The Antioxidant Enzyme Methionine Sulfoxide Reductase A (MsrA) Interacts with Jab1/CSN5 and Regulates Its Function
Abstract
1. Introduction
2. Material and Methods
2.1. Ethics Statement
2.2. The Yeast Two-Hybrid System (Y2H)
2.3. GST-MsrA “Pull-Down” Experiments
2.4. Immunoprecipitation (IP) Experiments
2.5. In Vivo Met Oxidation of Jab1 in Mouse Brain
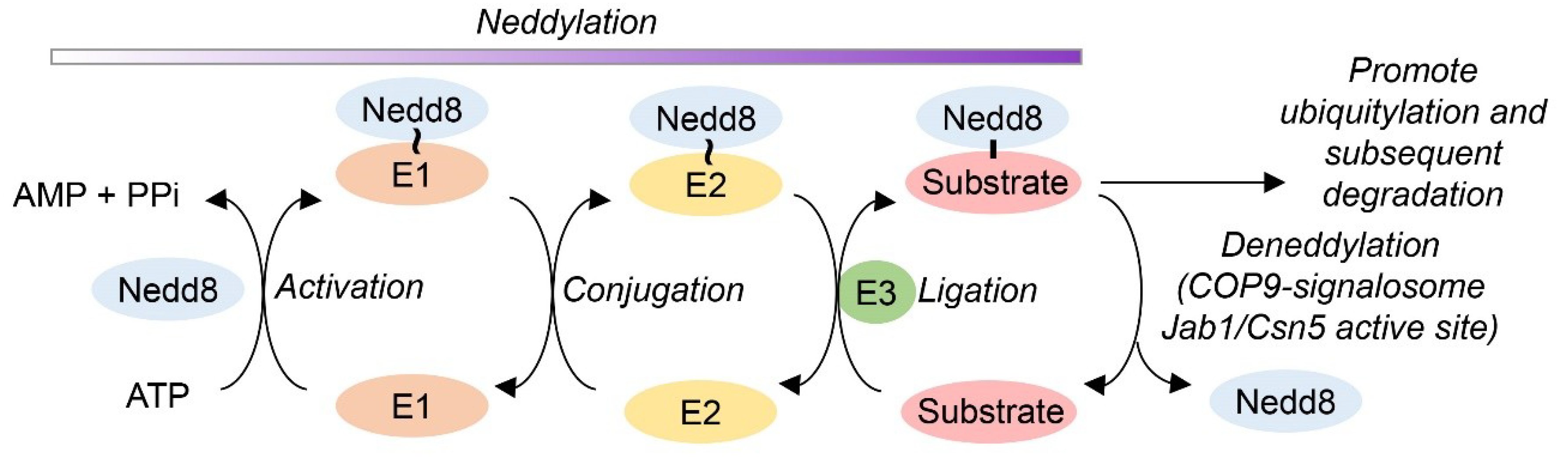
2.6. Deneddylase Activity of Jab1 on Cul-1
2.7. Recombinant Nedd8 (FHN8) Conjugate Synthesis and Purification
2.8. Deneddylation Activity Assay
2.9. Jab1 Binding to P27 and P27 Expression in Brain
3. Results
3.1. MsrA Interacting Proteins Identified by Yeast Two-Hybrid (Y2H)
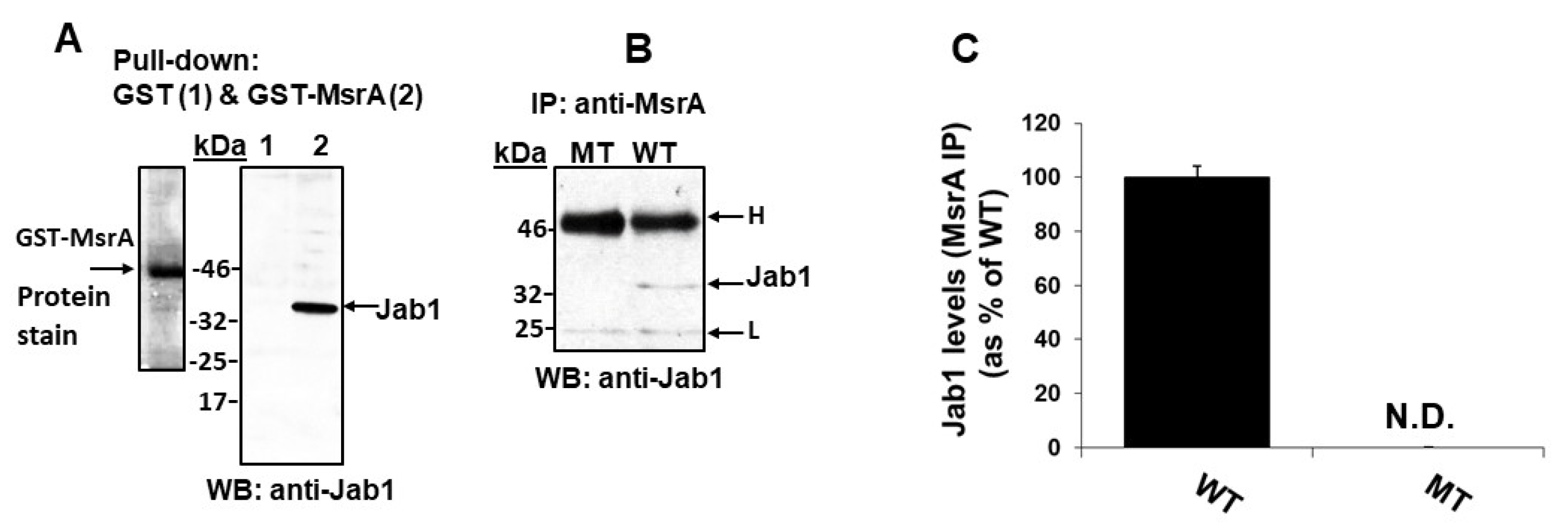
3.2. GST-MsrA “Pull-Down” of Jab1
3.3. Jab1 Co-Immunoprecipitates with MsrA
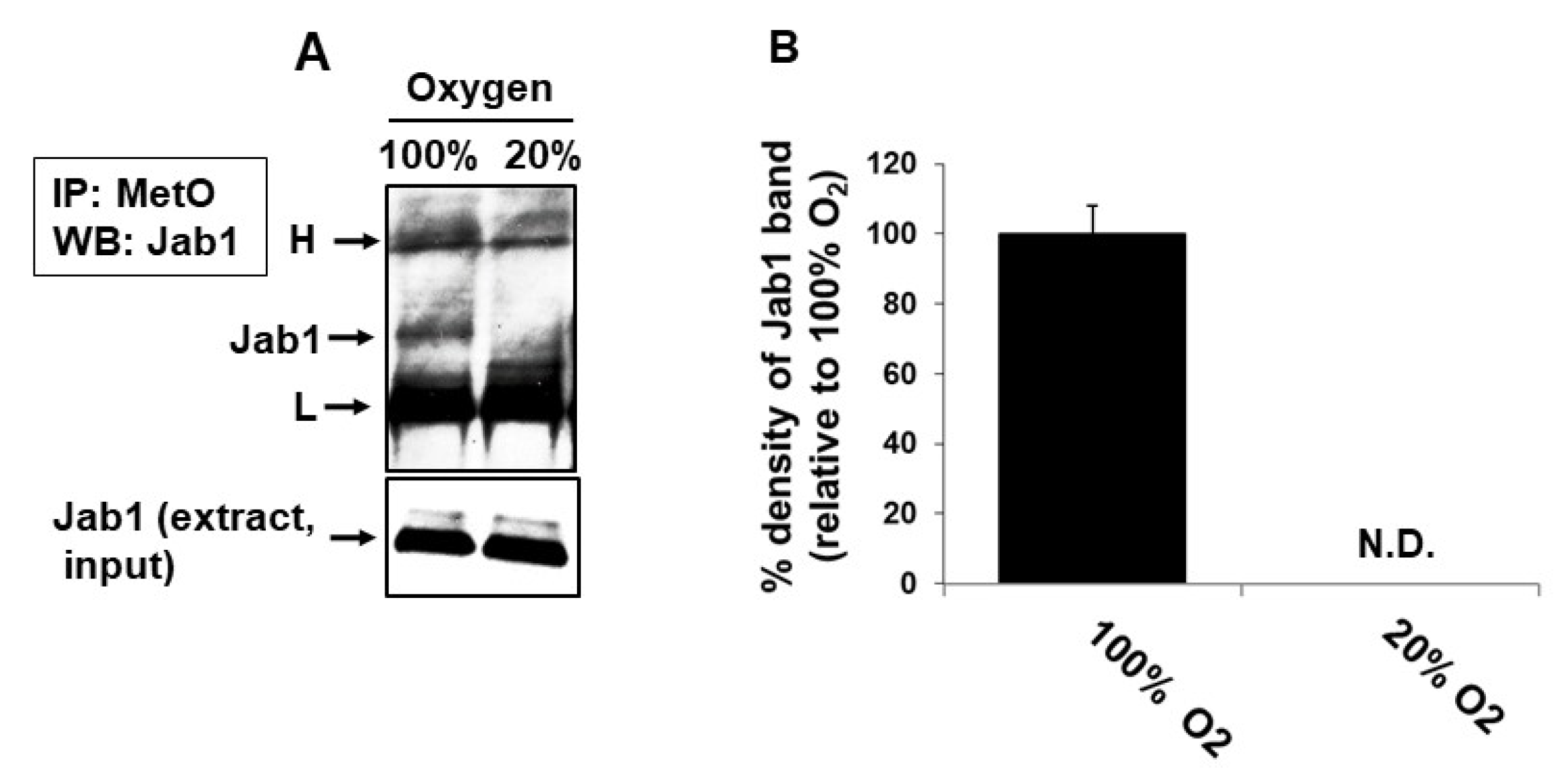
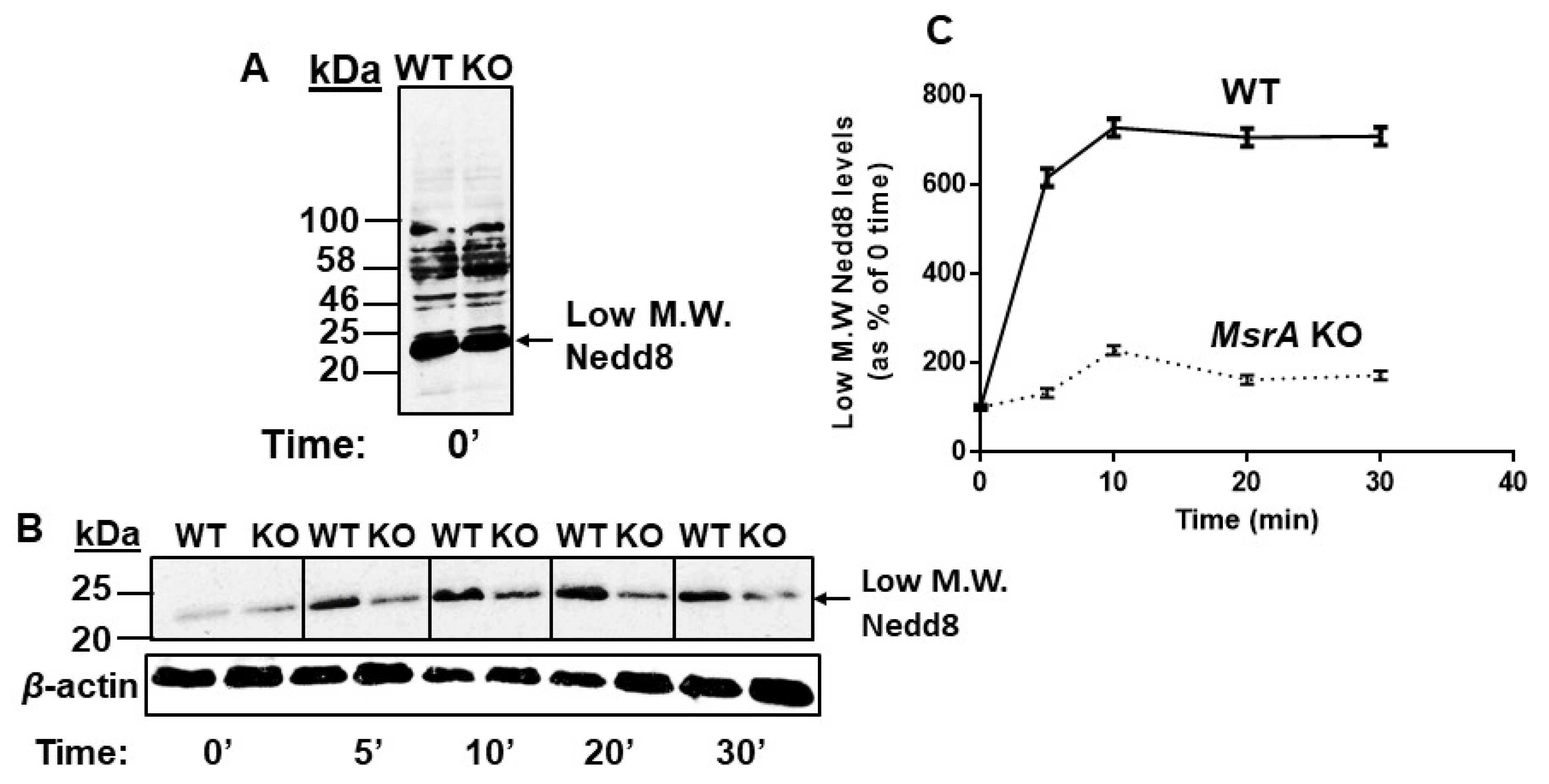
3.4. In Vivo Met Oxidation of Jab1 in Mouse Brain
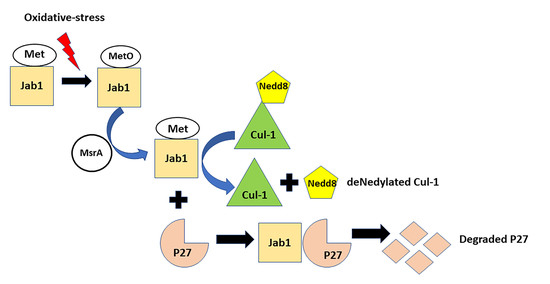
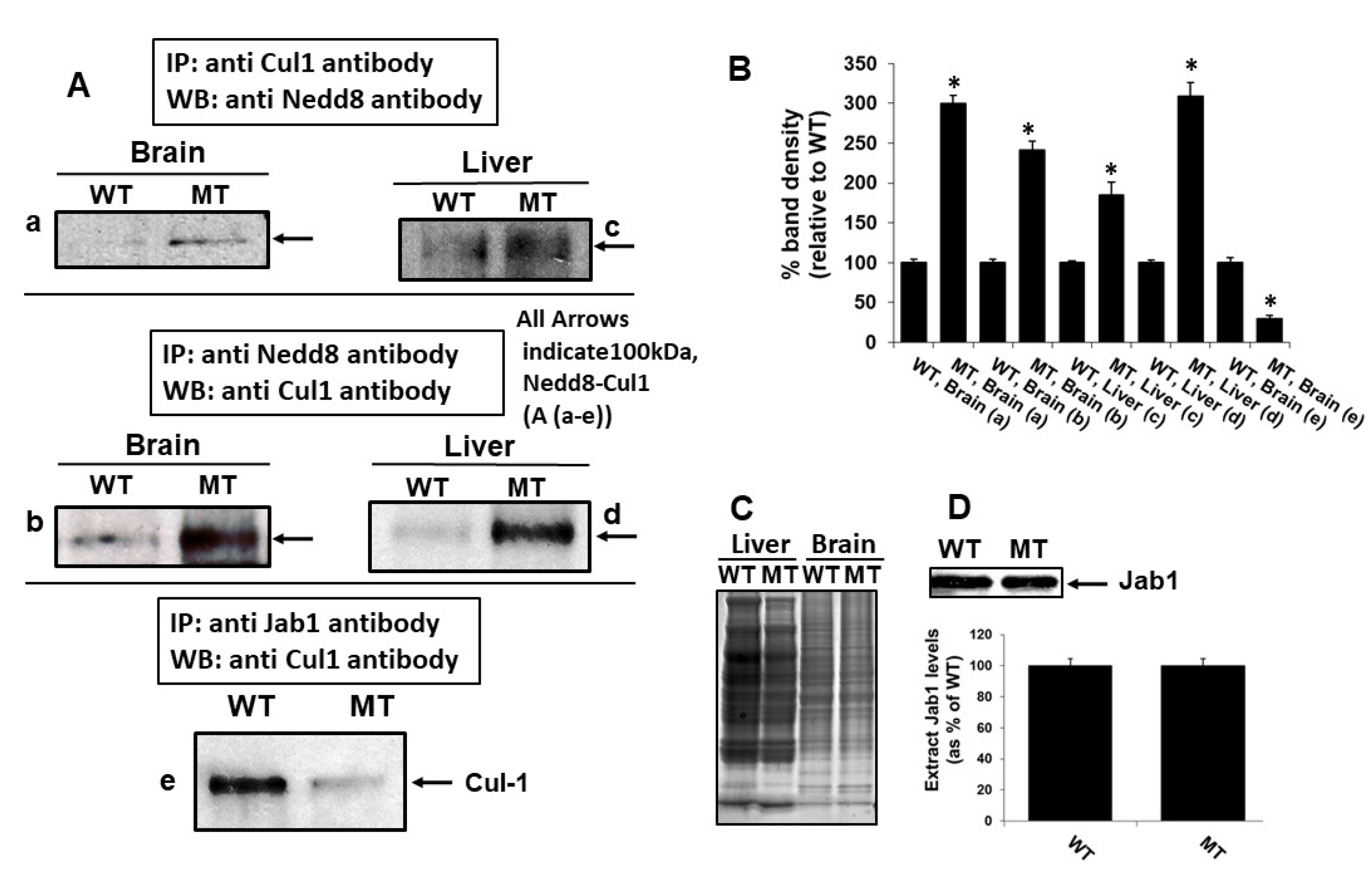
3.5. MsrA-Mediated Deneddylase Activity of Jab1 on Cul-1
3.6. MsrA Impact on Jab1 Binding to Cul-1
3.7. MsrA Impact on Jab1 Deneddylation Activity
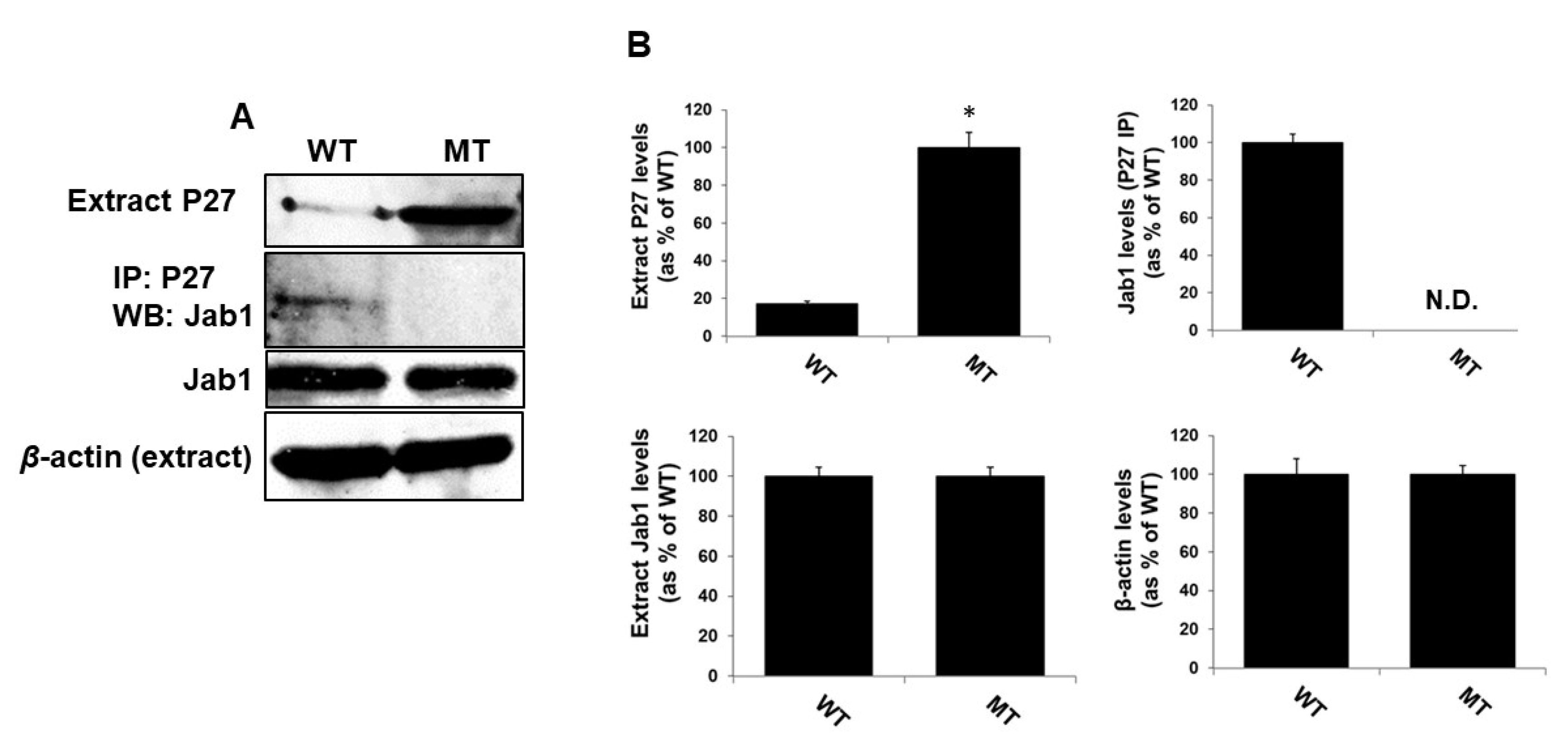
3.8. MsrA-Dependent Jab1 Binding to and Expression of P27
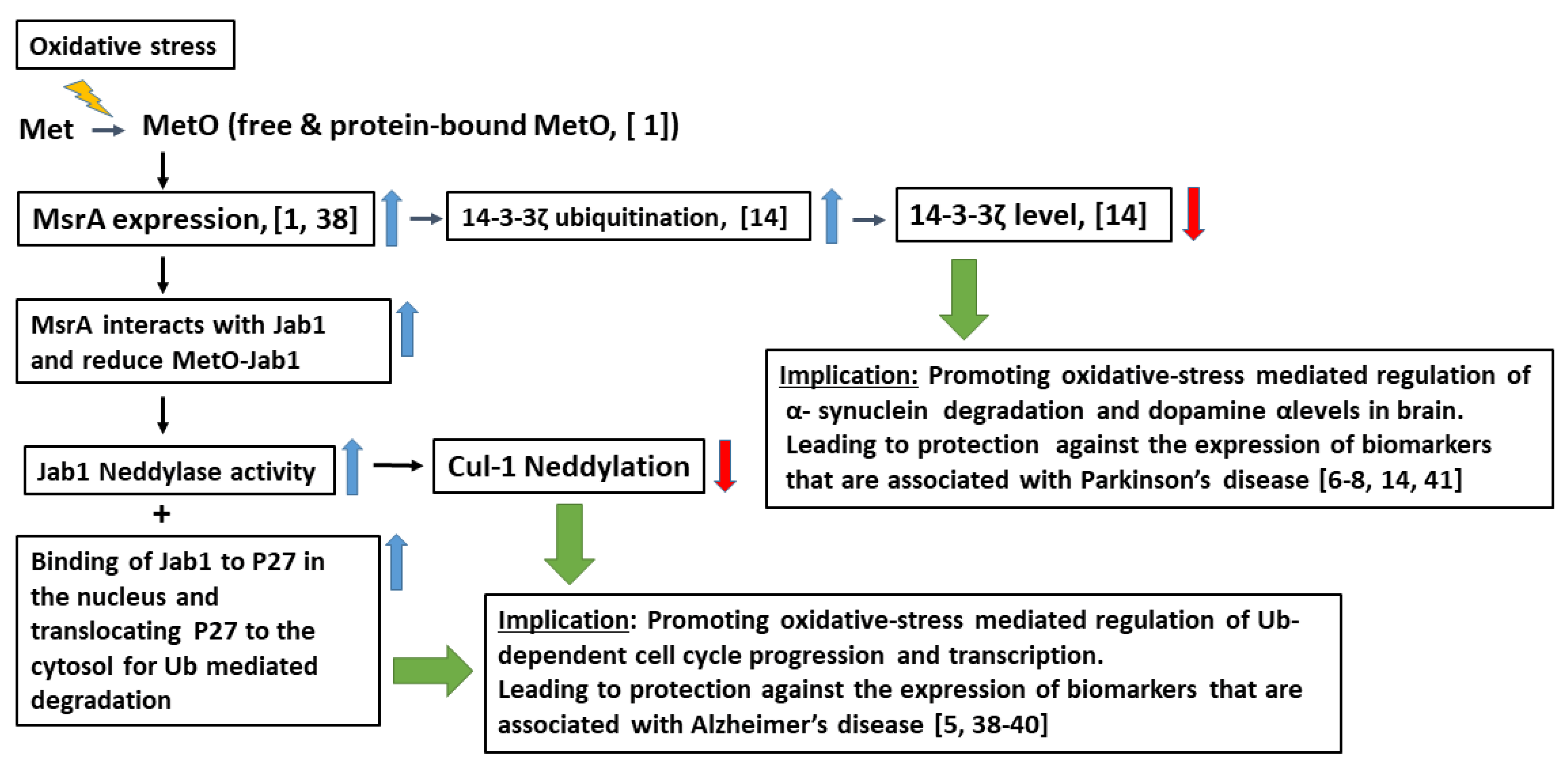
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Oien, D.B.; Moskovitz, J. Substrates of the methionine sulfoxide reductase system and their physiological relevance. Curr. Top. Dev. Biol. 2008, 80, 93–133. [Google Scholar] [PubMed]
