Abstract
The consensus that assisted reproduction technologies (ART), like in vitro fertilization, to induce oxidative stress (i.e., the known) belies how oocyte/zygote mitochondria—a major presumptive oxidative stressor—produce reactive oxygen species (ROS) with ART being unknown. Unravelling how oocyte/zygote mitochondria produce ROS is important for disambiguating the molecular basis of ART-induced oxidative stress and, therefore, to rationally target it (e.g., using site-specific mitochondria-targeted antioxidants). I review the known mechanisms of ROS production in somatic mitochondria to critique how oocyte/zygote mitochondria may produce ROS (i.e., the unknown). Several plausible site- and mode-defined mitochondrial ROS production mechanisms in ART are proposed. For example, complex I catalyzed reverse electron transfer-mediated ROS production is conceivable when oocytes are initially extracted due to at least a 10% increase in molecular dioxygen exposure (i.e., the intriguing). To address the term oxidative stress being used without recourse to the underlying chemistry, I use the species-specific spectrum of biologically feasible reactions to define plausible oxidative stress mechanisms in ART. Intriguingly, mitochondrial ROS-derived redox signals could regulate embryonic development (i.e., their production could be beneficial). Their potential beneficial role raises the clinical challenge of attenuating oxidative damage while simultaneously preserving redox signaling. This discourse sets the stage to unravel how mitochondria produce ROS in ART, and their biological roles from oxidative damage to redox signaling.
1. Mitochondrial ROS, Oxidative Stress, and Assisted Reproduction: An Introduction
From a historical perspective, the ability of mitochondria to produce superoxide—a key reactive oxygen species (ROS, see Box 1)—has been known since 1966 [1]. Two years later, Auerbach and Brinster [2] found that: exposing mouse zygotes to atmospheric ground-state molecular dioxygen (O2) levels (i.e., 21% O2) underlies the so-called “2-cell block” to embryo culture, wherein zygotes fail to progress to the 4-cell stage or exhibit severely delayed development. Importantly, lowering [O2] by 16% from 21 to 5%, a plausible in utero value, overcome the 2-cell block. A decade later, their findings would have important repercussions for assisted reproduction technologies (ART), like in vitro fertilization (IVF), when the first “test-tube” baby was born in 1978 [2]. In the subsequent ~40 years: (1) ART has become invaluable for treating infertility, which currently affects 15% of couples worldwide [3]; (2) Helmut Sies introduced the term oxidative stress [4] and the biochemistry of key species like superoxide is now well-understood [5]; (3) we now understand much of the mechanistic basis of oxidative phosphorylation (OXPHOS) and mitochondrial superoxide production [6] (e.g., beyond complex I and III, we now know that over 10 enzymes can produce ROS [7]); and (4) a nuanced view of the biological role of ROS has emerged (e.g., as opposed to being purely damaging agents, ROS are now considered to play beneficial biological roles [8]). In 2020, it is, therefore, surprising that little can be stated with any great confidence about how mitochondria produce ROS in ART. The “known” extends little further than: (1) oocyte/zygote mitochondria produce ROS [9,10,11]; (2) oocyte/zygote mitochondria derived from older females tend to produce more ROS [12,13,14]; and (3), in an ageing setting at least, certain mitochondria-targeted antioxidants seem protective [14,15]. Moreover, the wider chemical biology of ART-induced oxidative stress is unclear. Taking the 2-cell block as an example, mechanistic understanding is fragmentary, because chemically-defined source-target relationships are lacking. In the clinic, insufficient mechanistic understanding means that how culturing embryos at 5% O2 improves live birth rates by ~13% is unclear [16]. Unravelling the underlying mechanisms holds promise for increasing live birth rates by rationally targeting oxidative stress.
Against this backdrop, I critique how somatic mitochondria produce ROS (“the known”; Section 2) as a resource to address “the unknown” mechanisms of mitochondrial ROS production in ART (Section 3). Extending the knowns (e.g., the sites and modes will evolve with time) and making plausible experimentally testable hypotheses about the unknowns (e.g., novel oocyte extraction-induced ROS production mechanisms) advances current understanding. Finally, I consider “the intriguing” potential functional roles of mitochondrial ROS production in ART (Section 4). To do so, I present a chemically-defined framework to explain how mitochondrial ROS may cause oxidative stress. Importantly, the paradigm shifting ability of mitochondrial ROS-derived redox signals to regulate development is considered (i.e., their production could be beneficial [17]). Before proceeding, the present discourse focuses on mitochondria, because of their strategic importance [18], complex redox biology (i.e., there are over 10 differentially regulated sites of ROS production in mitochondria), and the mechanisms for how cytosolic ROS sources, like NADPH oxidase enzymes or xanthine oxidase, for example, produce ROS are well understood [19]. Section 4 remains relevant to any reader interested in understanding how cytosolic ROS cause oxidative stress. For the purposes of brevity, the present review is delimited to mammals, complex I-III, and oocyte/zygote mitochondria (sperm are considered elsewhere [20]).
Box 1. Oxygen, ROS and Oxidative stress: A primer.
Oxygen: Ground state molecular dioxygen (O2) is a free radical—a molecule capable of independent existence with at least one unpaired electron—because it contains two unpaired electrons with parallel spins [8]. Parallel spins (i.e., ↑↑) make aerobic life possible by spin-restricting the ability of O2 to react appreciably with most spin paired (i.e., ↑↓) biomolecules [21,22,23]. The importance of spin restriction is underlined by the ability of singlet oxygen (∆1O2, an electronically excited species) to oxidize several biomolecules, because the excitation energy spin pairs (i.e., ↑↓) the two electrons [24]. ∆1O2 is an excellent example of a non-radical species with greater chemical reactivity than the parent radical. Spin restriction limits O2 to single electron transfers [8]. Single electron transfers are essential in mitochondria, wherein cytochrome c oxidase (Complex IV) splits O2 to atomic oxygen, before reducing it to form water (H2O) without releasing catalytic radical intermediates [25].
ROS: The umbrella term reactive oxygen species (ROS) encompasses the free radicals superoxide anion (O2●−) and hydroxyl radical (•OH), as well as the non-radical hydrogen peroxide (H2O2). Their interrelationship is summarized below:
The chemistry and metabolism of each species are discussed elsewhere [8,26,27,28]. Relevant points are threefold. First, their chemical reactivity differs by orders of magnitude. For example, OH oxidizes DNA a billion times faster than O2●− or H2O2. Second, the biochemistry of O2●− and H2O2 is selective—they react rapidly with a few strategically important biomolecules—whereas OH is chemically promiscuous. Third, their intracellular concentrations seldom rise above nanomolar (i.e., 10−9 M) levels due to efficient metabolism or diffusion-controlled reactivity in the case of ●OH.
O2 → O2●− → H2O2→ ●OH ↔ H2O
Oxidative stress: Helmut Sies first coined the term oxidative stress in 1985 [4]; his subsequent work defines oxidative stress as increased oxidative damage and/or disrupted redox signaling [29,30,31]. Redox signaling refers to the ability of ROS to transduce intracellular signals, which is a central tenet of the redox code proposed by Sies and Jones [32]. Oxidative stress is a frequently used but seldom chemically qualified term [33]. That is, oxidative stress confers no useful mechanistic information unless the chemical details are disclosed [29,34]. While unravelling the underlying chemistry is a challenging task [35], it is required to rationally target oxidative stress. For example, insufficient mechanistic understanding played a key part in the failure of vitamin E and C to treat disease—they seem to have little ability to react with relevant species at the relevant time and place [8,36].
2. Mechanisms of Mitochondrial ROS Production: The Known
2.1. The Major and Minor Mitochondiral Electron Pathways
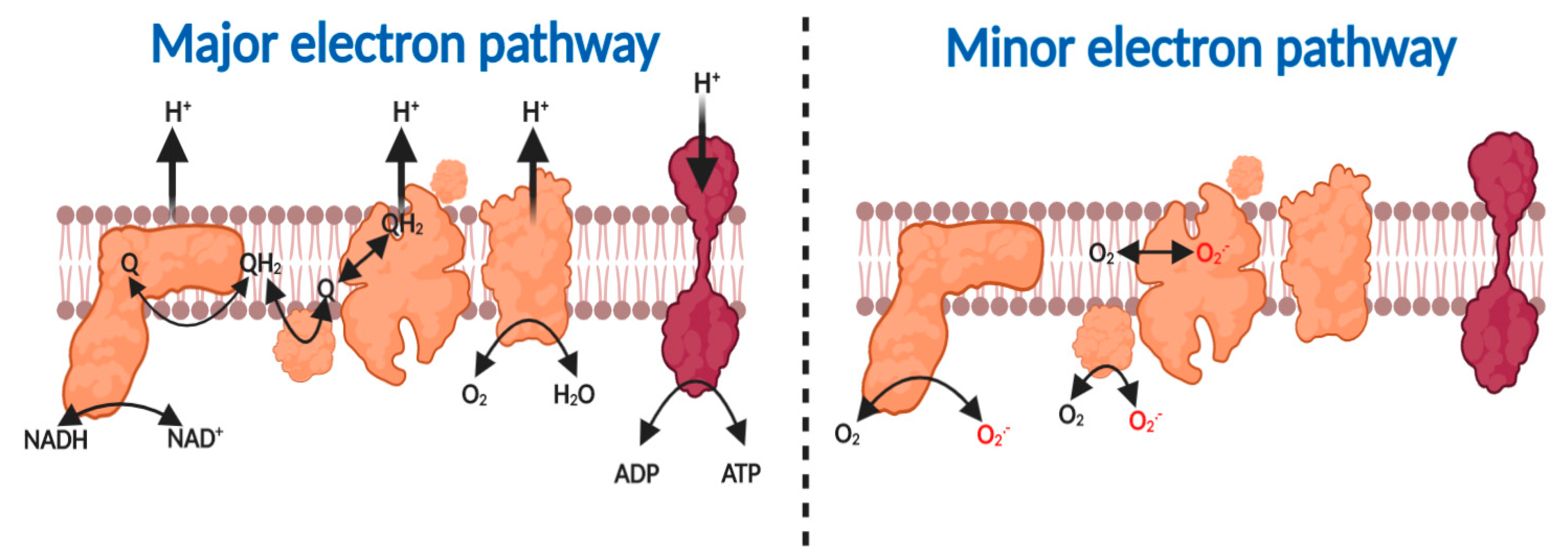
The major fate of substrate-derived electrons—free radicals—tunneling (a quantum mechanical property that enables electrons to penetrate a potential energy barrier without further input energy), according to the principles of quantum mechanics, through the redox centers nestled within the mitochondrial respiratory complexes is to reduce O2—a diradical—to H2O (reaction 1, see Figure 1) [21,25,37,38,39,40]. The free energy associated with the thermodynamically favorable electron transfer from NADH (Eo’ = −340 mV) or FADH (Eo’ = +31 mV) via ubiquinol (Eo’ = +45 mV) to O2 (Eo’ = +840 mV) is used by complex I, III and IV to pump protons, to create an electrochemical proton motive force (Δp), comprising a membrane potential (ΔΨm~−150–200 mV) and pH component (ΔpH = ~0.8), across the inner mitochondrial membrane [6,41,42,43]. Δp enables OXPHOS by forcing the F1-Fo ATP synthase (i.e., complex V) to synthesize, as opposed to hydrolyze, ATP (reaction 2) [44,45,46]. A minor fate of substrate-derived electrons in the respiratory chain is to support the univalent reduction of O2 to superoxide at complex I, II and III (reaction 3, see Box 1 and Figure 1) [47].
reaction 1: O2 + 8H+ +4 e –> 2H2O + Δp (4H+ pumped)
reaction 2: ADP + Pi + Δp <–> ATP
reaction 3: e + O2 –> O2− (superoxide)
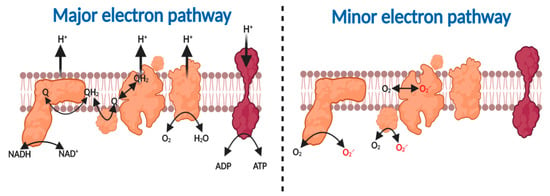
Figure 1.
Major and minor electron pathways in the mitochondrial respiratory chain. Major pathway (left to right). Complex I oxidizes NADH to NAD+ to reduce ubiquinone (Q) to ubiquinol (QH2). Complex II oxidizes succinate to fumarate to produce QH2. Complex III oxidizes QH2 to reduce cytochrome c. Reduced cytochrome c is then oxidized by complex IV to reduce O2 to H2O. The proton motive force generated by complex I, III and IV is harnessed by Complex V to synthesize ATP. Minor pathway left to right. Substrate-derived electrons can reduce O2 to O2●− at complex I, II and III.
2.2. How Mitochondria Produce Superoxide
The interested reader is referred to Murphy’s [47] classic account for a comprehensive overview of the field. At first glance, the thermodynamics of reaction 3 (E = −160 mV at pH 7) would appear to restrict superoxide production to highly reducing electron donors. When the Nernst equation is used to compute Eo’ at plausible O2 (~3–30 µM) and superoxide (~100–200 pM), levels values of 150–230 mV are obtained [47]. Many thermodynamically competent NADH (Eo’ = −340 mV), FADH (Eo’ = +31 mV) or ubiquinol (Eo’ = +45 mV) linked enzymes could, therefore, catalyze the univalent reduction of O2 to superoxide, provided a kinetic mechanism exists. Superoxide is typically produced via an outer sphere electron tunneling mechanism from the donor to acceptor [47,48,49]. The pKa of superoxide (4.8 [5]) means the bulk (~99%) exists as an anion (O2●−) as opposed to the hydroperoxyl radical (HO2●). In the mitochondrial matrix, the ratio of HO2● to O2●− is 1:1000 at pH 7.8. The reorganization energy for O2 to accept a single electron is simplified by the lack of proton transfer. Instead, outer sphere electron tunneling distance is key [37] (i.e., the rate decreases as the distance between donor and acceptor increases—explaining why ensconcing labile redox active iron-sulfur clusters deep within enzymes protects against facile superoxide production [50]). Mitochondrial superoxide production, for a given site, is set by the amount of the reduced enzyme in an O2 accessible form (ERED), the amount of O2, and the kinetics (k) of their second order bimolecular reaction [42,47,51]. The rate of total superoxide production over a set time interval can be calculated using Equation (1):
where k is the weighted mean of the second order biomolecular reaction of all mitochondrial superoxide with O2 and [ERED] is the sum of their redox state.
[O2−]/t = [O2] ∑(k [ERED])
Despite Chance’s group clearly stating that 1–2% of O2 produces superoxide in isolated mitochondria under defined conditions [52,53], their finding has often erroneously been taken to mean that a fixed immutable percentage of O2 uptake supports superoxide production. The amount of mitochondrial [O2] that gives rise to superoxide varies over time according to the prevailing conditions. That is, no invariant immutable percentage exists [54]. Much of the superoxide produced by mitochondria is rapidly converted to H2O2 by manganese superoxide dismutase (MnSOD) [55,56,57]. Since superoxide, H2O2, and several other species (e.g., OH) co-exist in mitochondria (and in biological systems per se), the term Reactive Oxygen Species (ROS, see Box 1) is used. As Sies and Jones [58], as well as Halliwell and Gutteridge [8] remark, ROS is an umbrella term—no molecule called ROS actually exists! It is most rewarding to keep this in mind, when appraising oxidative stress mechanisms. Before considering ART, I define the key sites and proposed operational modes of mitochondrial superoxide production.
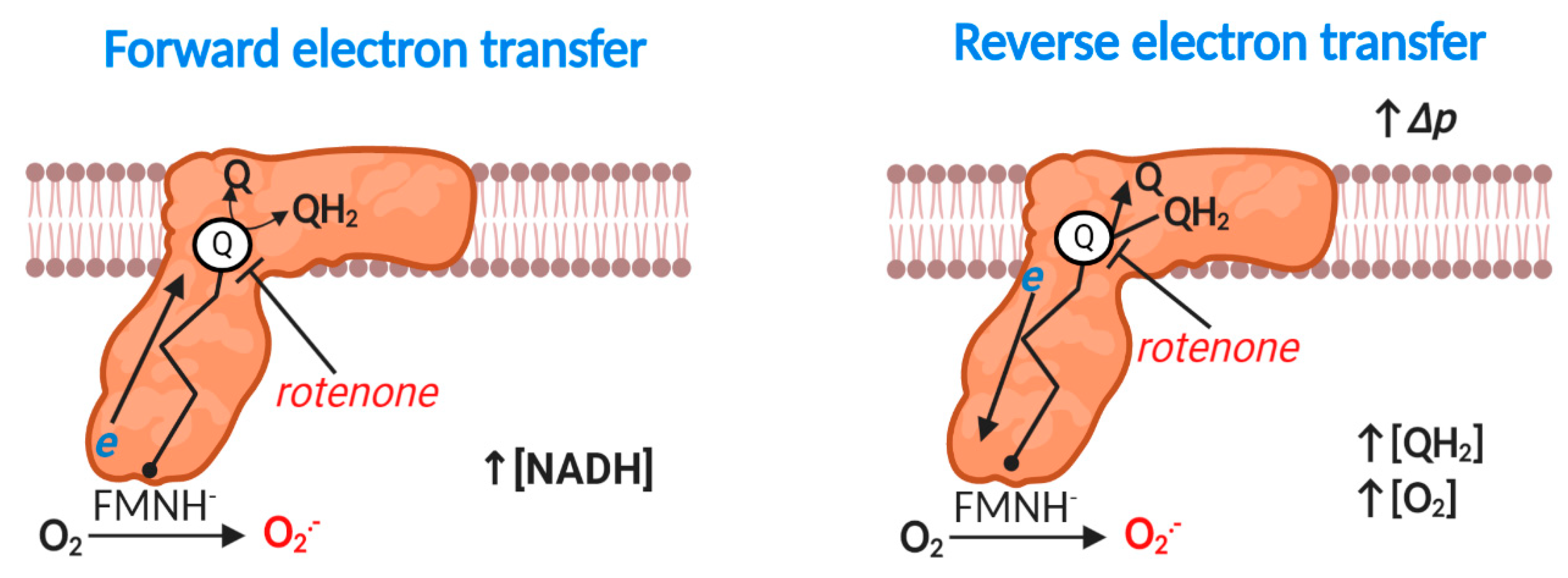
2.3. Complex I: Forward Mode
Eukaryotic mitochondrial complex I (NADH: ubiquinone oxidoreductase) is a 1 megadalton, multi-subunit (14 core and 31 accessory), transmembrane enzyme responsible for coupling NADH oxidation to ubiquinone reduction and vectoral proton transfer by an unresolved spatially delocalized mechanism [59,60,61,62,63,64] (reaction 4). Hirst’s group [65,66] have established that: a partially reduced prosthetic flavin mononucleotide species (FMNH-) reacts with O2 to produce superoxide at complex I (reaction 5). Bound NAD+/NADH can, therefore, sterically occlude FMN-mediated superoxide production [65] (i.e., increasing the distance between donor and acceptor). Upstream bi and tetranuclear iron sulfur clusters can also control superoxide production by limiting FMNH- (i.e., tERED) lifetime (i.e., tERED). An additional (i.e., bypassed in normal electron tunneling from FMN to the Q binding site) binuclear iron-sulfur cluster termed N1a may also modify superoxide production, potentially by sequestering electrons to decrease [FMNH-] or via a peptide bond gated switch [67,68]. When electron transfer stalls as occurs in the rotenone (a Q binding site inhibitor) inhibited complex, considerable superoxide production can occur (Figure 2). However, much superoxide can also emanate from tricarboxylic acid cycle (TCA) dehydrogenases at high NADH/NAD+ ratios (see Table 1), depending on the substrate supply [69].
reaction 4: NAD+ + Q + H+ (matrix) <–>NADH + QH2 + Δp
reaction 5: FMNH− + O2 –>FMNH + O2●−
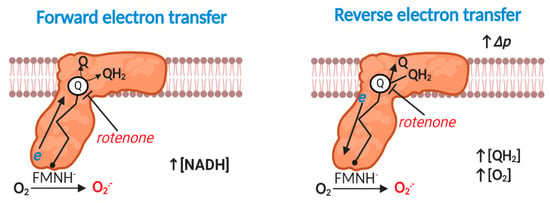
Figure 2.
