
|---|---|---|---|
| rxYFP | 2.2 (in vitro) | Midpoint potential = −261 mV | [162] |
| rxYFP-Grx1p | 2.1 (in vitro) | Midpoint potential = −267 mV | [163] |
| roGFP1 | ~6 (in vitro) | Midpoint potential = −294 mV | [151,152] |
| roGFP1-Rx family | 5.4–7.5 | Midpoint potential −263 mV – −284 mV | [153] |
| roGFP1-iX | 2.4–7.2 | Midpoint potential −229 mV – −246 mV | [154] |
| roGFP2 | ~6 (in vitro) | Midpoint potential = −287 mV | [151,152] |
| roUnaG | ~9 (in vitro) | Midpoint potential = −275 mV | [161] |
| Grx1-roGFP2 | ~4.4 (in living cells) | Midpoint potential = −280 mV | [155] |
| rxmRuby2 | ~2 (in vitro) | Midpoint potential = −265 ± 22 mV | [164] |
| rxRFP | 4 | Midpoint potential = −290 mV | [160] |
| Grx1-roCherry | 1.5 | Midpoint potential = −311 mV (pH = 7.0) | [158] |
| TrxRFP1 | ~6 (in vitro) | EC50 = 13.5 ± 0.7 μM (H2O2 in HEK293T cells) EC50 = 3.2 ± 1.1 μM (auranofin in HEK293T cells) | [159] |
| Name | Range Max | Sensitivity/Kinetics | Reference |
|---|---|---|---|
| TriPer | NM | NM | [170] |
| HyPer | 3.3 (in vitro) | Ks = 5 × 105 M−1 s−1 | [166,168] |
| HyPer-2 | ~6 (HeLa cells) | Ks = 1.2 × 105 M−1 s−1 | [167,168] |
| HyPer-3 | ~6 (HeLa cells) | Ks = 2.5 × 105 M−1 s−1 | [168] |
| HyPer7 | ~10 (in vitro) | i.v. = 26.9 ± 0.28 a.u./s (for HyPer3 it is 0.315 ± 0.007 a.u./s) | [173] |
| roGFP2-Orp1 | 4.8 (HeLa cells) | responds to low micromolar concentrations of exogenously applied H2O2 (HeLa cells) | [174] |
| roGFP2-Tsa2ΔCR | ~6 (in vitro) | low-nanomolar or high-picomolar endogenous H2O2 concentrations | [175] |
| HyPerRed | ~2 | Ks=3 × 105 M−1 s−1 | [169] |
| geNOps group (different colors) | 1.07–1.18 (HeLa cells) | EC50 = 50–94.1 nM | [178] |
| Name | Analyte | Range Max | Kd | Reference |
|---|---|---|---|---|
| Peredox | NADH/NAD+ | 2.5 | Kd to NADH < 5 nM for initial P0 construct | [183] |
| Frex | NADH | 9 (fl. increase) | Kd~3.7 μM at pH 7.4 | [187] |
| FrexH | NADH | 3 (fl. decrease) | Kd~ 40 nM | [187] |
| RexYFP | NADH/NAD+ | ~ 2 (in vitro) | K’(NADH) = 180 nM, K’(NADPH) = 6.2 μM | [184] |
| SoNar | NADH/NAD+ | 15 | Kd(NADH) ~0.2 μM, Kd(NAD+) ~5.0 μM at pH 7.4 | [185] |
| FiNad | NAD+/AXP | ~7 | Kd(NAD+) shifts from ~14 μM to ~1.3 mM in the presence of ATP or ADP | [189] |
| NAD+ sensor | NAD+ | ~2 | Kd~65 μM | [188] |
| iNaps | NADPH | 10 | Kd = 2.0–120 μM | [186] |
| Name | Analyte | Range Max | Kd | Reference |
|---|---|---|---|---|
| MaLionB | ATP | 1.9 | Kd = 0.46 mM | [193] |
| QUEEN-7μ | ATP | ~6 | Kd = 7.2 μM (25 °C) | [191] |