- Jiang, B.; Moskovitz, J. The Functions of the Mammalian Methionine Sulfoxide Reductase System and Related Diseases. Antioxidants 2018, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J.; Flescher, E.; Berlett, B.S.; Azare, J.; Poston, J.M.; Stadtman, E.R. Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc. Natl. Acad. Sci. USA 1998, 95, 14071–14075. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J.; Bar-Noy, S.; Williams, W.M.; Requena, J.; Berlett, B.S.; Stadtman, E.R. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA 2001, 98, 12920–12925. [Google Scholar] [CrossRef] [PubMed]
- Pal, R.; Oien, D.B.; Ersen, F.Y.; Moskovitz, J. Elevated levels of brain-pathologies associated with neurodegenerative diseases in the methionine sulfoxide reductase a knockout mouse. Exp. Brain Res. 2007, 180, 765–774. [Google Scholar] [CrossRef]
- Oien, D.B.; Osterhaus, G.L.; Latif, S.A.; Pinkston, J.W.; Fulks, J.; Johnson, M.A.; Fowler, S.C.; Moskovitz, J. MsrA knockout mouse exhibits abnormal behavior and brain dopamine levels. Free Radic. Biol. Med. 2008, 45, 193–200. [Google Scholar] [CrossRef]
- Oien, D.B.; Ortiz, A.N.; Rittel, A.G.; Dobrowsky, R.T.; Johnson, M.A.; Levant, B.; Fowler, S.C.; Moskovitz, J. Dopamine D(2) receptor function is compromised in the brain of the methionine sulfoxide reductase A knockout mouse. J. Neurochem. 2010, 114, 51–61. [Google Scholar] [CrossRef]
- Moskovitz, J.; Walss-Bass, C.; Cruz, D.A.; Thompson, P.M.; Bortolato, M. Methionine sulfoxide reductase regulates brain catechol-O-methyl transferase activity. Int. J. Neuropsychopharmacol. 2014, 17, 1707–1713. [Google Scholar] [CrossRef]