Superoxide production at mitochondrial complex I. Forward mode. At high NADH levels or when electron transfer between flavin mononucleotide radical (FMN) via seven iron sulfur (Fe-S) clusters to the Q reduction site stalls (e.g., rotenone inducible), reduced FMN transfers a single electron to O2 to produce O2●−. Reverse mode. When Δp is near maximal and the Q pool is highly reduced, complex I produces substantial O2●− by reverse electron transfer (RET). Rotenone blocks QH2 oxidation to inhibit RET-mediated O2●− production. The exact site of O2●− production is unclear (see text); the figure shows FMN catalyzed O2●− production for the purposes of clarity. A flavin mononucleotide radical (FMN●) may also contribute. RET-mediated O2●− production linearly depends on [O2].

Table 1.
Additional sites of superoxide production in mitochondria by enzyme, substrate, iso-potential group, source and topology. Commentary is provided as appropriate.
2.4. Complex I: Reverse Electron Transfer
Energetically degenerate catalytic steps [62] render reaction 4 fully reversible, provided a sufficient thermodynamic driving force exists, which occurs when the free energy released from electron transfer is insufficient to pump protons against the prevailing Δp [70] (Figure 2). Reverse electron transfer (RET) catalyzed superoxide production was discovered by Chance and co-workers in 1967 [71], and was considered irrelevant, until Murphy’s group showed that RET contributed to cardiac ischemia reperfusion injury (IRI) in 2013–2014 [72,73]. We now know that RET plays several important physiological (e.g., sleep, lifespan and O2 sensing) and pathological (e.g., in organ transplantation) roles [74,75,76,77,78,79,80]. Robb and colleagues [81] have identified the factors that govern RET: a highly reduced Q pool and near maximal Δp. A near maximal Δp necessitates low ATP synthesis (and limited activity of other Δp consumers; e.g., the transhydrogenase or the adenine nucleotide transporter [6]). RET-mediated superoxide production responds linearly to [O2] in isolated mitochondria [81]. The site of RET-mediated superoxide production is disputed: some favor FMN on structural (O2 may be unable to access bound Q) and dielectric (superoxide anion is unlikely to migrate to the negatively charged matrix) grounds, and others claim that O2 could access a prosthetic semiquinone radical (SQ●−) during dynamic catalysis (structures are static), or it could disengage [66,69,70]. Regardless of the exact site (s), RET is occluded by compounds able to bind the Q site and/or dissipate Δp [70].
2.5. Complex II
The uniquely entirely nuclear encoded and non-proton pumping complex II (i.e., succinate dehydrogenase) catalyzes succinate/fumarate and ubiquinone/ubiquinol oxidoreduction (i.e., succinate +UQ <–> fumarate + QH2). For many years, complex II was thought to only produce superoxide when it was damaged or mutated [82]. In 2012, Brand’s group [83] discovered that complex II can produce superoxide via its prosthetic flavin adenine nucleotide (FAD, reaction 6) moiety, in the absence of overt damage, provided key criteria are met. A flavin radical may also contribute (i.e., FAD● + O2 –> O2●−). Specifically, succinate (forward) or ubiquinol (reverse) is required to reduce FAD, and O2 must be able to access FADH [69]. The redox state of the Q pool and O2 availability are, therefore, important determinants of complex II-mediated superoxide production. Univalent electron transfer is blocked by FAD bound dicarboxylic acids (i.e., inhibited at high (succinate)) [84]. Analogous to certain fumarate dehydrogenases [85], the redox state of the upstream iron-sulfur clusters may favor direct H2O2 production, potentially via -OOH release. Mathematical modelling suggests that, in the absence of respiratory inhibitors (e.g., aptenin A5 which inhibits the Q binding site of complex II), that an iron–sulfur cluster may produce superoxide (i.e., [3Fe-4S] + O2 –> [3Fe-4S]− + O2●−) [86]. The 3Fe-4S cluster may, therefore, be a physiologically important source of complex II-derived superoxide [87]. Depending on the redox state of the complex, the 3-Fe-4S cluster and the flavin may operate to produce superoxide in parallel.
reaction 6: FADH− + O2 –> FAD + O2●−
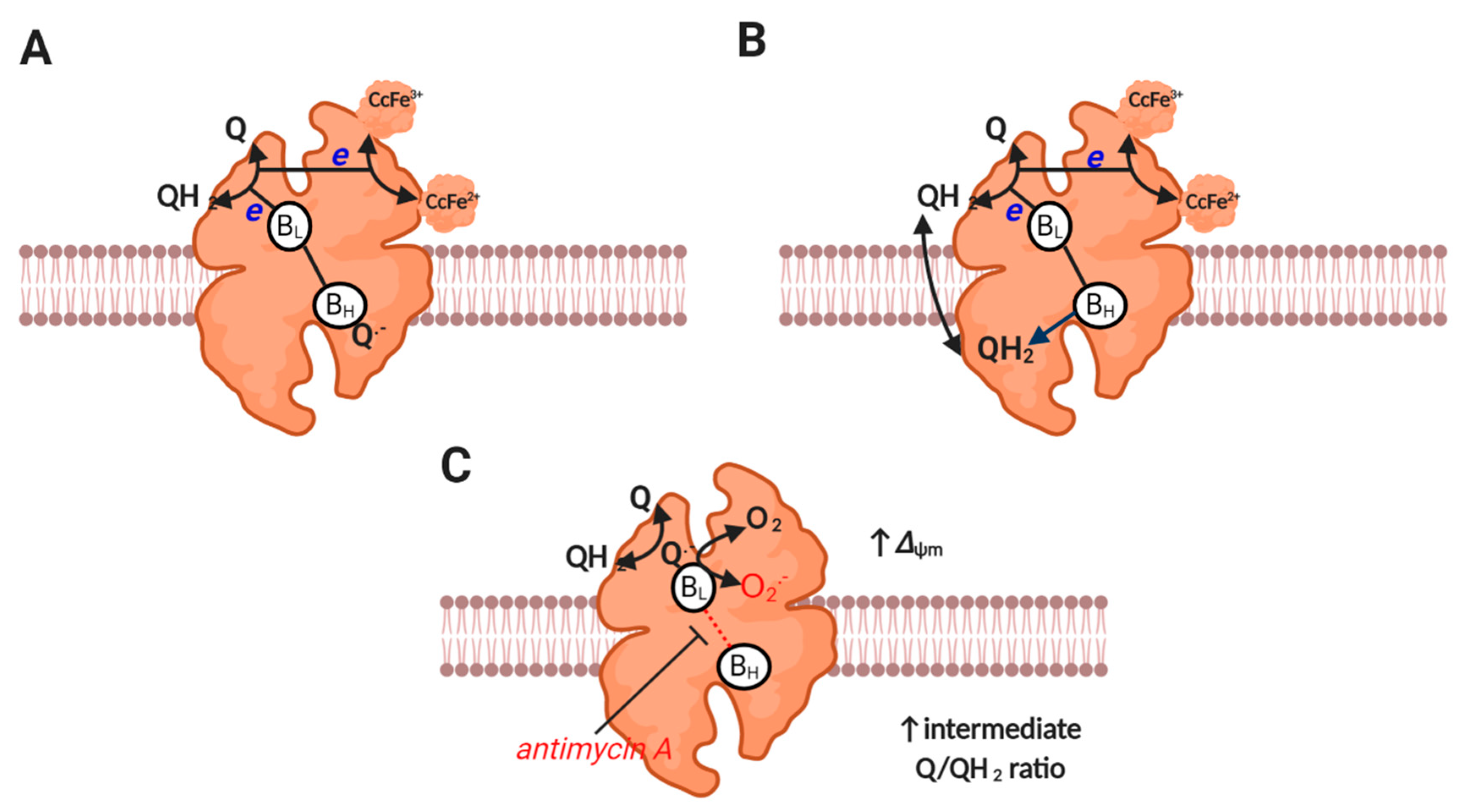
2.6. Complex III
Complex III (i.e., ubiquinol: cytochrome c oxidoreductase) is responsible for catalyzing Mitchell’s classic proton motive Q cycle [88], wherein an electron bifurcation pathway couples ubiquinol oxidation to cytochrome c reduction and proton pumping [6] (reaction 7, see Figure 3). One ubiquinol-derived electron is transferred to cytochrome c via the Reiske iron-sulfur protein and cytochrome c1, while the other electron is transferred via heme BL on the P (i.e., intermembrane space), to heme BH on the N (i.e., matrix) side [89]. Heme BH transfers an electron to a bound ubiquinone species at the Qi site to produce a stable (i.e., non-superoxide producing) SQ●− intermediate that is reduced to ubiquinol when electron bifurcation is repeated. While the pioneering work of Jensen, Cadenas and others established—many decades ago—that complex III can produce superoxide [1,90,91], the actual site and mechanism is still debated [92]. Debate concerns whether a prosthetic SQ●− formed near BL (termed the Qo site) is formed in forward mode (i.e., as a catalytic intermediate, reaction 8), or in reverse mode (i.e., a back reaction between heme BL and ubiquinone, reaction 9) [92]. Much will depend on whether the electron bifurcation pathway is sequential (permits forward or reverse) or concerted (prohibits forward). Whether HO2● is produced is unclear [93,94,95], but has implications for direct diffusion, given its uncharged nature from the P side to the matrix (i.e., site topology [96]). Superoxide directly released to the intermembrane space could diffuse to the matrix secondary to H2O2 production (mediated by HO2● or CuZnSOD), and may be favored by deeply folded cristae [92]. Obligate complex III dimers may also reduce superoxide production by permitting electron transfer between monomers [97]. Far from being trivialities, the mechanisms are important for discerning the factors that control superoxide production (e.g., ubiquinone would promote superoxide production in reverse mode when Heme BL is reduced) [98] and regulate function (e.g., HO2● within the inner membrane could abstract a proton from a methylene group to initiate lipid peroxidation by producing a chain propagating peroxyl radical [99,100]). A better understanding of enzyme catalysis (e.g., sequential or concerted) is required to dissect how complex III produces superoxide.
reaction 7: Ubiquinol + 2 cytochrome c Fe(III) + 2 H+ (matrix) –> ubiquinone + ubiquinol + 2 cytochrome c Fe2+ + 4 H+ (P side, Δp)
reaction 8: SQ.− + O2 –> Q + O2●−
reaction 9: Heme BL (red) + Q –> Heme BL (ox) + SQ –> SQ.− + O2 –> Q + O2●−
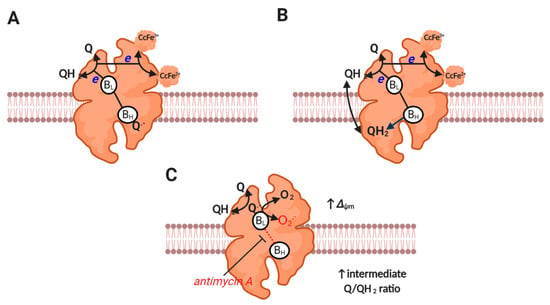
Figure 3.
Complex III catalysis and superoxide production. (A) Depicts the first step of the proton motive Q cycle. UQH2-derived electrons bifurcate to BL and Rieske protein to reduce a bound Q to Q− at BH and cytochrome c at cytochrome c1 reductase (both subunits are omitted for clarity). (B) Depicts the second step of the proton motive Q cycle wherein repeating step A results in the reduction of the bound Q to QH2 at BH. Release of QH2-derived protons on the P side of the membrane generates Δp. Electrons tunneling between HL and BH do so against the prevailing Δp (it is electrogenic). (C) Depicts superoxide production. A moderate to high ΔΨm slows electron transfer between BL and BH. Reduced BL reduces Q to Q−, which then reacts with O2 to produce superoxide (semi-reverse mechanism). Equally, a semi-forward mechanism may operate. Superoxide production seems to be favored by an intermediate Q/QH2 ratio (see main text).
Antimycin A induces complex III-mediated superoxide production by binding to the Qi site to prohibit electron transfer between the heme BL and BH [101], which favors superoxide production by increasing tERED. Inhibiting complex IV (e.g., with cyanide) can suppress complex III-mediated superoxide production by reducing cytochrome c to levels incompatible with catalysis (i.e., little to no oxidized cytochrome c available for reaction 7) [92,102,103]. Under physiological conditions, ΔΨm can sufficiently slow electron transfer between bL and bH, to induce superoxide production at complex III [92] (see Figure 3). Mathematical modelling reveals that complex III-mediated superoxide production is increased by a high ΔΨm (rising 4-fold as it increases from 150 to 200 mV), and is favored by a partially reduced Q pool [104]. Modelling also suggests that superoxide production is negligible at high ubiquinol levels, potentially due to restricting the availability of ubiquinone for a semi-reverse mechanism and BH binding (i.e., normal catalysis). The redox state of the Q pool and ΔΨm set physiological superoxide production at complex III [92].
2.7. Key Superoxide Production Modes
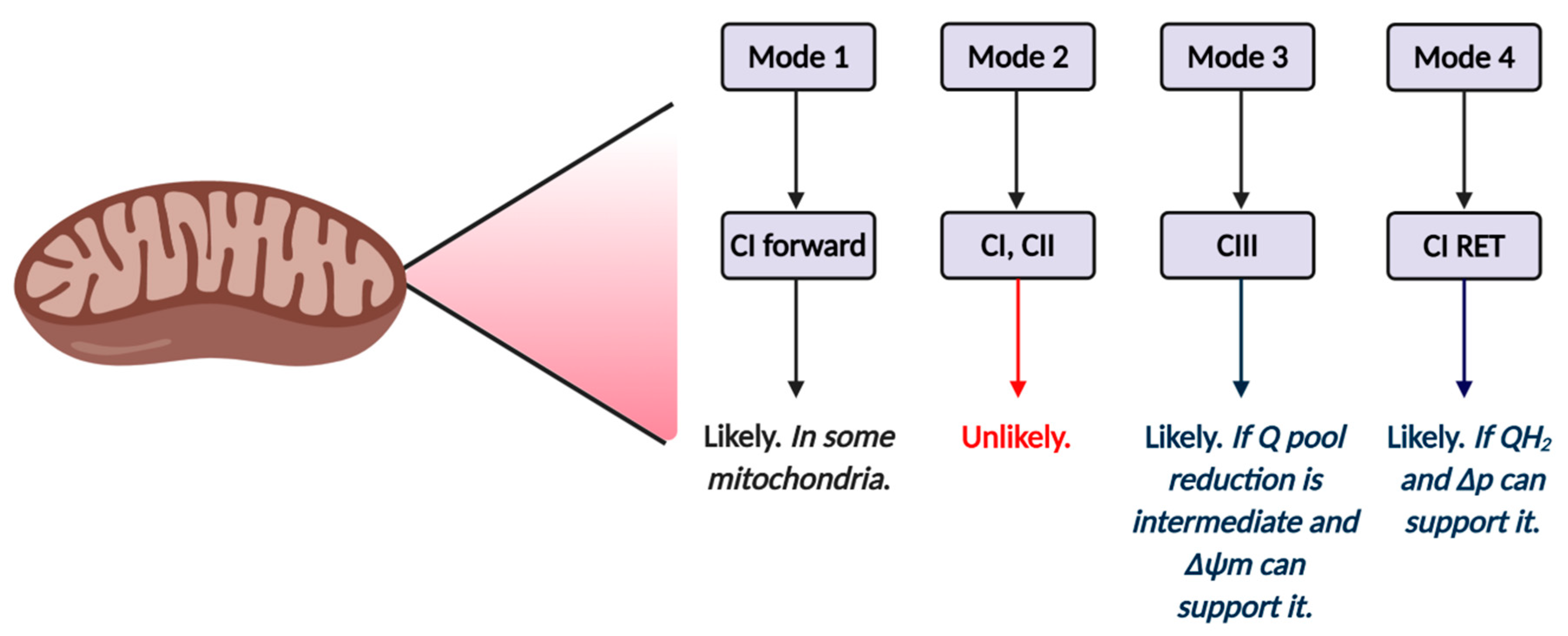
Four key modes of mitochondrial superoxide production exist (see Table 2). To qualify the terms high and low, a parameter (e.g., respiration) can dynamically occupy a granular modifiable (e.g., complex IV content can be increased) spectrum of allowed values between extremes. What constitutes a high and low value varies. Further, the relevant thresholds are insufficiently understood. For example, the threshold of low respiration required to induce superoxide cannot be stated absolutely, because it depends on several factors (e.g., substrate (s) oxidized). Assuming it could, [superoxide] produced would vary, potentially considerably (e.g., in mutated mitochondria), and would evolve over time (e.g., with substrate supply and ATP demand). Mode 1 is defined by comparatively low superoxide production, because OXPHOS and respiration are high due to ATP demand. Mode 1 occurs during intense skeletal muscle and neural activity [33,47,69,109,110]. Shorter electron residencies decrease tERED and superoxide production probability, even when O2 uptake is increased (i.e., less time is available for the reaction to occur). While the overall tendency is for decreased net superoxide production, some sites, particularly complex I in forward mode, can still produce superoxide at an appreciable rate, due to continued NADH availability [109].

Table 2.