| QUEEN-2m | ATP | ~5 | Kd = 4.5 mM (25 °C) | [191] |
| iATPSnFR1.0 | ATP | ~3.4 | EC50 ~ 120 μM | [192] |
| iATPSnFR1.1 | ATP | ~2.9 | EC50 ~ 50 μM | [192] |
| mRuby-iATPSnFR1.0 | ATP | Same as for iATPSnFR1.0 | Same as for iATPSnFR1.0 | [192] |
| MaLionG | ATP | 4.9 | Kd = 1.1 mM | [193] |
| Perceval | [ATP]:[ADP] | ~2 | KR~0.5 | [197] |
| PercevalHR | [ATP]:[ADP] | ~8 | KR~3.5 | [198] |
| MaLionR | ATP | 4.5 | Kd = 0.34 mM | [193] |
| ATeam1.03 | ATP | ~2.3 | Kd = 3.3 mM (37 °C) | [194] |
| ATeam3.10 | ATP | ~2 | Kd = 7.4 μM (37 °C) | [194] |
| GO-ATeam1 | ATP | ~2 | Kd = 7.1 mM (37 °C) | [195] |
| GO-ATeam2 | ATP | ~3 | Kd = 2.3 mM (37 °C) | [195] |
| BTeam (BRET) | ATP | ~3 | K0.5 = 1.7 mM (25 °C) | [196] |
| Laconic | Lactate | ~1.2 (in vitro) ~1.4 (in living cells) | NM | [200] |
| Name | Range Max | pKa | Reference |
|---|---|---|---|
| ratio-pHluorin | ~3 (pH 5.5–7.5) | 6.9 | [208] |
| pHluorin2 | ~3 (pH 5.8–7.4) in HEK293 | 7.1 | [209,213] |
| EYFP | NM | 7.1 | [211] |
| E2GFP | ~8 (pH 4.5–9.0) | ~7.0 | [212] |
| SypHer3s | >36 (pH 5.5–10.55) | 7.8 in HeLa Kyoto | [172] |
| pHoran4 | 17 (pH 5.5–7.5) | 7.5 | [203] |
| pHTomato | ~3 (pH 7.6–9.8) | 7.8 | [204] |
| pHuji | 22 (pH 5.5–7.5 ) | 7.7 | [203] |
| pHRed | >10 (pH 5.5–9.0) | 6.6 | [205] |
| mNectarine | >10 (pH 5.5–9.0) | 6.9 | [206] |
| deGFPs | NM | 6.8–8.0 | [210] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostyuk, A.I.; Kokova, A.D.; Podgorny, O.V.; Kelmanson, I.V.; Fetisova, E.S.; Belousov, V.V.; Bilan, D.S. Genetically Encoded Tools for Research of Cell Signaling and Metabolism under Brain Hypoxia. Antioxidants 2020, 9, 516. https://doi.org/10.3390/antiox9060516
Kostyuk AI, Kokova AD, Podgorny OV, Kelmanson IV, Fetisova ES, Belousov VV, Bilan DS. Genetically Encoded Tools for Research of Cell Signaling and Metabolism under Brain Hypoxia. Antioxidants. 2020; 9(6):516. https://doi.org/10.3390/antiox9060516
Chicago/Turabian StyleKostyuk, Alexander I., Aleksandra D. Kokova, Oleg V. Podgorny, Ilya V. Kelmanson, Elena S. Fetisova, Vsevolod V. Belousov, and Dmitry S. Bilan. 2020. "Genetically Encoded Tools for Research of Cell Signaling and Metabolism under Brain Hypoxia" Antioxidants 9, no. 6: 516. https://doi.org/10.3390/antiox9060516
APA StyleKostyuk, A. I., Kokova, A. D., Podgorny, O. V., Kelmanson, I. V., Fetisova, E. S., Belousov, V. V., & Bilan, D. S. (2020). Genetically Encoded Tools for Research of Cell Signaling and Metabolism under Brain Hypoxia. Antioxidants, 9(6), 516. https://doi.org/10.3390/antiox9060516