- Moskovitz, J.; Du, F.; Bowman, C.F.; Yan, S.S. Methionine sulfoxide reductase a affects β-amyloid solubility and mitochondrial function in a mouse model of Alzheimer’s disease. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E388–E393. [Google Scholar] [CrossRef]
- Erickson, J.R.; Joiner, M.L.; Guan, X.; Kutschke, W.; Yang, J.; Oddis, C.V.; Bartlett, R.K.; Lowe, J.S.; O’Donnell, S.E.; Aykin-Burns, N.; et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell 2008, 133, 462–474. [Google Scholar] [CrossRef]
- Singh, M.P.; Kim, K.Y.; Kim, H.Y. Methionine sulfoxide reductase a deficiency exacerbates acute liver injury induced by acetaminophen. Biochem. Biophys. Res. Commun. 2017, 484, 189–194. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Sanna, F.; Sallese, M.; Ruggiero, C.; Grossi, M.; Sacchetta, P.; Rossi, C.; De Laurenzi, V.; Di Ilio, C.; Favaloro, B. Methionine sulfoxide reductase A down-regulation in human breast cancer cells results in a more aggressive phenotype. Proc. Natl. Acad. Sci. USA 2010, 107, 18628–18633. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, S.; Chertoff, M.; Durham, D.; Moskovitz, J.; Staecker, H.; Peppi, M. Methionine Sulfoxide Reductase A Knockout Mice Show Progressive Hearing Loss and Sensitivity to Acoustic Trauma. Audiol. Neurotol. 2018, 23, 20–31. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, B.; Rankin, C.L.; Toyo-Oka, K.; Richter, M.L.; Maupin-Furlow, J.A.; Moskovitz, J. Methionine sulfoxide reductase A (MsrA) mediates the ubiquitination of 14-3-3 protein isotypes in brain. Free Radic. Biol. Med. 2018, 129, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Adams, Z.; Liu, R.; Hepowit, N.L.; Wu, Y.; Bowmann, C.F.; Moskovitz, J.; Maupin-Furlow, J.A. Methionine Sulfoxide Reductase A (MsrA) and Its Function in Ubiquitin-Like Protein Modification in Archaea. MBio 2017, 8. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, Y.; Luo, Q.; Li, L.; Jia, L. Neddylation: A novel modulator of the tumor microenvironment. Mol. Cancer 2019, 18, 77. [Google Scholar] [CrossRef]
- del Pozo, J.C.; Dharmasiri, S.; Hellmann, H.; Walker, L.; Gray, W.M.; Estelle, M. AXR1-ECR1–dependent conjugation of RUB1 to the Arabidopsis cullin At CUL1 is required for auxin response. Plant Cell 2002, 14, 421–433. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Giovanna, S.; Deng, X.W. Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 2002, 14, 2553–2563. [Google Scholar] [CrossRef]
- Scott, D.C.; Sviderskiy, V.O.; Monda, J.K.; Lydeard, J.R.; Cho, S.E.; Harper, J.W.; Schulman, B.A. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein Nedd8. Cell 2014, 157, 1671–1684. [Google Scholar] [CrossRef]
- Duda, D.M.; Borg, L.A.; Scott, D.C.; Hunt, H.W.; Hammel, M.; Schulman, B.A. Structural insights into Nedd8 activation of cullin-RING ligases: Conformational control of conjugation. Cell 2008, 134, 995–1006. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A.P. RING domain E3 ubiquitin ligases. Ann. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.I.; Nakayama, K. Ubiquitin ligases: Cell-cycle control and cancer. Nat. Rev. Cancer 2006, 6, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Claret, F.X.; Hibi, M.; Dhut, S.; Toda, T.; Karin, M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 1996, 383, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Lyapina, S.; Cope, G.; Shevchenko, A.; Serino, G.; Tsuge, T.; Zhou, C.; Wolf, D.A.; Wei, N.; Shevchenko, A.; Deshaies, R.J. Promotion of Nedd8-CUL1 conjugate cleavage by COP9 signalosome. Science 2001, 292, 1382–1385. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Serino, G.; Callis, J.; Crosby, W.L.; Lyapina, S.; Deshaies, R.J.; Gray, W.M.; Estelle, M.; Deng, X.W. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 2001, 292, 1379–1382. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.J.; Allen, M.D.; Löwe, J.; Bycroft, M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 2003, 42, 11460–11465. [Google Scholar] [CrossRef]
- Ning, W.; Serino, G.; Deng, X.W. The COP9 signalosome: More than a protease. Trends Biochem. Sci. 2008, 33, 592–600. [Google Scholar]
- Cope, G.A.; Deshaies, R.J. COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 2003, 114, 663–671. [Google Scholar] [CrossRef]
- Cope, G.A.; Suh, G.S.; Aravind, L.; Schwarz, S.E.; Zipursky, S.L.; Koonin, E.V.; Deshaies, R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 2002, 298, 608–611. [Google Scholar] [CrossRef]
- Ning, W.; Deng, X.W. The COP9 signalosome. Ann. Rev. Cell Dev. Biol. 2003, 19, 261–286. [Google Scholar]
- Deshaies, R.J. SCF and Cullin/Ring H2-based ubiquitin ligases. Ann. Rev. Cell Dev. Biol. 1999, 15, 435–467. [Google Scholar] [CrossRef] [PubMed]
- Kouvaraki, M.A.; Korapati, A.L.; Rassidakis, G.Z.; Tian, L.; Zhang, Q.; Chiao, P.; Ho, L.; Evans, D.B.; Claret, F.X. Potential role of jun activation domain–binding protein 1 as a negative regulator of p27kip1 in pancreatic adenocarcinoma. Cancer Res. 2006, 66, 8581–8589. [Google Scholar] [CrossRef] [PubMed]