Summary of the main modes of mitochondrial ROS production. The dominate electron transport chain site is listed for each mode. The site may vary according the prevailing conditions. In practice, when one considers Table 1, other sites will also operate depending on the substrates being oxidized (e.g., pyruvate dehydrogenase (PDH) in mode 2). X denotes that a given variable has no known appreciable role in governing the mode.
In mode 2, high [NADH] can drive superoxide production at complex I via forward electron transfer and TCA dehydrogenases (depending on substrate availability), provided that OXPHOS/respiration are comparatively low [47,111,112]. When Δp is low, a minimal role for RET and complex III would be expected. Mode 2 is sensitive to uncoupling (e.g., via uncoupling proteins [113,114,115]), because it would increase electron transfer to complex IV. Uncoupling decreases electron residency times (e.g., FMNH lifetime) to reduce the probability of superoxide production. A role for complex II could be envisaged if the reduced FAD or 3Fe-4s cluster were accessible to O2 [83]. In mode 3, the prevailing conditions are similar, except that a moderate to high ΔΨm drives superoxide production from complex III when the ubiquinol pool is low to moderately reduced and OXPHOS/respiration is low [92]. Mode 3 could occur alongside mode 2 when the NAD+/NADH pool is reduced and is sensitive to uncoupling. In mode 4, the Q pool is highly reduced, Δp is near maximal, and superoxide production occurs via complex I catalyzed RET, provided OXPHOS/respiration are low [47,81].
The exact sites that operate in each mode are dependent on contextual factors, including: substrate availability, enzyme content, and several regulatory factors. For example, high ATP, as well as reversible thiol oxidation, would constrain alpha keto glutarate dehydrogenase-mediated superoxide production in mode 2 to promote complex I-mediated superoxide production [69]. In mode 4, RET would be inhibited if complex I is locked in a structural inactive state termed the D-state [72,116,117,118,119]. Different modes and sites (even within the same mode) can operate in parallel.
3. How Mitochondria Produce Superoxide in ART: The Unknown
3.1. General Considerations
Given that oocytes contain ~100,000 mitochondria and their biogenesis is repressed until the 4–8 cell stage [120], the total amount of enzyme is fixed (at least for the respiratory chain, as a turnover of TCA enzymes may be possible) at a high level. Superoxide production could, therefore, be substantial if a near maximal state of reduction (i.e., ERED) at 21% O2 were achieved. Equally, distributing electrons amongst ~100,000 mitochondria could limit ERED. Substrate supply, as primarily set by the media lactate pyruvate ratio [121], and OXPHOS will set ERED. In general, culture at 21% O2 (actually 18.6% in a humidified incubator with 5% CO2 [122]) should favor superoxide production at a given ERED by increasing O2 by ~10–15% compared to in utero. The impact of decreasing O2 from 21 to 5% on mitochondrial ROS production in ART is, however, unknown. Dynamic interplay between [O2] and [ERED] means that both evolve over time with attendant implications of superoxide production, and that oxidative stress can only be rationally targeted by considering both determinants. Clinically, 5% O2 can improve live birth rates (+13%) [16], but its efficacy varies and is disputed [123]. Perhaps, differences in the media used and, therefore, substrate levels contributed. Suboptimal media (i.e., electron oversupply) could induce substantial mitochondrial superoxide production, even at 5% O2 by increasing [ERED]. Consistent with this, decreasing [O2] has, in some cases, failed to reduce ROS production [124]. Moreover, high pyruvate levels are sufficient to induce oxidative stress at a fixed O2 in zygotes [125]. Media changes could also induce IRI by resetting O2 gradients [122], with the importance of this factor for oxidative stress depending on the frequency of the media changes and whether the composition changes. That is, different substrate ratios (e.g., lactate:pyruvate ratios) could impact mitochondrial ROS production; with their effect potentially depending on the developmental stage. Additionally, the media pH and transition metal ion content can induce oxidative stress (see Section 4). The number of oocytes cultured is important because multicellularity by establishing O2 gradients is arguably the best defense against oxidative stress [8,126]. That is, oxidative stress should be greater in single oocytes compared to groups owing to increased [O2]. Additionally, light exposure induces oxidative stress due to flavin autoxidation and singlet dioxygen production in media (e.g., via riboflavin) and mitochondria (e.g., via FMN) [24,127,128,129,130].
3.2. Site and Mode-defined Mechanisms of Superoxide Production in ART
A popular way to infer the site of superoxide production is to treat cells/isolated mitochondria with a respiratory chain inhibitor (e.g., rotenone) to terminally arrest electron transfer at a strategic nexus, before assessing the impact on a proxy marker of superoxide production, typically a fluorescent probe (e.g., MitoSOX [131]) in vivo or ex vivo in isolated mitochondria (e.g., Amplex Red [132]). For example, if antimycin A increases the marker, then a role for complex III is assigned. That is, complex III is assumed to contribute to the superoxide production that occurs in the absence of antimycin A (i.e., the “native” rate). As Brand remarks [69,133], (1) terminally arresting electron transfer will block OXPHOS (e.g., taking mode 1 mitochondria to 2–4); (2) electron blockade will reduce upstream and oxidize downstream sites (e.g., artificially rerouting electron transfer could make an inoperative site make superoxide or vice versa); (3) certain inhibitors alter the properties of the site (e.g., antimycin A largely abolishes the effect of ΔΨm on complex III [104]); and (4) several have off-target effects (e.g., sodium azide can inhibit SOD). With the aforementioned caveats in mind, the sole study assessing the impact of respiratory chain inhibitors is judiciously critiqued. In 1991, Johnson’ group [134] showed that antimycin A and rotenone, decreased ROS levels, as inferred by cytosolic DCF fluorescence, by ~70 and 50%, respectively, at 52 h post-fertilization in murine oocytes at 21% O2. At the time, DCF was considered a valid H2O2 assay; but we now know that it is overtly flawed (e.g., the intermediate DCF radical can react with O2 to produce superoxide), and fails to appreciably react with H2O2 [135,136,137]. DCF caveats aside, their work, albeit tentatively, suggests that the Q pool—the major determinant of superoxide production at complex III under antimycin A treatment—is too oxidized or reduced to support superoxide production, which is maximal at ~50–70% Q pool reduction [104]. While chronic treatment did cause membrane blebbing [134], acute treatment with rotenone still decreased ROS levels. Their rotenone findings, again albeit with caveats, argues against a major role for forward electron transfer at the NADH linked sites (e.g., complex I). Recall, in forward mode, rotenone would be expected to approximate mode 2 by increasing [NADH]. We can, therefore, cautiously proceed on the basis that forward mode production is unlikely to play a dominant role, and the Q pool lies close to an extreme (highly oxidized or reduced), at least at the observed timepoint. Recontextualizing the original finding suggests that the unexpected inability of antimycin A and rotenone to increase ROS could reflect nuances in Q pool dependence and RET, respectively.
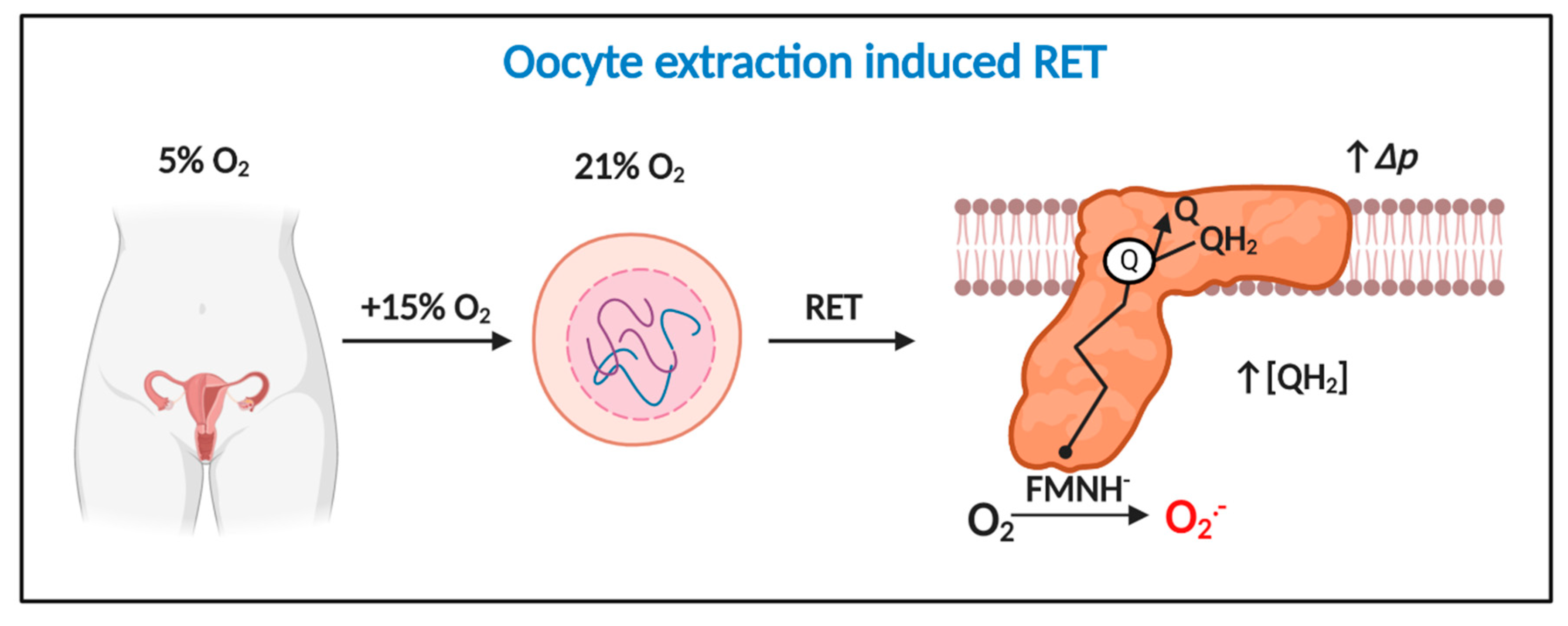
When Johnson’s group performed their pioneering study [134], RET was considered a physiologically irrelevant curiosity of isolated mitochondria. Murphy’s seminal work [73] suggests the ability of rotenone to sterically occlude ubiquinol oxidation-curtailed RET [70]. That is, zygote mitochondria may have been making superoxide via RET. If so, Δp must be near maximal and the Q pool must be highly reduced [47]. Two findings strengthen the appeal of RET as a superoxide production mechanism. First, RET linearly responds to O2 [81]; which helps tie the ability of 21% O2 to cause oxidative stress to a discrete mechanism (Figure 4). Second, oocyte/zygotes are known to contain a distinct pool of mitochondria with a high ΔΨm [138,139,140], which may enable them to support RET. Intriguingly, loss of highly polarized mitochondria impairs cell division, implying a potential regulatory role [141,142]. However, some studies find no evidence for pools of highly polarized mitochondria [143]. Discrepancies could reflect the species, timepoint, and/or probe used [143]. The plausibility of RET as an underlying mechanism rests on the redox state of the Q pool; which is unknown, in part, because it is difficult to assess (e.g., extracted ubiquinol rapidly oxidizes to ubiquinone [144]). If RET operates, then how did antimycin A reduce ROS production? Perhaps, antimycin A dissipated Δp secondary to arresting complex III and IV activity. RET may also be transiently abolished by fertilization-induced Δp-dependent Ca2+ uptake [145]. Conversely, RET could be enhanced by a nitric oxide (NO●)-dependent decrease in OXPHOS due to complex IV inhibition and a resultant increase in Δp [146,147]; the presence of cumulus cells is likely key to NO● dependent affects [148] as is the prevailing [O2] (NO● will exert a greater negative affect at lower [O2] levels). From a clinical perspective, significant RET catalyzed superoxide production can occur at comparatively low [O2] (e.g., ~40 nmol H2O2/min−1/mg−1 at 25 µM O2) [81]. That is, RET is still possible, especially at media pyruvate levels, when O2 is reduced from 21 to 5%.
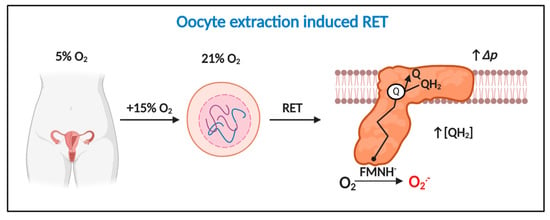
Figure 4.
Oocyte extraction-induced RET is a potential temporally defined mechanism of mitochondrial superoxide production in assisted reproduction technologies (ART). Extracting oocytes induces a 15% increase in O2 exposure, which drives considerable RET-dependent O2●− production provided Δp is near maximal and QH2 is abundant. Extraction-induced light and damage could potentiate RET.
Resolving the redox state of the Q pool is essential for appraising the veracity of an alternate possibility: rotenone by blocking ubiquinol production and decreasing Δp secondary to arresting complex I activity could have decreased complex III-mediated superoxide production. Reports of highly polarized mitochondria are also, therefore, relevant to complex III (i.e., mode 3). Additionally, the ability of antimycin A or rotenone to impact the levels of dicarboxylic acids may have attenuated complex II-mediated superoxide production. In discerning between the sites and modes experiments with uncouplers (to decrease Δp), malonate (sterically occlude the complex II FAD) and aptenin 5A (prevents ubiquinol driven superoxide production at complex II) may be informative [69,83,149]. For example, RET would be highly sensitive to uncouplers and malonate (to block ubiquinol production). To disambiguate the exact site (s), then selective inhibitors of mitochondrial superoxide production at complex I and complex III termed S1QEL and S3QEL, respectively, would be useful [150,151,152,153] (see Box 2). Unlike respiratory chain inhibitors (e.g., rotenone), S1QEL/S3QEL seem to selectively block superoxide production without terminally arresting electron transfer, which could make it possible to infer native sites. Disambiguating how the SQEL family inhibit superoxide production is required to interpret their effects [154].
Box 2. Experimental tools to help advance current knowledge.
Mitochondria-targeted antioxidants: Several mitochondria-targeted antioxidants are available to interrogate the importance of mitochondrial oxidative stress by manipulating the matrix redox environment [155,156]. Promising compounds include mitochondria-targeted vitamin E, vitamin C, and Q [157,158,159,160,161,162,163]. Redox-active compounds based on SOD (termed manganese porphyrins) with pleiotropic in vivo redox chemistry may prove useful [164,165,166]. Inherent chemical limits (e.g., inability to react with H2O2) mean that the failure of a single compound to improve ART cannot be taken as evidence against a role for oxidative stress: such a result is equally compatible with the inability of the compound to react with the relevant species [167]. For example, MitoVE would be relatively ineffective at attenuating matrix superoxide production (e.g., at the complex I flavin), because it concentrates in membranes and is unable to appreciably react with aqueous superoxide [168,169].
S1QEL and S3QEL: As discussed in text, S1QEL and S3QEL, when combined with an appropriate mitochondria-targeted probe, may prove useful for dissecting the site and mode of mitochondrial superoxide in an ART context.
Mitochondria-targeted probes: The ART literature is plagued by the use of flawed probes like DCF [136]. While expressing genetically encoded ratio-metric redox indicators may be problematic [170], several chemically well-understood next-generation small molecule ratio-metric probes can be used to measure mitochondrial superoxide and H2O2 (e.g., MitoNeoD and MitoB) [171,172,173]. If MitoSOX is used, it is important to assess the diagnostic superoxide specific product (2-OH-E) by HPLC [174].
Protein thiol oxidation: Redox proteomics approaches are useful to measure protein thiol oxidation in a systematic high-throughput manner [175,176,177,178,179]. Immunological techniques (e.g., Click PEGylation) can also be used to assess the redox state of a protein of interest by Western blotting [180,181,182,183].
Other: Ratio-metric mass spectrometry-based tools exist to measure the ΔΨm and the redox state of the Q pool [184,185]. Fluorescent approaches (e.g., TMRE) can also be used to measure ΔΨm [6]. Moreover, mitochondria-targeted pro-oxidants like mitochondria-targeted paraquat and CDNB may be useful for appraising the role of complex I-derived superoxide and thiol oxidation, respectively [186,187]. Additionally, several well-established methods exist to measure mitochondrial antioxidant defense parameters (e.g., glutathione and MnSOD) [188,189].
Reports of spatially segregated pools of highly and moderately polarized mitochondria [138], together with a fertilization-induced increase in OXPHOS (i.e., sensitivity of respiration to oligomycin [125,145,190]), are consistent with some mitochondria operating in mode 1 [191,192] (with mode 1 becoming more important in the blastocyst stage). If so, then complex I (forward mode) may also make a low but persistent contribution to total superoxide production in mode 1 mitochondria, provided that the results obtained with skeletal muscle mitochondria in a metabolic milieu mimicking exercise hold in the oocyte/zygote [109]. Given the structural differences between somatic and embryonic mitochondria, and the potential functional immaturity of the latter (see [193]), further research is required to determine whether the same mechanisms translate to oocytes/zygotes. To give a recent structural example, entrapment of some of the Q pool in supercomplexes in hypoxia supports Na+ regulated complex III-mediated superoxide production in somatic mitochondria [194]. Clearly, the structural immaturity of embryonic mitochondria (e.g., spherical morphology) could have profound consequences for superoxide production (e.g., a lack of supercomplexes may obviate complex III-mediated superoxide production in utero, due to homogenous Q pool distribution). Our understanding of how oocyte/zygote mitochondria produce ROS is, therefore, intimately tied to mitochondrial form and function.
Given the well-documented reliance of oocyte mitochondria on pyruvate [195,196], pyruvate dehydrogenase (PDH) would be expected to contribute to superoxide production in modes 1–3. Proximal superoxide production may be self-limiting, since it can inactivate PDH via the reversible thiol oxidation of the E2 subunit [105,197]. Likewise, redox regulation of the mitochondrial pyruvate carrier may constrain pyruvate uptake [198], albeit at the expense of OXPHOS, until it can be reduced by matrix antioxidant defense. Such oscillatory behavior may support cyclic superoxide fluxes. If PDH is cyclically inactivated, then the ability of mitochondria to support lactate and amino acid metabolism, particularly via the malate-aspartate shuttle, will be key [199]. A fascinating finding arguing against a role for PDH is that it migrates to the nucleus, along with several other TCA enzymes, to support epigenetic wiring and genome activation in mouse and human zygotes [200,201]. Their ability to migrate to the nucleus reinforces the inability of rotenone to stimulate forward mode production, potentially by limiting [NADH]. A stalled TCA cycle could increase Δp and [ubiquinol] to levels compatible with RET, as the rotenone finding suggests, in a significant subset of mitochondria.
3.3. How Oocyte/Zygote Mitochondria Produce Superoxide is Unknown
Further research using next-generation tools (see Box 2) to expand the current boundary of knowledge (see Figure 5) is required because the use of flawed probes and approaches means that how oocyte/zygote mitochondria produce superoxide, both in utero and in ART, is unknown. For example, ambiguity surrounds RET as a plausible superoxide production mechanism in ART, because the redox state of the Q pool is unknown and rotenone could have had off-target effects (see Section 3.3).
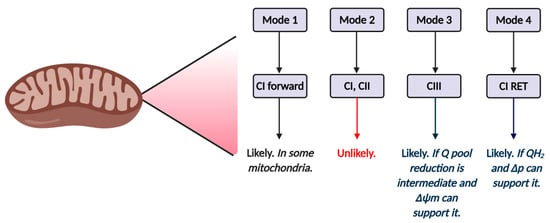
Figure 5.
Tentative mechanisms of mitochondrial reactive oxygen species (ROS) production in ART by mode and site. Mode 1 (see Table 2) is likely to operate in at least some mitochondria, and is likely driven by complex I forward electron transfer. It is possible that complex II also operates in mode 1. Inhibitor studies (see main text) reveal forward mode is unlikely to operate in mode 2, since rotenone fails to increase ROS production in zygotes. The relative importance of mode 3 and 4 will depend on Δp and the redox state of the Q pool. Inhibitor studies (see main text) support a role for RET.
Despite how oocyte/zygote mitochondria produce ROS being a known unknown, it is possible to help advance the field, because several general points will apply, regardless of the mechanism. First, superoxide production will change over time as O2 and ERED evolve; with the latter being sensitive to substrate supply (i.e., media formula and changes). Second, superoxide production will exhibit mode and site heterogeneity within and between mitochondria (potentially at different developmental timepoints). Third, oxidative damage could impact superoxide production (see Section 4). Fourth, superoxide production is likely to vary according to the species (e.g., the reliance of porcine oocytes on beta oxidation [202] will profoundly impact the nature of the superoxide production observed). What is more, the nature of mitochondrial superoxide production will differ from what occurs in utero (i.e., the prevailing O2 sets a lower potential superoxide production limit in vivo). Furthermore, the known structural immaturity of oocyte and early zygotic mitochondria (e.g., spherical morphology) could impact superoxide production [203] (see Section 3.3). Finally, several timepoints, including: initial extraction; media changes, fertilization, and the metabolic switch to aerobic glycolysis will be of interest [204,205]. It would be unwise to extrapolate findings from one timepoint to another (e.g., as the PDH example in Section 3.3 attests).
4. A Framework for Interpreting the Role of Mitochondrial ROS in ART: The Intriguing
4.1. Interpreting Mitochondrial Superoxide Production
Mitochondrial superoxide production is often viewed as an unwanted side-reaction usually referred to as a “leak”, which broadly approximates a short circuit in an electrical analogy. Within complex IV, bound O2 is very likely reduced to superoxide (the O-O bond is weaker in superoxide compared to O2 [5]) via electron dissociation from the heme to O2, before it is concertedly reduced to H2O via bound O-2 intermediates [206]. The potential intermediacy of a transient bound superoxide to mitochondrial respiration, may make the release of superoxide via defined off-target pathways at complex I, II and III an evolutionary success as opposed to failure [207]. A radical off-pathway reporting the process of electrons—the simplest free radical—being transferred to a diradical to produce H2O is appealing. Deliberate superoxide production off-pathways, may enable reaction 3 to report on reactions 1 and 2 (i.e., a minor pathway reporting a major one [208]). Exquisite sensitivity to key parameters of OXPHOS, substrate supply and respiration (e.g., Δp) could enable superoxide production to report mitochondrial function [81,207,209,210,211]. For example, RET responds to OXPHOS (i.e., Δp), substrate supply (ubiquinol) and O2, making it a plausible function linked redox signal [70,81]. In support, Allen [211] contends that mitochondria retain a genome to respond to respiratory chain-derived redox signals. Basic redox chemistry means deliberate and adventitious can co-exist. For example, superoxide emanating from a deliberate pathway (e.g., FMN at complex) could lead, via hydrogen peroxide and transition metal ions, to adventitious production via hydroxyl radical-mediated attack of glutathione to a radical, which then adventitiously produces superoxide via the Winterbourn sink pathway. Regardless of whether mitochondrial superoxide production is adventitious (leak) or intended (deliberate off-pathway), it is wise to interpret superoxide production using a context-dependent functionality framework. Biological context reconciles the fundamental duality at the heart of redox biology, by determining whether the net effect of superoxide production benefits or harms the cell. From a therapeutic perspective, context-dependent functionality means that it would be unwise to abolish superoxide production in ART (i.e., it could negate an adaptive response). Consistent with this, in the presence of cumulus cells, the short-term exposure of bovine oocytes to even supraphysiological levels of H2O2 (50–100 µM, as opposed to 1–100 nM) improved subsequent zygotic development [212].
4.2. A Two-Step Bifurcation Model to Interpret ART-induced Oxidative Stress
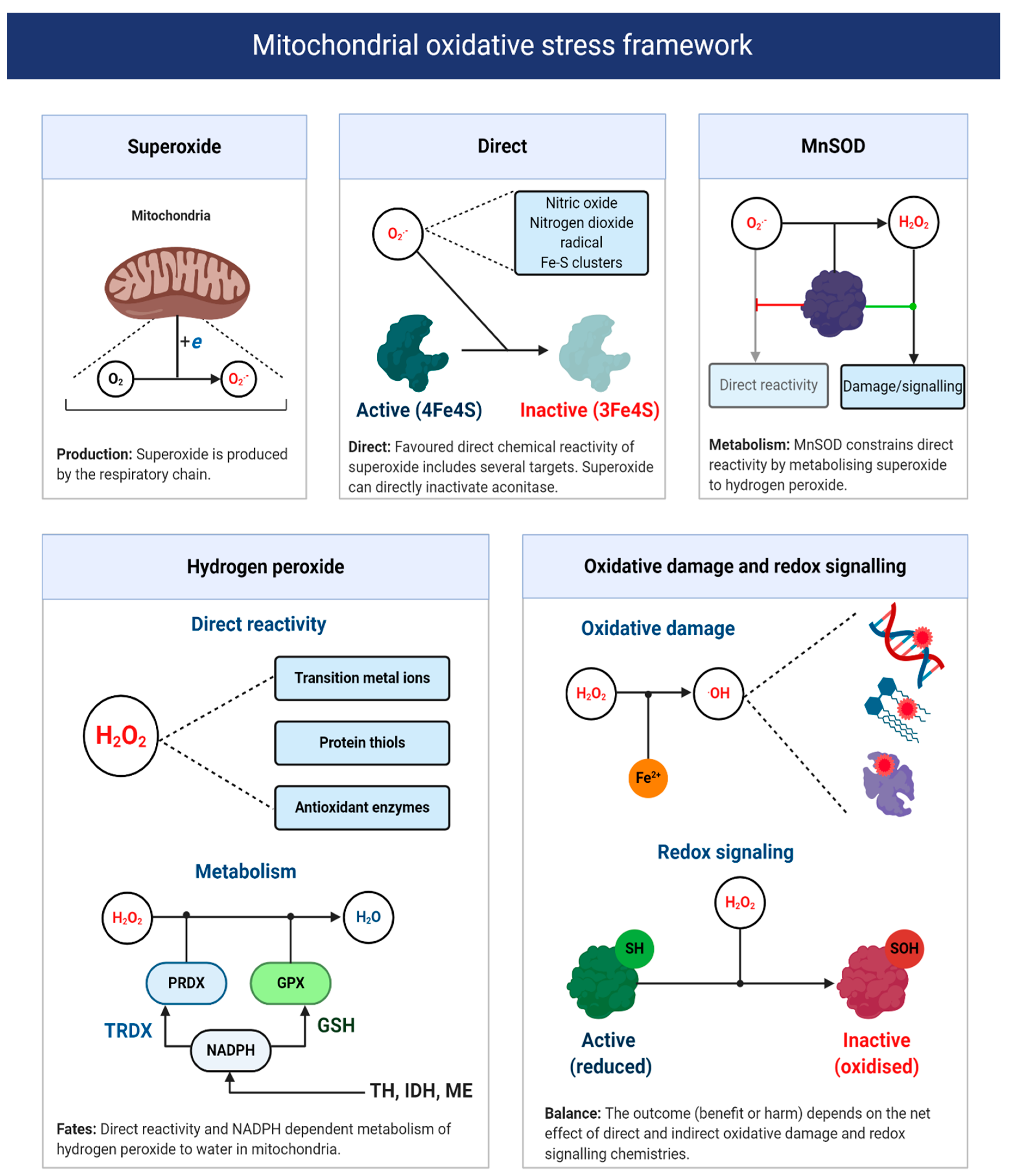
As Sies remarks [34], the term oxidative stress conveys no useful mechanistic information unless the underlying chemistry is defined. Recall one is unable to target ROS—no molecule called ROS exists [58]. A fragmentary understanding of ART-induced oxidative stress calls for a chemically defined two-step bifurcation model for ART-induced mitochondrial oxidative stress (see Figure 6). The first bifurcation stipulates that the nature of the oxidative stress observed depends, in part, on the ability of superoxide to evade MnSOD to directly react with its matrix interactome [208,213]. Specifically, matrix NO● to produce peroxynitrite (and subsequently free radicals, reactions 10–11), protein bound (e.g., aconitase) or “free” transition metal ions [214,215,216]. The alkaline pH of the mitochondrial matrix means that MnSOD controls the direct reactivity of superoxide, because uncatalyzed dismutation (reaction 12) proceeds at a negligible rate without HO2● (reaction 13), because two anions electrostatically repel one another [55,56,217,218,219]. HO2● is still relevant, because it can occur as a locally caged radical when a substrate protonates superoxide [5,95]. Active and abundant MnSOD will constrain superoxide to picomolar levels, to limit direct reactivity by catalyzing reaction 12 at a diffusion controlled rate (k~2 × 109 M−1 s−1) [220]; provided that it contains manganese as the iron ligated enzyme can produce ●OH [221]. As Imlay [220] calculates, even picomolar (10−11/12 M) levels of superoxide are sufficient to inactivate enzymes iron-sulfur proteins like aconitase, with a half-time of ~20 min in E. coli. By controlling the fate of superoxide, MnSOD exerts considerable control over the type of oxidative stress experienced [222], especially since, in the absence of an anion channel, superoxide will be retained in the matrix [223]. (reaction 11 in 33% yield [224,225,226]).
reaction 10: O2●− + NO● → ONOO−
reaction 11: ONOO− + CO2 → ONOOCO2→ CO3− + NO2●
reaction 12: O2●− + O2●− + 2H+ → H2O2 + O2
reaction 13: HO2● + O2●− +H+ → H2O2+ O2
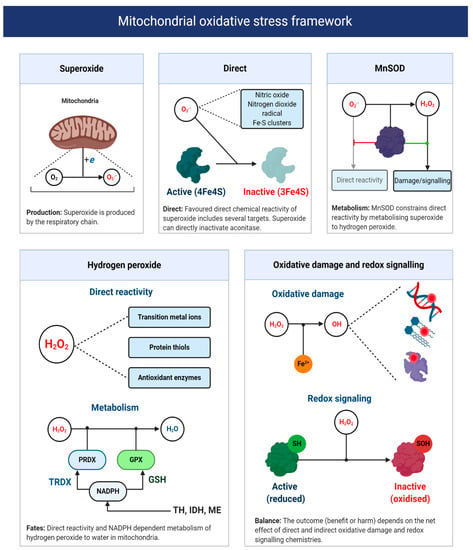
Figure 6.
A mitochondrial oxidative stress framework. Upper panels depict the direct chemical reactivity of superoxide, which is regulated by MnSOD activity. The lower left panel depicts the direct chemical reactivity and metabolism of H2O2. The lower right panel illustrates two H2O2-focused chemical defined examples of oxidative damage and redox signaling. The net function of mitochondrial superoxide production will depend on whether direct and indirect reactivity (via H2O2 and other species like hydroxyl radical) is beneficial or harmful. Abbreviations: TH = transhydrogenase; IDH = NADPH-dependent isocitrate dehydrogenase; and ME = NADPH-dependent malic enzyme.
The second bifurcation stipulates that the nature of the oxidative stress experienced depends on the partitioning of H2O2 between matrix peroxidases or other direct targets; most notably, transition metal ions and thiols. Matrix peroxidase metabolism (principally peroxiredoxin 3, 5, and glutathione peroxidase 1 [227]) is coupled to antioxidant defense, wherein H2O2 is converted to H2O in a process that oxidizes thioredoxin or glutathione—the reduced forms are then regenerated at the expense of NADPH by mitochondrial thioredoxin reductase and glutathione reductase, respectively [228]. NADPH can be produced by isocitrate dehydrogenase, malic enzyme, one carbon metabolism or the Δp consuming transhydrogenase (e.g., NADH + NADP+ + H+ (P side) –>NAD+ + NADPH + H+ (N side) [229]. NADPH-supported redox systems could couple the excessive production of H2O2 to antioxidant defense [228]. It could also be coupled to redox signaling (discussed below) [230]. Conversely, defective antioxidant defense will favor mitochondrial H2O2 release [231] or reactivity with transition metal ions to produce ●OH (reaction 14) [232]. Knowledge of oocyte/early zygote antioxidant defense is limited, so their capacity to rebuff an ART-induced increase in H2O2 is unclear. In zygotes, the migration of IDH to the nucleus could limit NADPH availability [200]. The attendant consequences for oxidative damage, given the diffusion controlled reactivity of ●OH (k ~ 109 M−1 s−1), are significant [233]. In particular, ●OH can impede OXPHOS by inactivating complex I–V [234] and cause mitochondrial DNA (mtDNA) heteroplasmy [235] by abstracting, adding and oxidizing nucleotides, notably guanine [236,237,238]. The need to maintain mtDNA homoplasmy to avoid bioenergetic defects and potentially increase superoxide production by a mismatched respiratory chain, is difficult to reconcile with active OXPHOS because, even in mode 1, it would sensitize oocytes/zygotes to DNA damage [193,209,239,240]. Perhaps, low [O2], together with cumulus cell-derived ATP supply limits OXPHOS in utero to protect mtDNA homoplasmy and/or any damage maintained, is repaired or prevented by DNA binding proteins. Spatial differences may segregate much of the damage into trophectoderm mitochondria, destined to become the placenta, to protect the inner cell mass.
reaction 14: H2O2+ Cu+ or Fe2+ –> OH + −OH + Cu2+ or Fe(III)
Redox signaling refers to the ability of ROS to signal changing protein activity, function, lifetime, binding partners, and location [241,242,243]. While redox signals can take many guises (e.g., direct reactivity of superoxide with aconitase [208,244]), a major form is defined by H2O2-mediated reversible oxidation of protein thiols (cysteine residues) and the resultant formation of distinct chemotypes from sulfenic acids (RSOH) to disulfide bonds (RSSR), depending on contextual factors (e.g., RSOH stability [58,241,245,246]). The mammalian proteome contains over 200,000 protein thiols, with many being located in the matrix (their matrix abundance is in the mM range), often in a deprotonated state (favoring direct H2O2 reactivity), due to the alkaline pH [228,247,248,249]. I propose that: ART-induced oxidative stress is causally linked to a pervasive H2O2-mediated increase in fractional protein thiol oxidation occupancies from adaptive (physiological) to maladaptive (pathological) levels. Reversible thiol oxidation can impact superoxide production directly, by altering respiratory chain activity [250]. For example, oxidizing thiols in subunits (e.g., NDUFS1) proximal to FMN in complex I can decrease superoxide production by sterically impeding NADH access [251]; which would curtail complex I-mediated superoxide production at the expense of OXPHOS (i.e., a negative feedback loop). Reversible oxidation of the alpha subunit of complex V can impact superoxide production indirectly by altering OXPHOS [252,253,254]. Impaired complex V activity could increase Δp to favor superoxide production, by reducing upstream complexes (i.e., ↑ [ERED]), which could promote RET or complex III-mediated superoxide production (i.e., a feedforward loop). Oxidative stress-induced defects in OXPHOS could impair ART by reducing the [ATP] available to support spindle formation, ion homeostasis, and protein synthesis [255].