- Tomoda, K.; Kubota, Y.; Arata, Y.; Mori, S.; Maeda, M.; Tanaka, T.; Yoshida, M.; Yoneda-Kato, N.; Kato, J.Y. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J. Biol. Chem. 2002, 277, 2302–2310. [Google Scholar] [CrossRef]
- Moskovitz, J.; Weissbach, H.; Brot, N. Cloning the expression of a mammalian gene involved in the reduction of methionine sulfoxide residues in proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 2095–2099. [Google Scholar] [CrossRef]
- Oien, D.B.; Canello, T.; Gabizon, R.; Gasset, M.; Lundquist, B.L.; Burns, J.M.; Moskovitz, J. Detection of oxidized methionine in selected proteins, cellular extracts and blood serums by novel anti-methionine sulfoxide antibodies. Arch. Biochem. Biophys. 2009, 485, 35–40. [Google Scholar] [CrossRef]
- Moskovitz, J.; Maiti, P.; Lopes, D.H.; Oien, D.B.; Attar, A.; Liu, T.; Mittal, S.; Hayes, J.; Bitan, G. Induction of methionine-sulfoxide reductases protects neurons from amyloid β-protein insults in vitro and in vivo. Biochemistry 2011, 50, 10687–10697. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Zhu, H.; Ma, C.; Dai, X.; Qin, C. Downregulated microRNA-222 is correlated with increased p27Kip¹ expression in a double transgenic mouse model of Alzheimer’s disease. Mol. Med. Rep. 2015, 12, 7687–7692. [Google Scholar] [CrossRef]
- Ogawa, O.; Lee, H.G.; Zhu, X.; Raina, A.; Harris, P.L.; Castellani, R.J.; Perry, G.; Smith, M.A. Increased p27, an essential component of cell cycle control in Alzheimer’s disease. Aging Cell 2003, 2, 105–110. [Google Scholar] [CrossRef]
- Chen, Y.; Neve, R.L.; Liu, H. Neddylation dysfunction in Alzheimer’s disease. J. Cell. Mol. Med. 2012, 16, 2583–2591. [Google Scholar] [CrossRef]
- Liu, F.; Hindupur, J.; Nguyen, J.L.; Ruf, K.J.; Zhu, J.; Schieler, J.L.; Bonham, C.C.; Wood, K.V.; Davisson, V.J.; Rochet, J.C. Methionine sulfoxide reductase A protects dopaminergic cells from Parkinson’s disease-related insults. Free Biol. Med. 2008, 45, 242–255. [Google Scholar] [CrossRef] [PubMed]
- DeTure, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Arendt, T. Synaptic plasticity and cell cycle activation in neurons are alternative effector pathways: The ‘Dr. Jekyll and Mr. Hyde concept’ of Alzheimer’s disease or the yin and yang of neuroplasticity. Prog. Neurobiol. 2003, 71, 83–248. [Google Scholar] [CrossRef] [PubMed]
- Bowser, R.; Smith, M.A. Cell cycle proteins in Alzheimer’s disease: Plenty of wheels but no cycle. J. Alzheimers Dis. 2002, 4, 249–254. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, Y.; Zhao, Y.; Shu, Y.; Liu, Z.; Zhou, H.; Wang, H.; Zhang, W. The pivotal oncogenic role of Jab1/CSN5 and its therapeutic implications in human cancer. Gene 2019, 687, 219–227. [Google Scholar] [CrossRef]
- Schlierf, A.; Altmann, E.; Quancard, J.; Jefferson, A.B.; Assenberg, R.; Renatus, M.; Jones, M.; Hassiepen, U.; Schaefer, M.; Kiffe, M.; et al. Targeted inhibition of the COP9 signalosome for treatment of cancer. Nat. Commun. 2016, 7, 13166. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.R. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
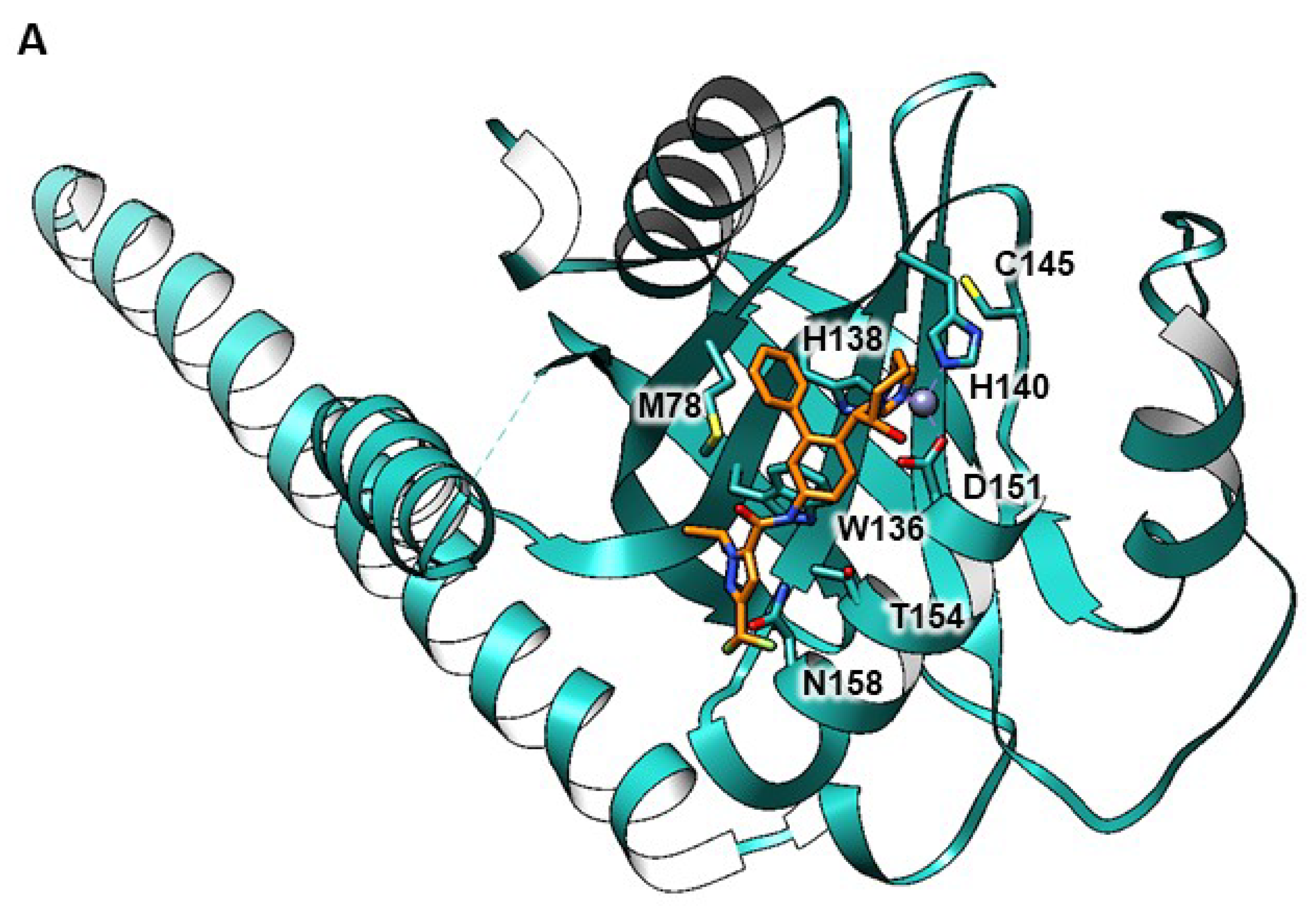
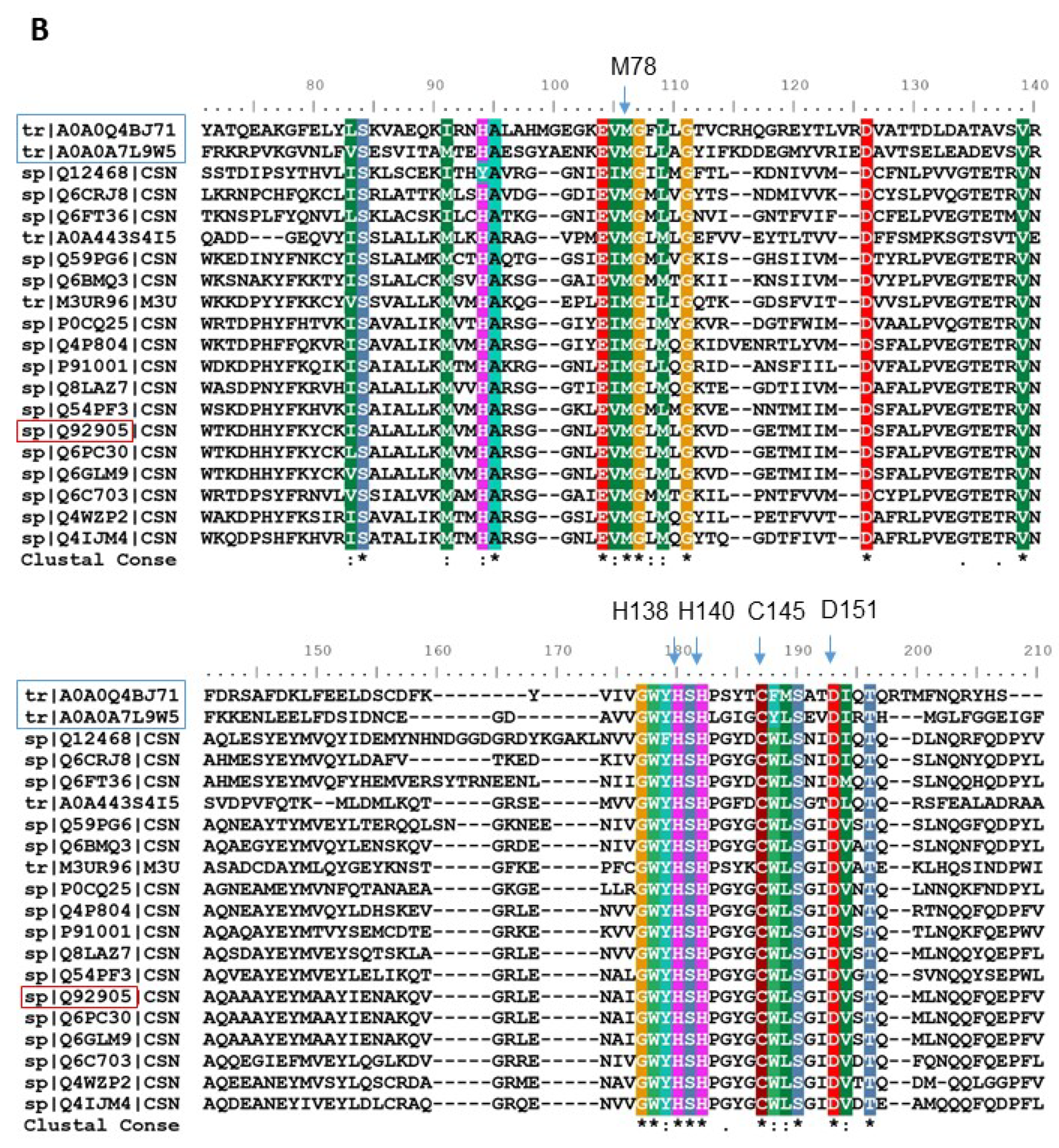
| No. of Hits | GenBank No. | Description [Homo Sapiens] | Met | Cys |
|---|---|---|---|---|
| 19 a | NP006828 | COP9 signalosome complex subunit 5 (Jab1/CSN5) | 3.9% b | 1.2% |