Kinetically, how reversible thiol oxidation proceeds is unclear, because H2O2 reacts at a slow rate (k~1–50 M−1 s−1) with most thiols compared to antioxidant enzymes (k~107−8 M−1 s−1) [256]. Several solutions to the kinetic conundrum have been proposed [257,258,259,260]. Redox relays represent an elegant solution wherein antioxidant enzymes transduce redox signals by transferring H2O2-derived electrons to target proteins (i.e., uncoupling peroxiredoxin 3 from thioredoxin 2). From a conceptual perspective, redox relays challenge the traditional view of antioxidant enzymes: they play a more nuanced role than merely removing ROS [222,230,258,261]. As Winterbourn enunciates [262], several parallel mechanisms may contribute (e.g., local inactivation of peroxiredoxins or locally caging H2O2 and the target). One ART relevant mechanism concerns bicarbonate (CO2/HCO3−), given its presence at relatively high levels (mM range) in oocyte/embryo media [121]. The equilibrium between H2O2 and HCO3− yields peroxomonocarbonate (HCO4−). HCO4− can react with protein thiols at a faster rate (sometimes a thousand times faster) than H2O2, and can be important for protein thiol oxidation (e.g., for protein tyrosine phosphatase 1B) in intact cells [263]. HCO4− could also yield carbonate radical, which can react with thiols (k~4.6 × 107 M−1 s−1) secondary to reacting with Cu+ (reaction 15). Another relevant possibility involves local thiol oxidation secondary to H2O2, reacting with a protein bound transition ion (e.g., Cu+) to produce ●OH; which could yield a thiyl radical (RS●) at a significant rate (k~7 × 109 M−1 s−1 for L-Cys) (reaction 16). ONOO−-derived radicals may also contribute [228,264]. Thiyl radicals react appreciably with NO● to form RSNO (reaction 17), or with another thiol (reaction 18) to yield a highly reducing radical disulfide (RSSR●). RSSR● reacts with O2 at an appreciable rate (k~5 × 108 M−1 s−1 for glutathione disulfide radical), to form superoxide (reaction 19), making it possible, in principle, for any protein thiol to produce superoxide [265]. RSSR● could be an adventitious source of superoxide in mitochondria (see Section 4.1 and footnote 3). MnSOD activity to remove superoxide is, therefore, important [266]. A role for free radicals in thiol-based redox signaling has been proposed [267] and evidenced for complex I [268].
reaction 15: HCO4− + Cu+ –> CO3− + Cu2+ + OH
reaction 16: OH + RSH –> RS● + H2O
reaction 17: RS + NO –> RSNO
reaction 18: RS + RSH –> RSSR● + H+
reaction 19: RSSR + O2 –> O2●− + RSSR
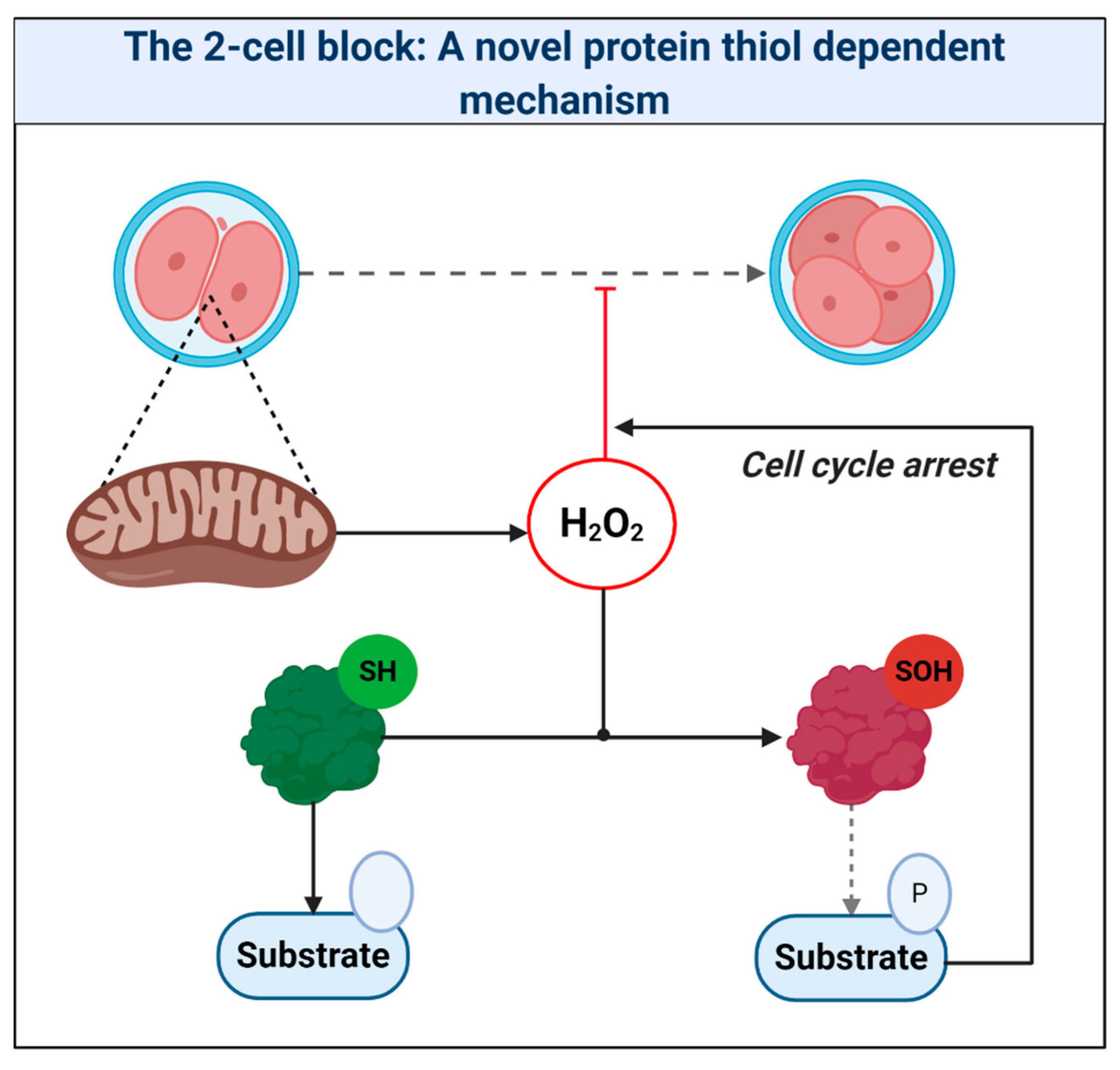
A pervasive increase in thiol oxidation is a novel cause of oxidative stress in ART. The mechanism reconciles the ability of the thiol oxidant diamide to disrupt OXPHOS in ART [269], and reductants like 1-4-dithiothreitol to improve ART [11]. The novel framework presented should provide a useful paradigm to interpret mitochondrial oxidative stress in ART. Research is, however, required to unravel the chemical nature of the oxidative stress and its relationship to proximal mitochondrial superoxide production. It is vital to understand the physiological role of any intrinsic fertilization-induced increase in ROS production [270]. While fertilization is well known to increase H2O2 production in sea urchins [271,272,273], Amaya’s group [274] recently tied fertilization triggered Ca2+-induced mitochondrial ROS release to the regulation of the embryonic cell cycle via reversible cdc25c oxidation in Xenopus laevis. By tying mitochondrial ROS to the early cell cycle via thiol oxidation, their work provides a precedent to reimagine the 2-cell block as increased thiol oxidation of regulatory cell cycle proteins [183] (see Figure 7). Consistent with this, reversing protein thiol oxidation may explain the ability of recombinant thioredoxin to release the 2-cell block [22,261,275,276]. To disambiguate the molecular details of ART-induced oxidative stress (e.g., source-target relationships), protein thiol oxidation of key regulatory proteins should be assessed [183,277]. Finally, intimate ties between mitochondria-regulated metabolites (e.g., succinate) and epigenetics raise the potential for far-reaching ART-induced oxidative stress, secondary to aberrant epigenetic wiring [278,279,280], which could impact imprinting [281]. Precedent exists: stochastic development fluxes in the redox state of the glutathione pool regulate lifespan and stress resistance in C. elegans [282,283]. Moreover, the enzymes responsible were regulated by reversible thiol oxidation [282]. In mammals, embryonic exposure to rotenone, which modifies complex I-mediated superoxide production, can produce lasting DNA methylation changes in offspring [284]. Future studies may wish to interrogate the potential regulatory role of ROS-mediated retrograde mito-nuclear epigenetic signaling [285].
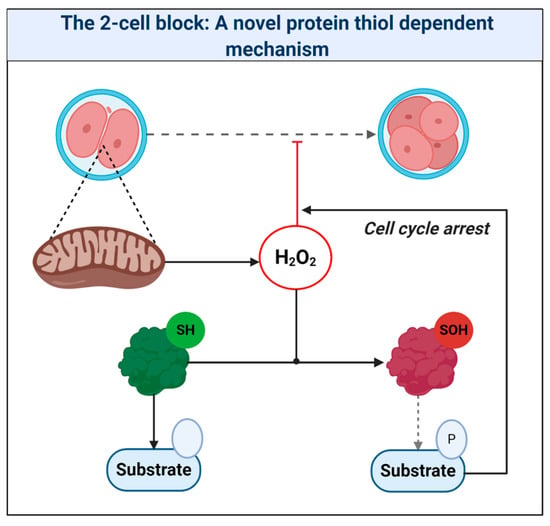
Figure 7.
The 2-cell block: A novel protein thiol-dependent mechanism. The scheme depicts mitochondria-derived hydrogen peroxide oxidizing the catalytic (Cys373) thiol of the tyrosine protein phosphatase cdc25c to a sulfenic acid (SOH). Thiol oxidation renders cdc25c unable to dephosphorylate target substrates. The resultant perturbation in zygotic phosphorylation tone could cause cell cycle arrest.
5. Conclusions
The known importance of ART-induced oxidative stress belies how oocyte/zygote mitochondria produce ROS being the unknown. Despite topical interest, current understanding of how oocyte/zygote mitochondria produce superoxide is unsatisfactory owing to the use of outmoded approaches (e.g., respiratory poisons) and methodologies (e.g., flawed probes). Considering how somatic mitochondria produce superoxide (i.e., the known [47]) makes several tractable and experimentally testable predictions about the unknown. For example, oocyte extraction may induce RET, owing to at least a 10% increase in O2 exposure, provided that Δp is near maximal and QH2 is abundant. Further research using next-generation tools is required to unequivocally dissect how oocyte/zygotic mitochondria produce ROS, with a view to understanding their likely nuanced biological roles. While some of the ROS produced by oocyte/zygote mitochondria will undoubtedly cause oxidative damage [228], their ability to transduce beneficial redox signals (e.g., to report OXPHOS [209]) raises the intriguing possibility that they regulate key developmental processes (e.g., the cell cycle). Irrespective of whether the net effect of mitochondria producing ROS is useful, neutral, or harmful, plausible explanatory source-target relationships are lacking. To address this unmet need, a chemically defined oxidative stress framework is presented. The intriguing possibility of redox signaling allied to several discrete sites and modes of superoxide production means that it has arguably never been more essential to understand the mechanistic basis of ART-induced oxidative stress to rationally target key enzymes/processes to preserve redox signaling while attenuating oxidative damage.
Author Contributions
Writing—original draft preparation, J.N.C.; writing—review and editing, J.N.C.; funding acquisition, J.N.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Highlands and Islands Enterprise (HMS 9353763), Tenovus Scotland (#G19.03), and Rosetrees Trust (#A2819).
Acknowledgments
All figures were created using BioRender.com. I am extremely grateful to Tenovus Scotland and Rostrees Trust for research funding.
Conflicts of Interest
The author declares no conflict of interest.
References
- Jensen, P. Antimycin-insensitive oxidation of succinate and reduced nicotinamide-adenine dinucleotide in electron-transport particles I. pH dependency and hydrogen peroxide formation. Biochim. Biophys. Acta 1966, 122, 157–166. [Google Scholar] [CrossRef]
- Steptoe, P.; Edwards, R. Birth after the reimplantation of a human embryo. Lancet 1978, 312, 366. [Google Scholar] [CrossRef]
- O’Flaherty, C. Reactive oxygen species and male fertility. Antioxidants 2020, 9, 287. [Google Scholar] [CrossRef]
- Sies, H. Oxidative Stress: Introductory Remarks. In Oxidative Stress; Academic Press: Cambridge, MA, USA, 1985; pp. 1–5. [Google Scholar]
- Sawyer, D.T.; Valentine, J.S. How super is superoxide? Acc. Chem. Res. 1981, 14, 393–400. [Google Scholar] [CrossRef]
- Nicholls, D.G.; Ferguson, S.J. Bioenergetics 4, 4th ed.; Academic Press: London, UK, 2013. [Google Scholar]
- Mailloux, R.J. An update on mitochondrial reactive oxygen species production. Antioxidants 2020, 9, 472. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology & Medicine, 5th ed.; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Johnson, M.H.; Nasresfahani, M.H. Radical solutions and cultural problems: Could free oxygen radicals be responsible for the impaired development of preimplantation mammalian embryos invitro? BioEssays 1994, 16, 31–38. [Google Scholar] [CrossRef]
- Guerin, P.; El Mouatassim, S.; Menezo, Y. Oxidative stress and protection against reactive oxygen species in the pre-implantation embryo and its surroundings. Hum. Reprod. Update 2001, 7, 175–189. [Google Scholar] [CrossRef]
- Agarwal, A.; Gupta, S.; Sekhon, L.; Shah, R. Redox considerations in female reproductive function and assisted reproduction: From molecular mechanisms to health implications. Antioxid. Redox Sign. 2008, 10, 1375–1404. [Google Scholar] [CrossRef]
- Lord, T.; Nixon, B.; Jones, K.T.; Aitken, R.J. Melatonin prevents postovulatory oocyte aging in the mouse and extends the window for optimal fertilization in vitro. Biol. Reprod. 2013, 88, 67. [Google Scholar] [CrossRef]
- Lord, T.; Aitken, R.J. Oxidative stress and ageing of the post-ovulatory oocyte. Reproduction 2013, 146, R217–R227. [Google Scholar] [CrossRef]
- Zhang, M.; Shiyang, X.; Zhang, Y.; Miao, Y.; Chen, Y.; Cui, Z.; Xiong, B. Coenzyme Q10 ameliorates the quality of postovulatory aged oocytes by suppressing DNA damage and apoptosis. Free. Radic. Biol. Med. 2019, 143, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-G.; Park, H.-J.; Kim, J.-W.; Jung, J.-M.; Kim, M.-J.; Jegal, H.-G.; Kim, I.-S.; Kang, M.-J.; Wee, G.; Yang, H.-Y.; et al. Mito-TEMPO improves development competence by reducing superoxide in preimplantation porcine embryos. Sci. Rep. 2018, 8, 10130. [Google Scholar] [CrossRef] [PubMed]
- Bontekoe, S.; Mantikou, E.; Van Wely, M.; Seshadri, S.; Repping, S.; Mastenbroek, S. Low oxygen concentrations for embryo culture in assisted reproductive technologies. Cochrane Database Syst. Rev. 2012, 19, 209. [Google Scholar] [CrossRef] [PubMed]
- Rampon, C.; Volovitch, M.; Joliot, A.; Vriz, S. Hydrogen Peroxide and Redox Regulation of Developments. Antioxidants 2018, 7, 159. [Google Scholar] [CrossRef]
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef]
- Bedard, K.; Krause, K.-H. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Aitken, R.J.; Drevet, J.R. The importance of oxidative stress in determining the functionality of mammalian spermatozoa: A two-edged sword. Antioxidants 2020, 9, 111. [Google Scholar] [CrossRef]
- Lane, N. Oxygen: The Molecule that Made the World; Oxford University Press: Oxford, UK, 2002. [Google Scholar]
- Fridovich, I. Oxygen: How do we stand it? Med. Princ. Pract. 2012, 22, 131–137. [Google Scholar] [CrossRef]
- Abele, D. Toxic oxygen: The radical life-giver. Nature 2002, 420, 2002. [Google Scholar] [CrossRef]
- Ogilby, P.R. Singlet oxygen: There is indeed something new under the sun. Chem. Soc. Rev. 2010, 39, 3181–3209. [Google Scholar] [CrossRef]
- Kaila, V.R.I.; Verkhovsky, M.I.; Wikström, M. Proton-coupled electron transfer in cytochrome oxidase. Chem. Rev. 2010, 110, 7062–7081. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.C.; Chang, C.J. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 2011, 7, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. The biology of oxygen radicals. Science 1978, 201, 875–880. [Google Scholar] [CrossRef]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Ann. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Sies, H. Oxidative Stress: Concept and some Practical Aspects. Antioxidants 2020, 9, 852. [Google Scholar] [CrossRef]
- Sies, H. Oxidative Stress: Eustress and Distress; Academic Press: London, UK, 2020. [Google Scholar]
- Jones, D.P.; Sies, H. The Redox Code. Antioxid. Redox Sign. 2015, 23, 734–746. [Google Scholar] [CrossRef]
- Cobley, J.N. How Exercise Induces Oxidative Eustress. In Oxidative Stress; Sies, H., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 447–462. [Google Scholar] [CrossRef]
- Sies, H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015, 4, 180–183. [Google Scholar] [CrossRef]
- Murphy, M.P.; Holmgren, A.; Larsson, N.-G.; Halliwell, B.; Chang, C.J.; Kalyanaraman, B.; Rhee, S.G.; Thornalley, P.J.; Partridge, L.; Gems, D.; et al. Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011, 13, 361–366. [Google Scholar] [CrossRef]
- Bjelakovic, G.; Nikolova, D.; Gluud, C.; Simonetti, R.G.; Gluud, C. Antioxidant supplements for prevention of mortality in healthy participants and patients with various diseases. Cochrane Database Syst. Rev. 2012, 3, CD007176. [Google Scholar] [CrossRef]
- Moser, C.C.; Farid, T.A.; Chobot, S.E.; Dutton, P.L. Electron tunneling chains of mitochondria. Biochim. Biophys. Acta 2006, 1757, 1096–1109. [Google Scholar] [CrossRef]
- Moser, C.C.; Page, C.C.; Dutton, P.L. Darwin at the molecular scale: Selection and variance in electron tunnelling proteins including cytochrome c oxidase. Philos. Trans. R. Soc. B Biol. Sci. 2006, 361, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Wikström, M.; Sharma, V.; Kaila, V.R.I.; Hosler, J.P.; Hummer, G. New perspectives on proton pumping in cellular respiration. Chem. Rev. 2015, 115, 2196–2221. [Google Scholar] [CrossRef]
- De Vault, D.; Chance, B. Studies of photosynthesis using a pulsed laser: I. temperature dependence of cytochrome oxidation rate in chromatium. evidence for tunneling. Biophys. J. 1966, 6, 825–847. [Google Scholar] [CrossRef]
- Vafai, S.B.; Mootha, V.K. Mitochondrial disorders as windows into an ancient organelle. Nature 2012, 491, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Mailloux, R.J. Teaching the fundamentals of electron transfer reactions in mitochondria and the production and detection of reactive oxygen species. Redox Biol. 2015, 4, 381–398. [Google Scholar] [CrossRef]
- Saraste, M. Oxidative Phosphorylation at the fin de siècle. Science 1999, 283, 1488–1493. [Google Scholar] [CrossRef]
- Mitchell, P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 1961, 192, 452–454. [Google Scholar] [CrossRef]
- Walker, J.E. The ATP synthase: The understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013, 41, 1–16. [Google Scholar] [CrossRef]
- Yoshida, M.; Muneyuki, E.; Hisabori, T. ATP synthase—A marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001, 2, 669–677. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2008, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Marcus, R.; Sutin, N. Electron transfers in chemistry and biology. Biochim. Biophys. Acta 1985, 811, 265–322. [Google Scholar] [CrossRef]
- Moser, C.C.; Keske, J.M.; Warncke, K.; Farid, R.S.; Dutton, P.L. Nature of biological electron transfer. Nature 1992, 355, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A.; Sethu, R.; Rohaun, S.K. Evolutionary adaptations that enable enzymes to tolerate oxidative stress. Free. Radic. Biol. Med. 2019, 140, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003, 552, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Chance, B.; Sies, H.; Boveris, A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979, 59, 527–605. [Google Scholar] [CrossRef]
- Boveris, A.; Chance, B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem. J. 1973, 134, 707–716. [Google Scholar] [CrossRef]
- Barja, G. Updating the mitochondrial free radical theory of aging: An integrated view, key aspects, and confounding concepts. Antioxid. Redox Sign. 2013, 19, 1420–1445. [Google Scholar] [CrossRef]
- Mccord, J.M.; Fridovich, I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar]
- Keele, B.B.; Mccord, J.M.; Fridovich, I. Superoxide dismutase from escherichia coli B. A new manganese-containing enzyme. J. Biol. Chem. 1970, 245, 6176–6181. [Google Scholar]