| 6 | NP006826.1 | Cyclin-I (Cyc-1) | 2.9% | 2.9% |
| 6 | NP002991.2 | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial precursor | 3.6% | 5.0% |
| 1 | NP689854.2 | AT-rich interactive domain-containing protein 2 | 1.9% | 1.6% |
| 1 | NP004757.1 | Coatomer subunit β’ | 2.0% | 1.7% |
| 1 | NP055093.2 | Heat shock 70 kDa protein 4L | 2.7% | 2.1% |
| 1 | NP005678.3 | Phenylalanyl-tRNA synthetase β chain | 2.0% | 1.9% |
| 1 | NP001171895.1 | WD repeat-containing protein 44 isoform 3 | 1.7% | 1.0% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, B.; Adams, Z.; Moonah, S.; Shi, H.; Maupin-Furlow, J.; Moskovitz, J. The Antioxidant Enzyme Methionine Sulfoxide Reductase A (MsrA) Interacts with Jab1/CSN5 and Regulates Its Function. Antioxidants 2020, 9, 452. https://doi.org/10.3390/antiox9050452
Jiang B, Adams Z, Moonah S, Shi H, Maupin-Furlow J, Moskovitz J. The Antioxidant Enzyme Methionine Sulfoxide Reductase A (MsrA) Interacts with Jab1/CSN5 and Regulates Its Function. Antioxidants. 2020; 9(5):452. https://doi.org/10.3390/antiox9050452
Chicago/Turabian StyleJiang, Beichen, Zachary Adams, Shannon Moonah, Honglian Shi, Julie Maupin-Furlow, and Jackob Moskovitz. 2020. "The Antioxidant Enzyme Methionine Sulfoxide Reductase A (MsrA) Interacts with Jab1/CSN5 and Regulates Its Function" Antioxidants 9, no. 5: 452. https://doi.org/10.3390/antiox9050452
APA StyleJiang, B., Adams, Z., Moonah, S., Shi, H., Maupin-Furlow, J., & Moskovitz, J. (2020). The Antioxidant Enzyme Methionine Sulfoxide Reductase A (MsrA) Interacts with Jab1/CSN5 and Regulates Its Function. Antioxidants, 9(5), 452. https://doi.org/10.3390/antiox9050452