- Loschen, G.; Azzi, A.; Richter, C.; Flohé, L. Superoxide radicals as precursors of mitochondrial hydrogen peroxide. FEBS Lett. 1974, 42, 68–72. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Hirst, J. Mitochondrial Complex I. Ann. Rev. Biochem. 2013, 82, 551–575. [Google Scholar] [CrossRef] [PubMed]
- Vinothkumar, K.R.; Zhu, J.; Hirst, J. Architecture of mammalian respiratory complex I. Nature 2014, 515, 80–84. [Google Scholar] [CrossRef]
- Brandt, U. A two-state stabilization-change mechanism for proton-pumping complex I. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 1364–1369. [Google Scholar] [CrossRef]
- Kaila, V.R.I. Long-range proton-coupled electron transfer in biological energy conversion: Towards mechanistic understanding of respiratory complex I. J. R. Soc. Interface 2018, 15. [Google Scholar] [CrossRef]
- Parey, K.; Wirth, C.; Vonck, J.; Zickermann, V. Respiratory complex I—structure, mechanism and evolution. Curr. Opin. Struct. Biol. 2020, 63, 1–9. [Google Scholar] [CrossRef]
- Zickermann, V.; Wirth, C.; Nasiri, H.; Siegmund, K.; Schwalbe, H.; Hunte, C.; Brandt, U. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 2015, 347, 44–49. [Google Scholar] [CrossRef]
- Kussmaul, L.; Hirst, J. The mechanism of superoxide production by NADH: Ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc. Natl. Acad. Sci. USA 2006, 103, 7607–7612. [Google Scholar] [CrossRef]
- Pryde, K.R.; Hirst, J. Superoxide is produced by the reduced flavin in mitochondrial complex I. J. Biol. Chem. 2011, 286, 18056–18065. [Google Scholar] [CrossRef]
- Birrell, J.A.; Morina, K.; Bridges, H.R.; Friedrich, T.; Hirst, J. Investigating the function of [2Fe–2S] cluster N1a, the off-pathway cluster in complex I, by manipulating its reduction potential. Biochem. J. 2013, 456, 139–146. [Google Scholar] [CrossRef]
- Schulte, M.; Frick, K.; Gnandt, E.; Jurkovic, S.; Burschel, S.; Labatzke, R.; Aierstock, K.; Fiegen, D.; Wohlwend, D.; Gerhardt, S.; et al. A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I. Nat. Commun. 2019, 10, 2551. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free. Radic. Biol. Med. 2016, 100, 14–31. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Pell, V.R.; James, A.M.; Work, L.M.; Saeb-Parsy, K.; Frezza, C.; Krieg, T.; Murphy, M.P. A unifying mechanism for mitochondrial superoxide production during ischemia-reperfusion injury. Cell Metab. 2016, 23, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Hinkle, P.C.; Butow, R.A.; Rackers, E.; Chance, B. Partial resolution of the enzymes catalyzing oxidative phosphorylation. XV. Reverse electron transfer in the flavin-cytochrome β region of the respiratory chain of beef heart submitochondrial particles. J. Biol. Chem. 1967, 242, 5169–5173. [Google Scholar]
- Chouchani, E.T.; Methner, C.; Nadtochiy, S.M.; Logan, A.; Pell, V.R.; Ding, S.; James, A.M.; Cochemé, H.M.; Reinhold, J.; Lilley, K.S.; et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat. Med. 2013, 19, 753–759. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Pell, V.R.; Gaude, E.; Aksentijević, D.; Sundier, S.Y.; Robb, E.L.; Logan, A.; Nadtochiy, S.M.; Ord, E.N.; Smith, A.C.; et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 2014, 515, 431–435. [Google Scholar] [CrossRef]
- Martin, J.L.; Costa, A.S.H.; Gruszczyk, A.V.; Beach, T.E.; Allen, F.M.; Prag, H.A.; Hinchy, E.C.; Mahbubani, K.T.; Hamed, M.; Tronci, L.; et al. Succinate accumulation drives ischaemia-reperfusion injury during organ transplantation. Nat. Metab. 2019, 1, 966–974. [Google Scholar] [CrossRef]
- Scialo’, F.; Sriram, A.; Fernández-Ayala, D.J.M.; Gubina, N.; Lõhmus, M.; Nelson, G.; Logan, A.; Cooper, H.M.; Navas, P.; Enriquez, J.A.; et al. Mitochondrial ROS produced via reverse electron transport extend animal lifespan. Cell Metab. 2016, 23, 725–734. [Google Scholar] [CrossRef]
- Onukwufor, J.O.; Berry, B.J.; Wojtovich, A.P. Physiologic implications of reactive oxygen species production by mitochondrial complex I reverse electron transport. Antioxidants 2019, 8, 285. [Google Scholar] [CrossRef]
- Fernández-Agüera, M.C.; Gao, L.; González-Rodríguez, P.; Pintado, C.O.; Arias-Mayenco, I.; García-Flores, P.; García-Pergañeda, A.; Pascual, A.; Ortega-Sáenz, P.; López-Barneo, J. Oxygen sensing by arterial chemoreceptors depends on mitochondrial complex I signaling. Cell Metab. 2015, 22, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Arias-Mayenco, I.; González-Rodríguez, P.; Torres-Torrelo, H.; Gao, L.; Fernández-Agüera, M.C.; Bonilla-Henao, V.; Ortega-Sáenz, P.; López-Barneo, J. Acute O2 sensing: Role of coenzyme QH2/Q ratio and mitochondrial ROS compartmentalization. Cell Metab. 2018, 28, 145–158. [Google Scholar] [CrossRef]
- Guarás, A.; Perales-Clemente, E.; Calvo, E.; Acín-Pérez, R.; Loureiro-López, M.; Pujol, C.; Martínez-Carrascoso, I.; Núñez, E.; García-Marqués, F.; Rodríguez-Hernández, A.; et al. The CoQH2/CoQ ratio serves as a sensor of respiratory chain efficiency. Cell Rep. 2016, 15, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Kempf, A.; Song, S.M.; Talbot, C.B.; Miesenböck, G. A potassium channel β-subunit couples mitochondrial electron transport to sleep. Nature 2019, 568, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Robb, E.L.; Hall, A.R.; Prime, T.A.; Eaton, S.; Szibor, M.; Viscomi, C.; James, A.M.; Murphy, M.P. Control of mitochondrial superoxide production by reverse electron transport at complex I. J. Biol. Chem. 2018, 293, 9869–9879. [Google Scholar] [CrossRef]
- Yankovskaya, V.; Horsefield, R.; Törnroth-Horsefield, S.; Luna-Chavez, C.; Miyoshi, H.; Léger, C.; Byrne, B.; Cecchini, G.; Iwata, S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 2003, 299, 700–704. [Google Scholar] [CrossRef]
- Quinlan, C.L.; Orr, A.L.; Perevoshchikova, I.V.; Treberg, J.R.; Ackrell, B.A.; Brand, M.D. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J. Biol. Chem. 2012, 287, 27255–27264. [Google Scholar] [CrossRef]
- Siebels, I.; Dröse, S. Q-site inhibitor induced ROS production of mitochondrial complex II is attenuated by TCA cycle dicarboxylates. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 1156–1164. [Google Scholar] [CrossRef]
- Messner, K.; Imlay, J.A. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J. Biol. Chem. 2002, 277, 42563–42571. [Google Scholar] [CrossRef]
- Grivennikova, V.G.; Kozlovsky, V.S.; Vinogradov, A.D. Respiratory complex II: ROS production and the kinetics of ubiquinone reduction. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 109–117. [Google Scholar] [CrossRef]
- Manhas, N.; Duong, Q.V.; Lee, P.; Richardson, J.D.; Robertson, J.D.; Moxley, M.A.; Bazil, J.N. Computationally modeling mammalian succinate dehydrogenase kinetics identifies the origins and primary determinants of ROS production. J. Biol. Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P. The protonmotive Q cycle: A general formulation. FEBS Lett. 1975, 59, 137–139. [Google Scholar] [CrossRef]
- Crofts, A.R. The Cytochromebc1Complex: Function in the Context of Structure. Ann. Rev. Physiol. 2004, 66, 689–733. [Google Scholar] [CrossRef] [PubMed]
- Boveris, A.; Cadenas, E.; Stoppani, A.O.M. Role of ubiquinone in the mitochondrial generation of hydrogen peroxide. Biochem. J. 1976, 156, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Cadenas, E.; Boveris, A.; Ragan, C.; Stoppani, A.O. Production of superoxide radicals and hydrogen peroxide by NADH-ubiquinone reductase and ubiquinol-cytochrome c reductase from beef-heart mitochondria. Arch. Biochem. Biophys. 1977, 180, 248–257. [Google Scholar] [CrossRef]
- Bleier, L.; Dröse, S. Superoxide generation by complex III: From mechanistic rationales to functional consequences. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 1320–1331. [Google Scholar] [CrossRef]
- Muller, F.L. The nature and mechanism of superoxide production by the electron transport chain: Its relevance to aging. J. Am. Aging Assoc. 2000, 23, 227–253. [Google Scholar] [CrossRef]
- Muller, F.L.; Liu, Y.; Van Remmen, H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J. Biol. Chem. 2004, 279, 49064–49073. [Google Scholar] [CrossRef]
- De Grey, A.D. HO2: The forgotten radical. DNA Cell Biol. 2002, 21, 251–257. [Google Scholar] [CrossRef]
- St-Pierre, J.; Buckingham, J.A.; Roebuck, S.J.; Brand, M.D. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 2002, 277, 44784–44790. [Google Scholar] [CrossRef]
- Świerczek, M.; Cieluch, E.; Sarewicz, M.; Borek, A.; Moser, C.C.; Dutton, P.L.; Osyczka, A. An electronic bus bar lies in the core of cytochrome bc1. Science 2010, 329, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Drose, S.; Brandt, U. The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. J. Biol. Chem. 2008, 283, 21649–21654. [Google Scholar] [CrossRef] [PubMed]
- Bielski, B.H.J.; Arudi, R.; Sutherland, M. A study of the reactivity of HO2/O2-with unsaturated fatty acids. J. Biol. Chem. 1983, 258, 4759–4761. [Google Scholar] [PubMed]
- Bielski, B.H.J. Reactivity of HO2/O2-radicals in aqueous solution. J. Phys. Chem. 1985, 14, 1041–1091. [Google Scholar]
- Quinlan, C.L.; Gerencser, A.A.; Treberg, J.R.; Brand, M.D. The Mechanism of Superoxide Production by the Antimycin-inhibited Mitochondrial Q-cycle. J. Biol. Chem. 2011, 286, 31361–31372. [Google Scholar] [CrossRef]
- Cadenas, E.; Boveris, A. Enhancement of hydrogen peroxide formation by protophores and ionophores in antimycin-supplemented mitochondria. Biochem. J. 1980, 188, 31–37. [Google Scholar] [CrossRef]
- Chen, Q.; Vazquez, E.J.; Moghaddas, S.; Hoppel, C.L.; Lesnefsky, E.J. Production of reactive oxygen species by mitochondria. J. Biol. Chem. 2003, 278, 36027–36031. [Google Scholar] [CrossRef]
- Guillaud, F.; Drose, S.; Kowald, A.; Brandt, U.; Klipp, E. Superoxide production by cytochrome bc1 complex: A mathematical model. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 1643–1652. [Google Scholar] [CrossRef][Green Version]
- Mailloux, R.J. Protein S-glutathionylation reactions as a global inhibitor of cell metabolism for the desensitization of hydrogen peroxide signals. Redox Biol. 2020, 32, 101472. [Google Scholar] [CrossRef]
- Orr, A.L.; Quinlan, C.L.; Perevoshchikova, I.V.; Brand, M.D. A refined analysis of superoxide production by mitochondrialsn-glycerol 3-phosphate dehydrogenase. J. Biol. Chem. 2012, 287, 42921–42935. [Google Scholar] [CrossRef]
- Hey-Mogensen, M.; Gonçalves, R.L.; Orr, A.L.; Brand, M.D. Production of superoxide/H2O2 by dihydroorotate dehydrogenase in rat skeletal muscle mitochondria. Free. Radic. Biol. Med. 2014, 72, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Perevoshchikova, I.V.; Quinlan, C.L.; Orr, A.L.; Gerencser, A.A.; Brand, M.D. Sites of superoxide and hydrogen peroxide production during fatty acid oxidation in rat skeletal muscle mitochondria. Free. Radic. Biol. Med. 2013, 61, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, R.L.; Quinlan, C.L.; Perevoshchikova, I.V.; Hey-Mogensen, M.; Brand, M.D. Sites of superoxide and hydrogen peroxide production by muscle mitochondria assessedex vivounder conditions mimicking rest and exercise. J. Biol. Chem. 2015, 290, 209–227. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Fiorello, M.L.; Bailey, D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018, 15, 490–503. [Google Scholar] [CrossRef]
- Sidlauskaite, E.; Gibson, J.W.; Megson, I.L.; Whitfield, P.D.; Tovmasyan, A.; Batinic-Haberle, I.; Murphy, M.P.; Moult, P.R.; Cobley, J.N. Mitochondrial ROS cause motor deficits induced by synaptic inactivity: Implications for synapse pruning. Redox Biol. 2018, 16, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N. Synapse pruning: Mitochondrial ROS with their hands on the shears. BioEssays 2018, 40, e1800031. [Google Scholar] [CrossRef]
- Echtay, K.S.; Roussel, D.; St-Pierre, J.; Jekabsons, M.B.; Cadenas, S.; Stuart, J.A.; Harper, J.A.; Roebuck, S.J.; Morrison, A.; Pickering, S.; et al. Superoxide activates mitochondrial uncoupling proteins. Nature 2002, 415, 96–99. [Google Scholar] [CrossRef]
- Berry, B.J.; Trewin, A.J.; Amitrano, A.M.; Kim, M.; Wojtovich, A.P. Use the protonmotive force: Mitochondrial uncoupling and reactive oxygen species. J. Mol. Biol. 2018, 430, 3873–3891. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D.; Affourtit, C.; Esteves, T.C.; Green, K.; Lambert, A.J.; Miwa, S.; Pakay, J.L.; Parker, N. Mitochondrial superoxide: Production, biological effects, and activation of uncoupling proteins. Free. Radic. Biol. Med. 2004, 37, 755–767. [Google Scholar] [CrossRef]
- Blaza, J.N.; Vinothkumar, K.R.; Hirst, J. Structure of the deactive state of mammalian respiratory complex I. Structure 2018, 26, 312–319. [Google Scholar] [CrossRef]
- Galkin, A.; Meyer, B.; Wittig, I.; Karas, M.; Schägger, H.; Vinogradov, A.; Brandt, U. Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J. Biol. Chem. 2008, 283, 20907–20913. [Google Scholar] [CrossRef] [PubMed]
- Babot, M.; Birch, A.; Labarbuta, P.; Galkin, A. Characterisation of the active/de-active transition of mitochondrial complex I. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Dröse, S.; Stepanova, A.; Galkin, A. Ischemic A/D transition of mitochondrial complex I and its role in ROS generation. Biochim. Biophys. Acta-Bioenerg. 2016, 1857, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Pikó, L.; Taylor, K.D. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev. Biol. 1987, 123, 364–374. [Google Scholar] [CrossRef]
- Chronopoulou, E.; Harper, J.C. IVF culture media: Past, present and future. Hum. Reprod. Update 2014, 21, 39–55. [Google Scholar] [CrossRef]
- Place, T.L.; Domann, F.E.; Case, A.J. Limitations of oxygen delivery to cells in culture: An underappreciated problem in basic and translational research. Free. Radic. Biol. Med. 2017, 113, 311–322. [Google Scholar] [CrossRef]
- Nastri, C.O.; Nóbrega, B.N.; Teixeira, D.M.; Amorim, J.; Diniz, L.M.; Barbosa, M.W.; Giorgi, V.S.I.; Pileggi, V.N.; Martins, W.P. Low versus atmospheric oxygen tension for embryo culture in assisted reproduction: A systematic review and meta-analysis. Fertil. Steril. 2016, 106, 95–104. [Google Scholar] [CrossRef]
- Nasr-Esfahani, M.H.; Winston, N.J.; Johnson, M.H. Effects of glucose, glutamine, ethylenediaminetetraacetic acid and oxygen tension on the concentration of reactive oxygen species and on development of the mouse preimplantation embryo in vitro. J. Reprod. Fertil. 1992, 96, 219–231. [Google Scholar] [CrossRef]
- Dumollard, R.; Carroll, J.; Duchen, M.R.; Campbell, K.; Swann, K. Mitochondrial function and redox state in mammalian embryos. Semin. Cell Dev. Biol. 2009, 20, 346–353. [Google Scholar] [CrossRef]
- Jain, I.H.; Zazzeron, L.; Goli, R.; Alexa, K.; Schatzman-Bone, S.; Dhillon, H.; Goldberger, O.; Peng, J.; Shalem, O.; Sanjana, N.E.; et al. Hypoxia as a therapy for mitochondrial disease. Science 2016, 352, 54–61. [Google Scholar] [CrossRef]
- Kelley, R.L.; Gardner, D. Individual culture and atmospheric oxygen during culture affect mouse preimplantation embryo metabolism and post-implantation development. Reprod. Biomed. Online 2019, 39, 3–18. [Google Scholar] [CrossRef]
- Di Mascio, P.; Martinez, G.R.; Miyamoto, S.; Ronsein, G.E.; Medeiros, M.; Cadet, J. Singlet molecular oxygen reactions with nucleic acids, lipids, and proteins. Chem. Rev. 2019, 119, 2043–2086. [Google Scholar] [CrossRef]
- Takenaka, M.; Horiuchi, T.; Yanagimachi, R. Effects of light on development of mammalian zygotes. Proc. Natl. Acad. Sci. USA 2007, 104, 14289–14293. [Google Scholar] [CrossRef]
- Hockberger, P.E.; Skimina, T.A.; Centonze, V.E.; Lavin, C.; Chu, S.; Dadras, S.; Reddy, J.K.; White, J.G. Activation of flavin-containing oxidases underlies light-induced production of H2O2 in mammalian cells. Proc. Natl. Acad. Sci. USA 1999, 96, 6255–6260. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.M.; Janes, M.S.; Pehar, M.; Monette, J.S.; Ross, M.F.; Hagen, T.M.; Murphy, M.P.; Beckman, J.S. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc. Natl. Acad. Sci. USA 2006, 103, 15038–15043. [Google Scholar] [CrossRef] [PubMed]
- Kalyanaraman, B.; Darley-Usmar, V.M.; Davies, K.P.; Dennery, P.A.; Forman, H.J.; Grisham, M.B.; Mann, G.E.; Moore, K.; Roberts, L.J.; Ischiropoulos, H. Measuring reactive oxygen and nitrogen species with fluorescent probes: Challenges and limitations. Free. Radic. Biol. Med. 2011, 52, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D. The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 2010, 45, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Nasr-Esfahani, M.M.; Johnson, M.H. The origin of reactive oxygen species in mouse embryos cultured in vitro. Development 1991, 113, 551–560. [Google Scholar] [PubMed]
- Bonini, M.G.; Rota, C.; Tomasi, A.; Mason, R.P. The oxidation of 2′,7′-dichlorofluorescin to reactive oxygen species: A self-fulfilling prophesy? Free. Radic. Biol. Med. 2006, 40, 968–975. [Google Scholar] [CrossRef]
- Wardman, P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: Progress, pitfalls, and prospects. Free. Radic. Biol. Med. 2007, 43, 995–1022. [Google Scholar] [CrossRef]
- Zielonka, J.; Kalyanaraman, B. Small-molecule luminescent probes for the detection of cellular oxidizing and nitrating species. Free. Radic. Biol. Med. 2018, 128, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Van Blerkom, J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion 2011, 11, 797–813. [Google Scholar] [CrossRef] [PubMed]
- Van Blerkom, J.; Davis, P.; Mathwig, V.; Alexander, S. Domains of high-polarized and low-polarized mitochondria may occur in mouse and human oocytes and early embryos. Hum. Reprod. 2002, 17, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Wilding, M.; Dale, B.; Marino, M.; Di Matteo, L.; Alviggi, C.; Pisaturo, M.L.; Lombardi, L.; De Placido, G. Mitochondrial aggregation patterns and activity in human oocytes and preimplantation embryos. Hum. Reprod. 2001, 16, 909–917. [Google Scholar] [CrossRef]
- Van Blerkom, J.; Davis, P. High-polarized (ΔΨmHIGH) mitochondria are spatially polarized in human oocytes and early embryos in stable subplasmalemmal domains: Developmental significance and the concept of vanguard mitochondria. Reprod. Biomed. Online 2006, 13, 246–254. [Google Scholar] [CrossRef]
- Van Blerkom, J.; Davis, P. Mitochondrial signaling and fertilization. Mol. Hum. Reprod. 2007, 13, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Dumollard, R.; Duchen, M.R.; Carroll, J. The role of mitochondrial function in the oocyte and embryo. Curr. Top. Dev. Biol. 2007, 77, 21–49. [Google Scholar] [CrossRef]
- Barshop, B.A.; Gangoiti, J.A. Analysis of coenzyme Q in human blood and tissues. Mitochondrion 2007, 7, S89–S93. [Google Scholar] [CrossRef]
- Dumollard, R.; Marangos, P.; Fitzharris, G.; Swann, K.; Duchen, M.R.; Carroll, J. Sperm-triggered [Ca2+] oscillations and Ca2+ homeostasis in the mouse egg have an absolute requirement for mitochondrial ATP production. Development 2004, 131, 3057–3067. [Google Scholar] [CrossRef]
- Clementi, E.; Brown, G.C.; Feelisch, M.; Moncada, S. Persistent inhibition of cell respiration by nitric oxide: Crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc. Natl. Acad. Sci. USA 1998, 95, 7631–7636. [Google Scholar] [CrossRef]
- Toledo, J.O.; Augusto, O. Connecting the chemical and biological properties of nitric oxide. Chem. Res. Toxicol. 2012, 25, 975–989. [Google Scholar] [CrossRef] [PubMed]
- Van Blerkom, J.; Davis, P.; Thalhammer, V. Regulation of mitochondrial polarity in mouse and human oocytes: The influence of cumulus derived nitric oxide. Mol. Hum. Reprod. 2008, 14, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Dröse, S. Differential effects of complex II on mitochondrial ROS production and their relation to cardioprotective pre- and postconditioning. Biochim. Biophys. Acta-Bioenerg. 2013, 1827, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Orr, A.L.; Vargas, L.; Turk, C.N.; Baaten, J.E.; Matzen, J.T.; Dardov, V.J.; Attle, S.J.; Li, J.; Quackenbush, D.C.; Goncalves, R.L.S.; et al. Suppressors of superoxide production from mitochondrial complex III. Nat. Chem. Biol. 2015, 11, 834–836. [Google Scholar] [CrossRef]
- Brand, M.D.; Goncalves, R.L.; Orr, A.L.; Vargas, L.; Gerencser, A.A.; Jensen, M.B.; Wang, Y.T.; Melov, S.; Turk, C.N.; Matzen, J.T.; et al. Suppressors of superoxide-H2O2 production at site I Q of mitochondrial complex i protect against stem cell hyperplasia and ischemia-reperfusion injury. Cell Metab. 2016, 24, 582–592. [Google Scholar] [CrossRef]
- Wong, H.-S.; Monternier, P.-A.; Brand, M.D. S1QELs suppress mitochondrial superoxide/hydrogen peroxide production from site IQ without inhibiting reverse electron flow through Complex I. Free. Radic. Biol. Med. 2019, 143, 545–559. [Google Scholar] [CrossRef]
- Goncalves, R.L.; Watson, M.A.; Wong, H.-S.; Orr, A.L.; Brand, M.D. The use of site-specific suppressors to measure the relative contributions of different mitochondrial sites to skeletal muscle superoxide and hydrogen peroxide production. Redox Biol. 2020, 28, 101341. [Google Scholar] [CrossRef]
- Banba, A.; Tsuji, A.; Kimura, H.; Murai, M.; Miyoshi, H. Defining the mechanism of action of S1QELs, specific suppressors of superoxide production in the quinone-reaction site in mitochondrial complex I. J. Biol. Chem. 2019, 294, 6550–6561. [Google Scholar] [CrossRef]
- Margaritelis, N.; Cobley, J.; Paschalis, V.; Veskoukis, A.; Theodorou, A.; Kyparos, A.; Nikolaidis, M.G. Going retro: Oxidative stress biomarkers in modern redox biology. Free. Radic. Biol. Med. 2016, 98, 2–12. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Cobley, J.N.; Paschalis, V.; Veskoukis, A.S.; Theodorou, A.A.; Kyparos, A.; Nikolaidis, M.G. Principles for integrating reactive species into in vivo biological processes: Examples from exercise physiology. Cell. Sign. 2016, 28, 256–271. [Google Scholar] [CrossRef]
- James, A.M.; Cochemé, H.M.; Smith, R.A.J.; Murphy, M.P. Interactions of Mitochondria-targeted and Untargeted Ubiquinones with the mitochondrial respiratory chain and reactive oxygen species. J. Biol. Chem. 2005, 280, 21295–21312. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. Antioxidants as therapies: Can we improve on nature? Free. Radic. Biol. Med. 2014, 66, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Finichiu, P.G.; Larsen, D.S.; Evans, C.; Larsen, L.; Bright, T.P.; Robb, E.L.; Trnka, J.; Prime, T.A.; James, A.M.; Smith, R.A.; et al. A mitochondria-targeted derivative of ascorbate: MitoC. Free. Radic. Biol. Med. 2015, 89, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Murphy, M.P. Animal and human studies with the mitochondria-targeted antioxidant MitoQ. Ann. N. Y. Acad. Sci. USA 2010, 1201, 96–103. [Google Scholar] [CrossRef]
- Smith, R.A.J.; Hartley, R.C.; Murphy, M.P. Mitochondria-targeted small molecule therapeutics and probes. Antioxid. Redox Sign. 2011, 15, 3021–3038. [Google Scholar] [CrossRef]
- Kelso, G.F.; Porteous, C.M.; Coulter, C.V.; Hughes, G.; Porteous, W.K.; Ledgerwood, E.C.; Smith, R.A.J.; Murphy, M.P. Selective targeting of a redox-active ubiquinone to mitochondria within cells. J. Biol. Chem. 2001, 276, 4588–4596. [Google Scholar] [CrossRef]
- Smith, R.A.J.; Porteous, C.M.; Coulter, C.V.; Murphy, M.P. Selective targeting of an antioxidant to mitochondria. Eur. J. Chem. 1999, 263, 709–716. [Google Scholar] [CrossRef]
- Batinic-Haberle, I.; Rebouças, J.S.; Spasojevic, I. superoxide dismutase mimics: Chemistry, pharmacology, and therapeutic potential. Antioxid. Redox Sign. 2010, 13, 877–918. [Google Scholar] [CrossRef]
- Batinic-Haberle, I.; Tovmasyan, A.; Roberts, E.R.H.; Vujaskovic, Z.; Leong, K.W.; Spasojevic, I. SOD therapeutics: Latest insights into their structure-activity relationships and impact on the cellular redox-based signaling pathways. Antioxid. Redox Sign. 2014, 20, 2372–2415. [Google Scholar] [CrossRef]
- Batinic-Haberle, I.; Tovmasyan, A.; Spasojevic, I. An educational overview of the chemistry, biochemistry and therapeutic aspects of Mn porphyrins–From superoxide dismutation to H2O2-driven pathways. Redox Biol. 2015, 5, 43–65. [Google Scholar] [CrossRef]
- Gutteridge, J.M.; Halliwell, B. Antioxidants: Molecules, medicines, and myths. Biochem. Biophys. Res. Commun. 2010, 393, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Niki, E. Do antioxidants impair signaling by reactive oxygen species and lipid oxidation products? FEBS Lett. 2012, 586, 3767–3770. [Google Scholar] [CrossRef] [PubMed]
- Niki, E. Role of vitamin E as a lipid-soluble peroxyl radical scavenger: In vitro and in vivo evidence. Free. Radic. Biol. Med. 2014, 66, 3–12. [Google Scholar] [CrossRef]
- Meyer, A.J.; Dick, T.P. Fluorescent Protein-Based Redox Probes. Antioxid. Redox Sign. 2010, 13, 621–650. [Google Scholar] [CrossRef] [PubMed]
- Cochemé, H.M.; Quin, C.; McQuaker, S.J.; Cabreiro, F.G.; Logan, A.; Prime, T.A.; Abakumova, I.; Patel, J.V.; Fearnley, I.M.; James, A.M.; et al. Measurement of H2O2 within living drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011, 13, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Logan, A.; Cochemé, H.M.; Pun, P.B.L.; Apostolova, N.; Smith, R.A.; Larsen, L.; Larsen, D.S.; James, A.M.; Fearnley, I.M.; Rogatti, S.; et al. Using exomarkers to assess mitochondrial reactive species in vivo. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2014, 1840, 923–930. [Google Scholar] [CrossRef]
- Shchepinova, M.M.; Cairns, A.G.; Prime, T.A.; Logan, A.; James, A.M.; Hall, A.R.; Vidoni, S.; Arndt, S.; Caldwell, S.T.; Prag, H.A.; et al. MitoNeoD: A mitochondria-targeted superoxide probe. Cell Chem. Biol. 2017, 24, 1285–1298. [Google Scholar] [CrossRef]
- Zielonka, J.; Kalyanaraman, B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: Another inconvenient truth. Free. Radic. Biol. Med. 2010, 48, 983–1001. [Google Scholar] [CrossRef]
- Held, J.M. Redox Systems Biology: Harnessing the Sentinels of the Cysteine Redoxome. Antioxid. Redox Sign. 2020, 32, 659–676. [Google Scholar] [CrossRef]
- Xiao, H.; Jedrychowski, M.P.; Schweppe, D.K.; Huttlin, E.L.; Yu, Q.; Heppner, D.E.; Li, J.; Long, J.; Mills, E.L.; Szpyt, J.; et al. A Quantitative tissue-specific landscape of protein redox regulation during aging. Cell 2020, 180, 968–983.e24. [Google Scholar] [CrossRef]
- Cobley, J.N.; Sakellariou, G.K.; Husi, H.; McDonagh, B. Proteomic strategies to unravel age-related redox signalling defects in skeletal muscle. Free. Radic. Biol. Med. 2019, 132, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Carroll, K.S.; Liebler, D.C. The expanding landscape of the thiol redox proteome. Mol. Cell. Proteom. 2015, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gould, N.S.; Evans, P.; Martínez-Acedo, P.; Marino, S.M.; Gladyshev, V.N.; Carroll, K.S.; Ischiropoulos, H. Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Chem. Biol. 2015, 22, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, J.R.; Oviosu, O.; Eaton, P. The PEG-switch assay: A fast semi-quantitative method to determine protein reversible cysteine oxidation. J. Pharmacol. Toxicol. Methods 2013, 68, 297–301. [Google Scholar] [CrossRef]
- Van Leeuwen, L.A.; Hinchy, E.C.; Murphy, M.P.; Robb, E.L.; Cochemé, H.M. Click-PEGylation - A mobility shift approach to assess the redox state of cysteines in candidate proteins. Free. Radic. Biol. Med. 2017, 108, 374–382. [Google Scholar] [CrossRef]
- Cobley, J.N.; Noble, A.; Fernández, E.J.; Moya, M.-T.V.; Guille, M.; Husi, H. Catalyst-free Click PEGylation reveals substantial mitochondrial ATP synthase sub-unit alpha oxidation before and after fertilisation. Redox Biol. 2019, 26, 101258. [Google Scholar] [CrossRef]
- Cobley, J.N.; Husi, H. Immunological techniques to assess protein thiol redox state: Opportunities, challenges and solutions. Antioxidants 2020, 9, 315. [Google Scholar] [CrossRef]
- Logan, A.; Pell, V.R.; Shaffer, K.J.; Evans, C.; Stanley, N.J.; Robb, E.L.; Prime, T.A.; Chouchani, E.T.; Cochemé, H.M.; Fearnley, I.M.; et al. Assessing the mitochondrial membrane potential in cells and in vivo using targeted click chemistry and mass spectrometry. Cell Metab. 2016, 23, 379–385. [Google Scholar] [CrossRef]
- Burger, N.; Logan, A.; Prime, T.A.; Mottahedin, A.; Caldwell, S.T.; Krieg, T.; Hartley, R.C.; James, A.M.; Murphy, M.P. A sensitive mass spectrometric assay for mitochondrial CoQ pool redox state in vivo. Free. Radic. Biol. Med. 2020, 147, 37–47. [Google Scholar] [CrossRef]
- Robb, E.L.; Gawel, J.M.; Aksentijević, D.; Cochemé, H.M.; Stewart, T.; Shchepinova, M.M.; Qiang, H.; Prime, T.A.; Bright, T.P.; James, A.M.; et al. Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free. Radic. Biol. Med. 2015, 89, 883–894. [Google Scholar] [CrossRef]
- Booty, L.M.; Gawel, J.M.; Cvetko, F.; Caldwell, S.T.; Hall, A.R.; Mulvey, J.F.; James, A.M.; Hinchy, E.C.; Prime, T.A.; Arndt, S.; et al. Selective disruption of mitochondrial thiol redox state in cells and in vivo. Cell Chem. Biol. 2019, 26, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Giustarini, D.; Dalle-Donne, I.; Milzani, A.D.G.; Fanti, P.; Rossi, R. Analysis of GSH and GSSG after derivatization with N-ethylmaleimide. Nat. Protoc. 2013, 8, 1660–1669. [Google Scholar] [CrossRef] [PubMed]
- Weydert, C.J.; Cullen, J.J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 2010, 5, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Dumollard, R.; Duchen, M.R.; Sardet, C. Calcium signals and mitochondria at fertilisation. Semin. Cell Dev. Biol. 2006, 17, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Muller, B.; Lewis, N.; Adeniyi, T.; Leese, H.J.; Brison, D.R.; Sturmey, R.G. Application of extracellular flux analysis for determining mitochondrial function in mammalian oocytes and early embryos. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Van Blerkom, J.; Davis, P.W.; Lee, J. Fertilization and early embryolgoy: ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum. Reprod. 1995, 10, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Allen, J. Separate Sexes and the Mitochondrial Theory of Ageing. J. Theor. Biol. 1996, 180, 135–140. [Google Scholar] [CrossRef]
- Hernansanz-Agustín, P.; Choya-Foces, C.; Carregal-Romero, S.; Ramos, E.; Oliva, T.; Villa-Piña, T.; Moreno, L.; Izquierdo-Álvarez, A.; Cabrera-García, J.D.; Cortés, A.; et al. Na+ controls hypoxic signalling by the mitochondrial respiratory chain. Nature 2020. [Google Scholar] [CrossRef]
- Biggers, J.D.; Whittingham, D.G.; Donahue, R.P. The pattern of energy metabolism in the mouse oocyte and zygote. Proc. Nat. Acad. Sci. USA 1967, 58, 560–567. [Google Scholar] [CrossRef]
- Houghton, F.D.; Christopher, G.T.; Leese, H.J. Oxygen consumption and energy metabolism of the early mouse embryo. Mol. Reprod. Dev. 1996, 485, 476–485. [Google Scholar] [CrossRef]
- O’Brien, M.; Chalker, J.; Slade, L.; Gardiner, D.; Mailloux, R.J. Protein S-glutathionylation alters superoxide/hydrogen peroxide emission from pyruvate dehydrogenase complex. Free. Radic. Biol. Med. 2017, 106, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Gill, R.M.; O’Brien, M.; Young, A.; Gardiner, D.; Mailloux, R.J. Protein S-glutathionylation lowers superoxide/hydrogen peroxide release from skeletal muscle mitochondria through modification of complex I and inhibition of pyruvate uptake. PLoS ONE 2018, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lane, M.; Gardner, D. Mitochondrial malate-aspartate shuttle regulates mouse embryo nutrient consumption. J. Biol. Chem. 2005, 280, 18361–18367. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, R.; Sharpley, M.S.; Chi, F.; Braas, D.; Zhou, Y.; Kim, R.; Clark, A.T.; Banerjee, U. Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell 2017, 168, 210–223.e11. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, J.; Dahan, P.; Lu, V.; Zhang, C.; Li, H.; Teitell, M.A. Metabolism in pluripotent stem cells and early mammalian development. Cell Metab. 2018, 27, 332–338. [Google Scholar] [CrossRef]
- Sturmey, R.; Leese, H.; Sturmey, R.G. Energy metabolism in pig oocytes and early embryos. Reproduction 2003, 126, 197–204. [Google Scholar] [CrossRef]
- Motta, P.M.; Nottola, S.A.; Makabe, S.; Heyn, R. Mitochondrial morphology in human fetal and adult female germ cells. Hum. Reprod. 2000, 15 (Suppl. S2), 129–147. [Google Scholar] [CrossRef]
- Petrova, B.; Liu, K.; Tian, C.; Kitaoka, M.; Freinkman, E.; Yang, J.; Orr-Weaver, T.L. Dynamic redox balance directs the oocyte-to-embryo transition via developmentally controlled reactive cysteine changes. Proc. Natl. Acad. Sci. USA 2018, 115, E7978–E7986. [Google Scholar] [CrossRef]
- Smith, D.G.; Sturmey, R.G. Parallels between embryo and cancer cell metabolism. Biochem. Soc. Trans. 2013, 41, 664–669. [Google Scholar] [CrossRef]
- Muramoto, K.; Ohta, K.; Shinzawa-Itoh, K.; Kanda, K.; Taniguchi, M.; Nabekura, H.; Yamashita, E.; Tsukihara, T.; Yoshikawa, S. Bovine cytochromecoxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate. Proc. Natl. Acad. Sci. USA 2010, 107, 7740–7745. [Google Scholar] [CrossRef]
- Barja, G. Oxygen radicals, a failure or sucess of evolution. Free Radic. Res. 1993, 18, 63–70. [Google Scholar]
- Buettner, G.R. Superoxide dismutase in redox biology: The roles of superoxide and hydrogen peroxide. Anti-Cancer Agents Med. Chem. 2011, 11, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Lane, N. The Vital Question: Energy, Evolution, and the Origins of Complex Life; Profile Books Limited: London, UK, 2015. [Google Scholar]
- Finkel, T. Signal transduction by mitochondrial oxidants. J. Biol. Chem. 2011, 287, 4434–4440. [Google Scholar] [CrossRef] [PubMed]
- Allen, J. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, 10231–10238. [Google Scholar] [CrossRef] [PubMed]
- Vandaele, L.; Thys, M.; Bijttebier, J.; Van Langendonckt, A.; Donnay, I.; Maes, D.; Meyer, E.; Van Soom, A. Short-term exposure to hydrogen peroxide during oocyte maturation improves bovine embryo development. Reproduction 2010, 139, 505–511. [Google Scholar] [CrossRef]
- Buettner, G.R.; Ng, C.F.; Wang, M.; Rodgers, V.G.; Schafer, F.Q. A new paradigm: Manganese superoxide dismutase influences the production of H2O2 in cells and thereby their biological state. Free. Radic. Biol. Med. 2006, 41, 1338–1350. [Google Scholar] [CrossRef]
- Brown, G.C. Nitric oxide and mitochondrial respiration. Biochim. Biophys. Acta (BBA)-Bioenerg. 1999, 1411, 351–369. [Google Scholar] [CrossRef]
- Vásquez-Vivar, J.; Kalyanaraman, B.; Kennedy, M.C. Mitochondrial Aconitase Is a Source of Hydroxyl Radical. J. Biol. Chem. 2000, 275, 14064–14069. [Google Scholar] [CrossRef]
- Gardner, P.R.; Fridovich, I. Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 1991, 266, 19328–19333. [Google Scholar]
- Abreu, I.A.; Cabelli, D.E. Superoxide dismutases—A review of the metal-associated mechanistic variations. Biochim. Biophys. Acta-Proteins Proteom. 2010, 1804, 263–274. [Google Scholar] [CrossRef]
- Sheng, Y.; Abreu, I.A.; Cabelli, D.E.; Maroney, M.J.; Miller, A.-F.; Teixeira, M.; Valentine, J.S. Superoxide Dismutases and Superoxide Reductases. Chem. Rev. 2014, 114, 3854–3918. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Biological chemistry of superoxide radicals. ChemTexts 2020, 6, 1–13. [Google Scholar] [CrossRef]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Ganini, D.; Santos, J.H.; Bonini, M.G.; Mason, R.P. Switch of mitochondrial superoxide dismutase into a prooxidant peroxidase in manganese-deficient cells and mice. Cell Chem. Biol. 2018, 25, 413–425. [Google Scholar] [CrossRef]
- Palma, F.R.; He, C.; Danes, M.J.M.; Paviani, V.; Coelho, D.R.; Gantner, B.N.; Bonini, M.G.; Sampaio, V.P. Mitochondrial superoxide dismutase: What the established, the intriguing, and the novel reveal about a key cellular redox switch. Antioxid. Redox Sign. 2020, 32, 701–714. [Google Scholar] [CrossRef]
- Lustgarten, M.S.; Bhattacharya, A.; Muller, F.L.; Jang, Y.C.; Shimizu, T.; Shirasawa, T.; Richardson, A.; Van Remmen, H. Complex I generated, mitochondrial matrix-directed superoxide is released from the mitochondria through voltage dependent anion channels. Biochem. Biophys. Res. Commun. 2012, 422, 515–521. [Google Scholar] [CrossRef]
- Beckman, J.S.; Beckman, T.W.; Chen, J.; Marshall, P.A.; Freeman, B.A. Apparent hydroxyl radical production by peroxynitrite: Implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci. USA 1990, 87, 1620–1624. [Google Scholar] [CrossRef]
- Radi, R. Peroxynitrite, a stealthy biological oxidant. J. Biol. Chem. 2013, 288, 26464–26472. [Google Scholar] [CrossRef]
- Carballal, S.; Bartesaghi, S.; Radi, R. Kinetic and mechanistic considerations to assess the biological fate of peroxynitrite. Biochim. Biophys. Acta-Gen. Subj. 2014, 1840, 768–780. [Google Scholar] [CrossRef]
- Cox, A.G.; Winterbourn, C.C.; Hampton, M.B. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 2010, 425, 313–325. [Google Scholar] [CrossRef]
- Murphy, M.P. Mitochondrial thiols in antioxidant protection and redox signaling: Distinct roles for glutathionylation and other thiol modifications. Antioxid. Redox Sign. 2012, 16, 476–495. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.D.; Schmidt, C.A.; Lin, C.-T.; Fisher-Wellman, K.H.; Neufer, P. Flux through mitochondrial redox circuits linked to nicotinamide nucleotide transhydrogenase generates counterbalance changes in energy expenditure. J. Biol. Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sobotta, M.C.; Liou, W.; Stöcker, S.; Talwar, D.; Oehler, M.; Ruppert, T.; Scharf, A.N.D.; Dick, T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015, 11, 64–70. [Google Scholar] [CrossRef]
- Pak, V.V.; Ezeriņa, D.; Lyublinskaya, O.G.; Pedre, B.; Tyurin-Kuzmin, P.A.; Mishina, N.M.; Thauvin, M.; Young, D.; Wahni, K.; Gache, S.A.M.; et al. Ultrasensitive genetically encoded indicator for hydrogen peroxide identifies roles for the oxidant in cell migration and mitochondrial function. Cell Metab. 2020, 31, 642–653.e6. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Biological Production, Detection, and Fate of Hydrogen Peroxide. Antioxid. Redox Sign. 2018, 20, 541–551. [Google Scholar] [CrossRef]
- Sies, H. Biochemistry of oxidative stress. Biochem. Soc. Trans. 2007, 35, 1147–1151. [Google Scholar] [CrossRef]
- Zhang, Y.; Marcillat, O.; Giulivi, C.; Ernster, L.; Davies, K.J. The oxidative inactivation of mitochondrial electron transport chain components and ATPase. J. Biol. Chem. 1990, 265, 16330–16336. [Google Scholar]
- Shoubridge, E.A.; Wai, T. Mitochondrial DNA and the mammalian oocyte. Curr. Top. Dev. Biol. 2007, 77, 87–111. [Google Scholar] [CrossRef]
- Cobley, J.N.; Sakellariou, G.K.; Murray, S.; Waldron, S.; Gregson, W.; Burniston, J.G.; Morton, J.P.; Iwanejko, L.A.; Close, G.L. Lifelong endurance training attenuates age-related genotoxic stress in human skeletal muscle. Longev. Health 2013, 2, 11. [Google Scholar] [CrossRef]
- Cobley, J.N.; Margaritelis, N.V.; Morton, J.P.; Close, G.L.; Nikolaidis, M.G.; Malone, J.K. The basic chemistry of exercise-induced DNA oxidation: Oxidative damage, redox signaling, and their interplay. Front. Physiol. 2015, 6, 1–8. [Google Scholar] [CrossRef]
- Williamson, J.; Hughes, C.M.; Cobley, J.N.; Davison, G.W. The mitochondria-targeted antioxidant MitoQ, attenuates exercise-induced mitochondrial DNA damage. Redox Biol. 2020, 36, 101673. [Google Scholar] [CrossRef] [PubMed]
- Lane, N. Mitonuclear match: Optimizing fitness and fertility over generations drives ageing within generations. BioEssays 2011, 33, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Latorre-Pellicer, A.; Moreno-Loshuertos, R.; Lechuga-Vieco, A.V.; Sánchez-Cabo, F.; Torroja, C.; Acín-Pérez, R.; Calvo, E.; Aix, E.; González-Guerra, A.; Logan, A.; et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature 2016, 535, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, C.E.; Carroll, K.S. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef]
- Parvez, S.; Long, M.J.C.; Poganik, J.R.; Aye, Y. Redox signaling by reactive electrophiles and oxidants. Chem. Rev. 2018, 118, 8798–8888. [Google Scholar] [CrossRef]
- Janssen-Heininger, Y.M.; Mossman, B.T.; Heintz, N.H.; Forman, H.J.; Kalyanaraman, B.; Finkel, T.; Stamler, J.S.; Rhee, S.G.; Van Der Vliet, A. Redox-based regulation of signal transduction: Principles, pitfalls, and promises. Free. Radic. Biol. Med. 2008, 45, 1–17. [Google Scholar] [CrossRef]
- Cobley, J.N.; McHardy, H.; Morton, J.P.; Nikolaidis, M.G.; Close, G.L. Influence of vitamin C and vitamin E on redox signaling: Implications for exercise adaptations. Free. Radic. Biol. Med. 2015, 84, 65–76. [Google Scholar] [CrossRef]
- Bersweiler, A.; D’Autréaux, B.; Mazon, H.; Kriznik, A.; Belli, G.; Delaunay-Moisan, A.; Toledano, M.B.; Rahuel-Clermont, S. A scaffold protein that chaperones a cysteine-sulfenic acid in H2O2 signaling. Nat. Chem. Rev. Biol. 2017, 13, 909–915. [Google Scholar] [CrossRef]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef]
- Go, Y.-M.; Chandler, J.D.; Jones, D.P. The cysteine proteome. Free. Radic. Biol. Med. 2015, 84, 227–245. [Google Scholar] [CrossRef]
- Collins, Y.; Chouchani, E.T.; Menger, K.E.; Murphy, M.P.; Cochemé, H.M.; James, A.M. Mitochondrial redox signalling at a glance. J. Cell Sci. 2012, 125, 1837. [Google Scholar] [CrossRef]
- Requejo, R.; Hurd, T.R.; Costa, N.J.; Murphy, M.P. Cysteine residues exposed on protein surfaces are the dominant intramitochondrial thiol and may protect against oxidative damage. FEBS J. 2010, 277, 1465–1480. [Google Scholar] [CrossRef] [PubMed]
- Dröse, S.; Brandt, U.; Wittig, I. Mitochondrial respiratory chain complexes as sources and targets of thiol-based redox-regulation. Biochim. Biophys. Acta-Proteins Proteom. 2014, 1844, 1344–1354. [Google Scholar] [CrossRef] [PubMed]
- Hurd, T.R.; Requejo, R.; Filipovska, A.; Brown, S.; Prime, T.A.; Robinson, A.J.; Fearnley, I.M.; Murphy, M.P. Complex I within oxidatively stressed bovine heart mitochondria is glutathionylated on Cys-531 and Cys-704 of the 75-kDa subunit. J. Biol. Chem. 2008, 283, 24801–24815. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-B.; Foster, D.B.; Rucker, J.; O’Rourke, B.; Kass, D.A.; Van Eyk, J.E. Redox regulation of mitochondrial ATP synthase: Implications for cardiac resynchronization therapy. Circ. Res. 2011, 109, 750–757. [Google Scholar] [CrossRef]
- Wang, S.-B.; Murray, C.I.; Chung, H.S.; Van Eyk, J. Redox regulation of mitochondrial ATP synthase. Trends Cardiovasc. Med. 2013, 23, 18. [Google Scholar] [CrossRef]
- Cobley, J.N.; Noble, A.; Bessell, R.; Guille, M.; Husi, H. Reversible thiol oxidation inhibits the mitochondrial ATP Synthase in Xenopus laevis Oocytes. Antioxidants 2020, 9, 215. [Google Scholar] [CrossRef]
- Ramalho-Santos, J.; Varum, S.; Amaral, S.; Mota, P.C.; Sousa, A.P.M.; Amaral, A. Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 2009, 15, 553–572. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Hampton, M.B. Thiol chemistry and specificity in redox signaling. Free. Radic. Biol. Med. 2008, 45, 549–561. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Flohé, L. Basic principles and emerging concepts in the redox control of transcription factors. Antioxid. Redox Sign. 2011, 15, 2335–2381. [Google Scholar] [CrossRef]
- Stöcker, S.; Van Laer, K.; Mijuskovic, A.; Dick, T.P. The conundrum of hydrogen peroxide signaling and the emerging role of peroxiredoxins as redox relay hubs. Antioxid. Redox Sign. 2018, 28, 558–573. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.; Yim, S.H.; Shin, D.H.; Kang, D.; Yu, D.-Y.; Rhee, S.G. Inactivation of peroxiredoxin i by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell 2010, 140, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Reczek, C.R.; Chandel, N.S. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015, 33, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Close, G.L.; Bailey, D.M.; Davison, G.W. Exercise redox biochemistry: Conceptual, methodological and technical recommendations. Redox Biol. 2017, 12, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Hydrogen peroxide reactivity and specificity in thiol-based cell signalling. Biochem. Soc. Trans. 2020, 48, 1–10. [Google Scholar] [CrossRef]
- Dagnell, M.; Cheng, Q.; Rizvi, S.H.M.; Pace, P.E.; Boivin, B.; Winterbourn, C.C.; Arnér, E.S.J. Bicarbonate is essential for protein-tyrosine phosphatase 1B (PTP1B) oxidation and cellular signaling through EGF-triggered phosphorylation cascades. J. Biol. Chem. 2019, 294, 12330–12338. [Google Scholar] [CrossRef] [PubMed]
- Augusto, O.; Bonini, M.G.; Amanso, A.M.; Linares, E.; Santos, C.C.; De Menezes, S.L. Nitrogen dioxide and carbonate radical anion: Two emerging radicals in biology. Free. Radic. Biol. Med. 2002, 32, 841–859. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Revisiting the reactions of superoxide with glutathione and other thiols. Arch. Biochem. Biophys. 2016, 595, 68–71. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Superoxide as an intracellular radical sink. Free. Radic. Biol. Med. 1993, 14, 85–90. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Are free radicals involved in thiol-based redox signaling? Free. Radic. Biol. Med. 2015, 80, 164–170. [Google Scholar] [CrossRef]
- Kang, P.T.; Zhang, L.; Chen, C.-L.; Chen, J.; Green, K.B.; Chen, Y.-R. Protein thiyl radical mediates S-glutathionylation of complex I. Free. Radic. Biol. Med. 2012, 53, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Trimarchi, J.R.; Liu, L.; Porterfield, D.M.; Smith, P.J.; Keefe, D.L. Oxidative phosphorylation-dependent and -independent oxygen consumption by individual preimplantation mouse embryos1. Biol. Reprod. 2000, 62, 1866–1874. [Google Scholar] [CrossRef] [PubMed]
- Nasr-Esfahani, M.H.; Aitken, J.R.; Johnson, M.H. Hydrogen peroxide levels in mouse oocytes and early cleavage stage embryos developed in vitro or in vivo. Development 1990, 109, 501–507. [Google Scholar] [PubMed]
- Foerder, C.A.; Klebanoff, S.J.; Shapiro, B.M. Hydrogen peroxide production, chemiluminescence, and the respiratory burst of fertilization: Interrelated events in early sea urchin development. Proc. Natl. Acad. Sci. USA 1978, 75, 3183–3187. [Google Scholar] [CrossRef]
- Heinecke, J.W.; Shapiro, B.M. Respiratory burst oxidase of fertilization. Proc. Natl. Acad. Sci. USA 1989, 86, 1259–1263. [Google Scholar] [CrossRef]
- Wong, J.L.; Creton, R.; Wessel, G. The oxidative burst at fertilization is dependent upon activation of the dual oxidase Udx1. Dev. Cell 2004, 7, 801–814. [Google Scholar] [CrossRef]
- Han, Y.; Ishibashi, S.; Iglesias-Gonzalez, J.; Chen, Y.; Love, N.R.; Amaya, E. Ca 2+ Induced mitochondrial ros regulate the early embryonic cell cycle. Cell Rep. 2018, 22, 218–231. [Google Scholar] [CrossRef]
- Natsuyama, S.; Noda, Y.; Narimoto, K.; Umaoka, Y.; Mori, T. Release of two-cell block by reduction of protein disulfide with thioredoxin from Escherichia coli in mice. J. Reprod. Fertil. 1992, 95, 649–656. [Google Scholar] [CrossRef]
- Natsuyama, S.; Noda, Y.; Yamashita, M.; Nagahama, Y.; Mori, T. Superoxide dismutase and thioredoxin restore defective p34cdc2 kinase activation in mouse two-cell block. BBA-Mol. Cell Res. 1993, 1176, 90–94. [Google Scholar] [CrossRef]
- Ufer, C.; Wang, C.C.; Borchert, A.; Heydeck, D.; Kuhn, H. Redox control in mammalian embryo development. Antioxid. Redox Sign. 2010, 13, 833–875. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Frezza, C. Oncometabolites: Unconventional triggers of oncogenic signalling cascades. Free Radic. Biol. Med. 2016, 100, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.G.; Murphy, M.P.; Frezza, C.; Prag, H.A.; Chouchani, E.T.; O’Neill, L.A.; Mills, E.L. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat. Metab. 2018, 1, 16–33. [Google Scholar] [CrossRef]
- Harvey, A.J. Mitochondria in early development: Linking the microenvironment, metabolism and the epigenome. Reproduction 2019, 157, R159–R179. [Google Scholar] [CrossRef] [PubMed]
- Hitchler, M.J.; Domann, F.E. An epigenetic perspective on the free radical theory of development. Free Radic. Biol. Med. 2007, 43, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Bazopoulou, D.; Knoefler, D.; Zheng, Y.; Ulrich, K.; Oleson, B.J.; Xie, L.; Kim, M.; Kaufmann, A.; Lee, Y.-T.; Dou, Y.; et al. Developmental ROS individualizes organismal stress resistance and lifespan. Nature 2019, 576, 301–305. [Google Scholar] [CrossRef]
- Knoefler, D.; Thamsen, M.; Koniczek, M.; Niemuth, N.J.; Diederich, A.-K.; Jakob, U. Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol. Cell 2012, 47, 767–776. [Google Scholar] [CrossRef]
- Lozoya, O.A.; Xu, F.; Grenet, D.; Wang, T.; Grimm, S.A.; Godfrey, V.; Waidyanatha, S.; Woychik, R.P.; Santos, J.H. Single nucleotide resolution analysis reveals pervasive, long-lasting DNA methylation changes by developmental exposure to a mitochondrial toxicant. Cell Rep. 2020, 32. [Google Scholar] [CrossRef]
- Chowdhury, A.R.; Zielonka, J.; Kalyanaraman, B.; Hartley, R.C.; Murphy, M.P.; Avadhani, N.G. Mitochondria-targeted paraquat and metformin mediate ROS production to induce multiple pathways of retrograde signaling: A dose-dependent phenomenon. Redox Biol. 2020, 36, 101606. [Google Scholar] [CrossRef]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).